Significance
Cyanobacteria are an important group of photosynthetic bacteria that rely upon light energy for growth but frequently must adapt to darkness. Cells stop growing and decrease overall rates of gene expression and protein synthesis in the dark, but the molecular mechanisms behind these observations remain unknown. We find that a widespread bacterial stress response, the stringent response, helps cells conserve resources during darkness. In the dark, cells produce higher levels of the stringent response signaling molecule guanosine 3′-diphosphate 5′-diphosphate (ppGpp), thereby altering gene expression patterns and affecting the protein synthesis machinery. These results help explain previous observations in the cyanobacterial literature and extend our knowledge of how the same signaling pathway has been adapted to different bacterial lifestyles and metabolisms.
Keywords: cyanobacteria, Synechococcus, stringent response, (p)ppGpp, hibernation promoting factor
Abstract
The cyanobacterium Synechococcus elongatus relies upon photosynthesis to drive metabolism and growth. During darkness, Synechococcus stops growing, derives energy from its glycogen stores, and greatly decreases rates of macromolecular synthesis via unknown mechanisms. Here, we show that the stringent response, a stress response pathway whose genes are conserved across bacteria and plant plastids, contributes to this dark adaptation. Levels of the stringent response alarmone guanosine 3′-diphosphate 5′-diphosphate (ppGpp) rise after a shift from light to dark, indicating that darkness triggers the same response in cyanobacteria as starvation in heterotrophic bacteria. High levels of ppGpp are sufficient to stop growth and dramatically alter many aspects of cellular physiology, including levels of photosynthetic pigments and polyphosphate, DNA content, and the rate of translation. Cells unable to synthesize ppGpp display pronounced growth defects after exposure to darkness. The stringent response regulates expression of a number of genes in Synechococcus, including ribosomal hibernation promoting factor (hpf), which causes ribosomes to dimerize in the dark and may contribute to decreased translation. Although the metabolism of Synechococcus differentiates it from other model bacterial systems, the logic of the stringent response remains remarkably conserved, while at the same time having adapted to the unique stresses of the photosynthetic lifestyle.
The conversion of solar light energy to chemical energy through photosynthesis ultimately supports the majority of life on Earth. Light harvesting by photosynthetic antenna complexes and photosystems is directly tied to light availability, which can fluctuate greatly over the course of a single day (1). The growth and reproduction of photosynthetic organisms therefore depend upon their ability to capture light efficiently, and they have evolved several mechanisms that allow them to adapt to changing light conditions (2).
Cyanobacteria comprise a diverse bacterial phylum that oxygenated the atmosphere, gave rise to the plant chloroplast, and that performs 10 to 25% of global photosynthesis today. Synechococcus elongatus PCC 7942 (hereafter, Synechococcus) is a model cyanobacterium that relies exclusively upon photosynthesis and carbon assimilation to grow. Its obligate photoautotrophic lifestyle makes Synechococcus a useful system in which to investigate the coordination and regulation of these inherently essential metabolic processes.
Cyanobacteria frequently encounter transitions between light and dark in their environment, and these transitions fall into two distinct categories. One is due to the rising and setting of the sun, which yields predictable transitions from light to dark and back to light again. Synechococcus has a circadian rhythm that anticipates the timing of dawn and dusk and regulates the expression of a majority of its genes in a time-dependent manner (3, 4). The second type of light/dark transition is unpredictable and can occur due to transient cloud cover or shade cast by other organisms or geological features, for example. These transitions cannot be anticipated, and may require a rapid restructuring of metabolism. Studies of circadian rhythm in Synechococcus have provided insight into regulatory strategies that persist in constant light, but few studies have addressed the mechanisms by which cells adapt to darkness, predictable or otherwise.
Nearly all aspects of cyanobacterial physiology are affected by a shift from light to dark: cells stop elongating and dividing, cease DNA replication, and exhibit decreased rates of transcription and translation (5–7). Proteins produced in the dark differ from those produced in the light (8), and recent studies have identified genes that are differentially expressed between light and dark (9, 10). How the cell coordinates these transcriptional and translational changes remains largely unknown.
Not only does Synechococcus physiology change a great deal between light and dark, but its metabolism also shifts dramatically. Photosynthetically active cells reduce carbon dioxide into carbohydrates, which they accumulate as glycogen. When light is no longer available, cells catabolize their glycogen stores through respiration. Metabolism must be tightly controlled in the dark because total energy supply is finite and must be rationed (5), which raises the question of whether dark periods are analogous to starvation for Synechococcus and whether bacterial stress response mechanisms mediate adaptation to darkness.
Here, we show that the stringent response—a stress response pathway whose enzymes are conserved in nearly all bacteria, as well as plant plastids—is involved in dark adaptation in Synechococcus. We find that this pathway is active in Synechococcus and that it exerts dramatic effects on cellular physiology. Furthermore, this response is required for cells to adapt properly to darkness, as cells lacking the stringent response display pronounced growth defects in diurnal light/dark cycles and loss of viability after prolonged exposure to darkness. We investigate which genes are regulated by the stringent response in Synechococcus and find that one of them, ribosomal hibernation promoting factor, causes ribosomes to dimerize in the dark. Altogether, these results suggest that the stringent response mediates a coordinated transcriptional and translational reaction to periods of darkness.
Results
Darkness presents a major metabolic challenge for cyanobacteria, depriving them of their primary energy source—photosynthetically active radiation. In model heterotrophic bacteria, like Escherichia coli, metabolic stress by starvation leads to precipitous increases in levels of the alarmones ppGpp (guanosine 3′-diphosphate 5′-diphosphate) and pppGpp (guanosine 3′-diphosphate 5′-triphosphate) (11). Hereafter, we use the notation “(p)ppGpp” to refer to both molecules simultaneously when appropriate. Increased concentrations of (p)ppGpp, the nucleotide second messengers of the stringent response, cause polyphosphate (polyP)—a linear polymer of orthophosphates—to accumulate in E. coli (12). We hypothesized that increased levels of (p)ppGpp might also be responsible for increased polyP levels recently observed in Synechococcus in the dark (13). Although previous work has shown that (p)ppGpp is synthesized by Synechococcus (8, 14), mechanistic explanations for targets of the stringent response and its physiological effects are lacking, which led us to investigate the role of this pathway more broadly in Synechococcus.
ppGpp Levels Increase in the Dark in Synechococcus and Can Be Genetically Manipulated.
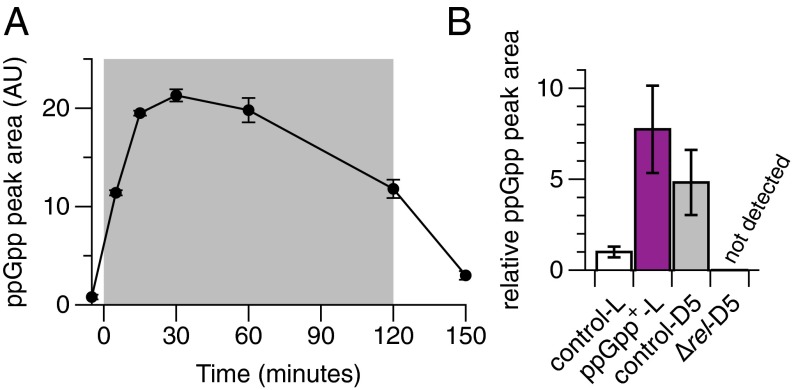
We hypothesized that the metabolic and physiological changes observed in Synechococcus cultures between light and dark could be due to altered levels of (p)ppGpp. We therefore shifted cells grown in the light into the dark, harvested cultures at defined time points, and analyzed cell extracts by high-performance liquid chromatography (HPLC) to determine ppGpp levels (Fig. 1A and SI Appendix, Fig. S1A). Cells growing in the light had low intracellular levels of ppGpp, which we estimated at around 5 μM (see Materials and Methods for details). When cells were shifted into the dark, ppGpp levels increased rapidly, peaked after ∼30 min, and remained elevated until cells were shifted back into the light. These results are consistent with those reported previously (8). Peak ppGpp levels in Synechococcus (30 min after onset of darkness) were ∼150 μM. We find, therefore, that ppGpp levels increase rapidly in Synechococcus in response to darkness.
Fig. 1.
ppGpp levels increase in the dark in Synechococcus and can be genetically manipulated. (A) Synechococcus cultures were shifted from the light (white background) to the dark (gray background) at 0 min and were harvested at the time points shown. Extracts were analyzed by anion exchange HPLC (AU, arbitrary units). Peaks eluting at the same time as a ppGpp standard were integrated to determine relative ppGpp levels. (B) Analysis of ppGpp levels from Synechococcus strains in the light (L) and the dark (D). Cultures harvested in the light were induced with IPTG for 17 h (control, WT-CmR). Cultures in the dark were harvested 5 min after the light-to-dark shift. ppGpp levels in the ∆rel mutant were below the limit of detection (∼1 μM). When analyzed using a one-tailed t test, ppGpp+-L vs. control-L, P = 0.0469; control-D5 vs. control-L, P = 0.0669. Data are presented as mean ± SEM (n = 3).
To artificially increase (p)ppGpp levels, we constructed a Synechococcus strain that inducibly expresses a small (p)ppGpp synthetase from Bacillus subtilis, yjbM/SAS1. This gene has been heterologously expressed in E. coli and resulted in high levels of (p)ppGpp (15). We refer to this strain as “ppGpp+.” As expected, the ppGpp+ strain had significantly higher ppGpp levels in the light than a control harboring an empty plasmid, as measured by HPLC and 32P-TLC (Fig. 1B and SI Appendix, Fig. S1 B–D). Based on the crystal structure and mutational studies of the homologous Rel (p)ppGpp synthetase from Streptococcus equisimilus (16), we generated a point mutant, D72G, to abrogate the activity of the B. subtilis synthetase. A recent crystal structure of yjbM/SAS1 revealed that Asp72 coordinates a magnesium ion required for catalytic activity (17). The D72G mutation did indeed inactivate (p)ppGpp synthetic activity, restoring ppGpp levels to those of the empty plasmid control (SI Appendix, Fig. S1 C and D). We refer to this strain as “ppGpp+ D72G.” Using these strains, we can control (p)ppGpp synthesis in Synechococcus, allowing us to investigate the cellular effects of manipulating (p)ppGpp levels.
High (p)ppGpp Levels Stop Growth and Dramatically Alter Synechococcus Physiology.
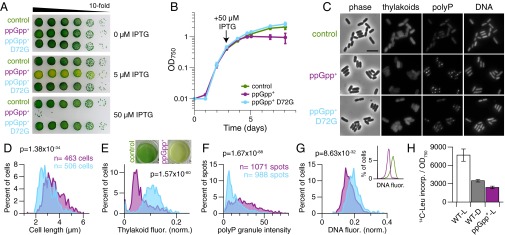
We next studied the effects of high (p)ppGpp levels on Synechococcus growth and physiology. When uninduced, the ppGpp+ strain grows as well as either an empty plasmid control or ppGpp+ D72G (Fig. 2A). Strikingly, upon addition of the inducer isopropyl β-d-1-thiogalactopyranoside (IPTG), a dramatic decrease in colony-forming units was observed in the ppGpp+ strain (Fig. 2A). When growth in liquid cultures was monitored by absorbance at 750 nm, a similar trend was apparent: ppGpp+ cultures stopped growing, whereas ppGpp+ D72G cultures grew as well as the empty plasmid control (Fig. 2B). Therefore, high (p)ppGpp levels are sufficient to stop growth of Synechococcus. When these cells were imaged by microscopy at 26 h after induction, a number of physiological changes were visible (Fig. 2C). For one, ppGpp+ cells were elongated compared with control cells, indicating that cell growth and division are misregulated when (p)ppGpp levels are high (Fig. 2D).
Fig. 2.
High (p)ppGpp levels stop growth and dramatically alter Synechococcus physiology. (A and B) Inducing high (p)ppGpp levels stops growth of Synechococcus. (A) When induced with IPTG, viability of the ppGpp+ strain decreases. Images are representative of three independent experiments. (B) Cultures of the indicated strains (control, WT-KanR) were grown in constant light, induced with IPTG where indicated, and monitored by absorbance at 750 nm (OD750). Data are presented as mean ± SD (n = 3). (C) At 26 h after IPTG induction, cultures were stained with DAPI and imaged by fluorescence microscopy. Representative images from three independent experiments are shown. (Scale bar: 5 μm.) (D–G) Microscopy images from two independent cultures at 26 h after induction were analyzed using MicrobeTracker and SpotFinder. Histogram colors match those in A, B, and C: purple, ppGpp+; light blue, ppGpp+ D72G. The same number of cells (ppGpp+, n = 463 cells; ppGpp+ D72G, n = 506 cells) was used for each analysis shown. Data were analyzed with the Mann–Whitney U test, and P values are indicated on the histograms. (D) ppGpp+ cells are longer than control cells. (E) Levels of light-harvesting pigments are lower in ppGpp+ cells than in control cells. Natural fluorescence from thylakoid membranes was normalized to cell area. (Inset) Pigmentation differences between control and ppGpp+ strains are striking. Cultures were imaged ∼48 h after induction. (F) Intensities of polyphosphate (polyP) granules are higher in ppGpp+ cells than in control cells. (G) DNA content is lower in ppGpp+ cells than in control cells. Histograms of DAPI-DNA fluorescence (AU; normalized to cell area) in ppGpp+ and ppGpp+ D72G cultures. (Inset) Analysis of DNA content by flow cytometry confirms that ppGpp+ cells contain less DNA per cell than control cells. Cultures were stained with Vybrant DyeCycle Green at 26 h after induction with IPTG. A total of 10,000 events were analyzed by flow cytometry for each condition. Histogram colors match those in A, B, and E: green, control; purple, ppGpp+. (H) Translation rates decrease in the dark and in ppGpp+ cells. Incorporation of 14C-Leu into trichloroacetic acid-precipitated proteins was measured by scintillation counting and is plotted as 14C counts per min/OD750. Labeling was performed for 1 h with 0.2 μCi 14C-Leu (for WT D, after a 2-h dark pulse; for ppGpp+, at 12 h after IPTG induction). Data are presented as mean ± SD (n = 6).
We noted striking differences in pigmentation between ppGpp+ and control cultures after induction, indicating that cells with high (p)ppGpp levels undergo chlorosis (bleaching) (Fig. 2E, Inset). Microscopy revealed that natural fluorescence from photosynthetic pigments was much lower in ppGpp+ cells than in either control condition (Fig. 2 C and E, SI Appendix, Fig. S2, and Materials and Methods). Furthermore, absorbance scans indicated that levels of pigments in phycobilisomes, major light-harvesting complexes, were lower in ppGpp+ cells and that chlorophyll absorbance also decreased (SI Appendix, Fig. S3). The lower levels of photosynthetic pigments in ppGpp+ cells could be due to increased pigment degradation, decreased pigment production, or a combination of the two. Based on these results, it is likely that ppGpp+ cells are less photosynthetically active than control cells (18, 19).
We tested whether (p)ppGpp affects levels of polyP, by staining cells with DAPI and using an imaging method that distinguishes between DAPI-polyP and DAPI-DNA fluorescence (Fig. 2 C and F, SI Appendix, Fig. S4, and Materials and Methods). In ppGpp+ cells, the intensity of polyP granules greatly increased (Fig. 2F), and numbers of polyP granules per cell were slightly higher than controls (SI Appendix, Fig. S4). Overall, these data show that (p)ppGpp regulates polyP granule formation in Synechococcus. Considering the finding that polyP granule size and number increase in the dark in Synechococcus (13), it is likely that (p)ppGpp stimulates polyP granule formation in the dark.
In ppGpp+ cells, DNA content per cell decreased, as measured by DAPI-DNA fluorescence (Fig. 2 C and G). We confirmed these results by staining cells with a different DNA-specific dye and analyzing cells by flow cytometry (Fig. 2G, Inset). These results suggest that (p)ppGpp decreases DNA replication in Synechococcus, which is consistent with the finding that (p)ppGpp regulates DNA replication in other bacteria. (p)ppGpp directly inhibits DNA primase from B. subtilis (20) and causes degradation of the replication initiation protein DnaA in Caulobacter crescentus (21).
To investigate whether the stringent response decreases translation rates, we measured incorporation of 14C-leucine into proteins under (p)ppGpp-varying conditions. We first verified that translation rates decrease in the dark in WT cells, as has been reported in the literature (22). After a 2-h incubation in the dark, translation rates decreased approximately twofold (Fig. 2H). Triggering high (p)ppGpp production in the light (ppGpp+) reduced translation rates to a level comparable with that of WT cells in the dark (Fig. 2H). Thus, (p)ppGpp is sufficient to markedly reduce translation rates in Synechococcus. Taken together, these results demonstrate a dramatic reshaping of cellular physiology and growth mediated by (p)ppGpp in this cyanobacterium.
(p)ppGpp Is Important for Maintaining Viability in Darkness.
The phenotypes exhibited by the ppGpp+ strain led us to investigate the endogenous stringent response pathway in Synechococcus. We deleted the rel gene, which encodes a bifunctional (p)ppGpp synthetase/hydrolase and is the only protein predicted to synthesize (p)ppGpp in the Synechococcus genome. As expected, this strain had very low ppGpp levels that were undetectable using our HPLC method, which has a limit of detection of ∼1 μM (Fig. 1B and SI Appendix, Fig. S1B).
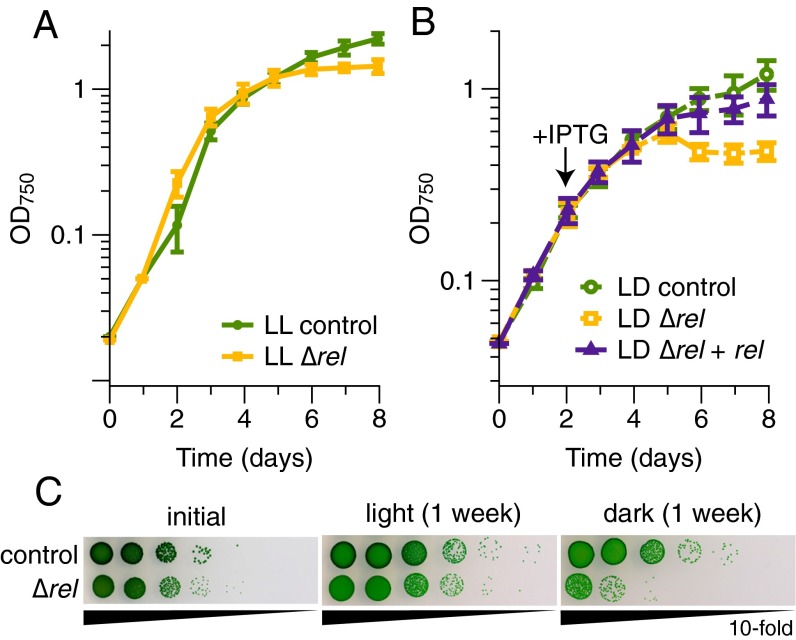
We postulated that (p)ppGpp would be more important for growth under changing environmental conditions than constant conditions. When grown in constant light, the control strain (WT harboring the same antibiotic resistance cassette as the mutant) and the ∆rel mutant grew similarly until they approached stationary phase, when growth of the ∆rel mutant became impaired (Fig. 3A). In 12-h light/12-h dark cycling conditions, however, the growth of the ∆rel mutant was severely impaired, and this phenotype could be rescued by expressing the rel gene from a neutral site in the genome (Fig. 3B).
Fig. 3.
(p)ppGpp is important for maintaining viability during darkness. (A) Growth of the control (WT-CmR) and the ∆rel mutant is similar in constant light (LL) until cells reach stationary phase. (B) Growth of the ∆rel mutant is impaired in 12-h light/12-h dark (LD) cycles, but complementation of the ∆rel mutant restores nearly WT growth (control, WT-CmR/KanR). An IPTG-inducible copy of the rel gene was reintroduced into the ∆rel mutant at a neutral site, and IPTG was added to all cultures when indicated by the arrow. Data are presented as mean ± SD (A, n = 3; B, n = 4). (C) Viability of the ∆rel strain decreases greatly after incubation in constant darkness. Tenfold serial dilutions of cultures were plated at the beginning of the experiment, after a week in constant light, or after a week in constant darkness. Images are representative of two independent experiments.
These phenotypes became even more pronounced when cells were maintained in darkness for an extended period of time. After a week in constant darkness, the viability of the ∆rel strain decreased several orders of magnitude relative to the control strain, whereas a week in constant light did not affect viability of the ∆rel strain (Fig. 3C). We find, therefore, that the stringent response is of limited import in constant light but helps Synechococcus respond to periods of darkness, highlighting the relevance of this pathway in environmental adaptation.
(p)ppGpp Regulates the Expression of Many Genes in Synechococcus.
To determine cellular targets downstream of (p)ppGpp in Synechococcus, we compared transcriptional profiles under (p)ppGpp-varying conditions using RNA-sequencing (RNA-seq). The reference conditions for this experiment were control cells (WT harboring an antibiotic resistance cassette) in the light or in the dark. We chose a 2-h dark pulse to study the effects of (p)ppGpp on gene regulation because this length of time would give the cells sufficient time to both increase their (p)ppGpp levels and to make resulting changes in gene expression.
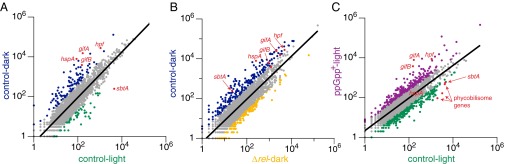
We found that expression of many genes changes between light and dark in WT cells (Fig. 4A), as has been seen previously (9, 10), and that some of these gene expression changes are (p)ppGpp-dependent (SI Appendix, Fig. S5). A comparison of gene expression between control-dark and ∆rel-dark revealed genes that are (p)ppGpp-regulated in the dark (Fig. 4B). At the same time, comparing gene expression between ppGpp+-light vs. control-light uncovered genes for which high (p)ppGpp levels were sufficient to alter regulation (Fig. 4C). In Fig. 4, each scatter plot is oriented such that the higher (p)ppGpp condition is on the y axis and the lower (p)ppGpp condition is on the x axis, to facilitate comparison. Points more than one SD above the regression line are considered up-regulated by (p)ppGpp, and points more than one SD below the regression line are considered down-regulated by (p)ppGpp. This stringent cutoff resulted in seven genes consistently up-regulated by (p)ppGpp across the three comparisons presented in Fig. 4 (listed in SI Appendix, Table S1). SI Appendix, Tables S2 and S3 list genes up-regulated (75 genes) or down-regulated (37 genes) by (p)ppGpp, respectively, in at least two comparisons. Expression data for all genes can be found in Dataset S1.
Fig. 4.
(p)ppGpp regulates the expression of many genes in Synechococcus. RNA-seq was performed for four different conditions: control (WT-CmR)-light; ppGpp+-light, ppGpp+ cultures in the light after 18 h of IPTG induction; control-dark, WT-CmR cultures after a 2-h dark pulse; ∆rel-dark, ∆rel cultures after a 2-h dark pulse. RNA-seq data were analyzed and normalized using Rockhopper. For all conditions except ppGpp+, n = 4 biological replicates; for ppGpp+, n = 3 biological replicates. All panels show scatter plots of expression values based on upper-quartile normalization. The regression line used to identify differentially expressed genes is shown (A, R2 = 0.81224; B, R2 = 0.77495; C, R2 = 0.7258). Genes with expression values at least one SD higher or lower than the values predicted by the regression line are colored according to the condition under which they are more highly expressed. Genes not considered differentially expressed are colored in gray. All plots are shown such that the higher (p)ppGpp condition is on the y axis and the lower (p)ppGpp condition is on the x axis: (p)ppGpp–up-regulated genes are above the regression line whereas (p)ppGpp–down-regulated genes are below the regression line. Selected differentially expressed genes discussed in (p)ppGpp Regulates the Expression of Many Genes in Synechococcus are colored in red, with the gene name indicated.
Several (p)ppGpp-up-regulated genes were of particular interest (Fig. 4 and SI Appendix, Table S1). Two genes strongly induced by (p)ppGpp were gifA and gifB, both of which encode glutamine synthetase inactivating factors. As a key link between carbon and nitrogen metabolism, glutamine synthetase is often a target of complex regulation (23). The gifA and gifB genes encode IF7 and IF17, respectively, both of which are small proteins that bind directly to glutamine synthetase and down-regulate its activity (24). When cultures of the cyanobacterium Synechocystis sp. PCC 6803 are shifted from one nitrogen source (nitrate) to another (ammonium), gifA and gifB help tune glutamine synthetase activity and improve cellular growth as a result (25). An analogous balancing could occur in the dark when metabolic changes necessitate tuning of enzyme activities.
A chaperone-encoding gene known to be highly expressed in the dark, hspA, required (p)ppGpp for its up-regulation (Fig. 4 and ref. 10). However, high (p)ppGpp levels in ppGpp+-L were not sufficient to induce high levels of the hspA transcript. It is likely that additional dark-induced but (p)ppGpp-independent factors are required for induction of hspA and other genes with similar expression patterns. (p)ppGpp–down-regulated genes were less consistent across comparisons, but one notable example encoded the bicarbonate transporter sbtA.
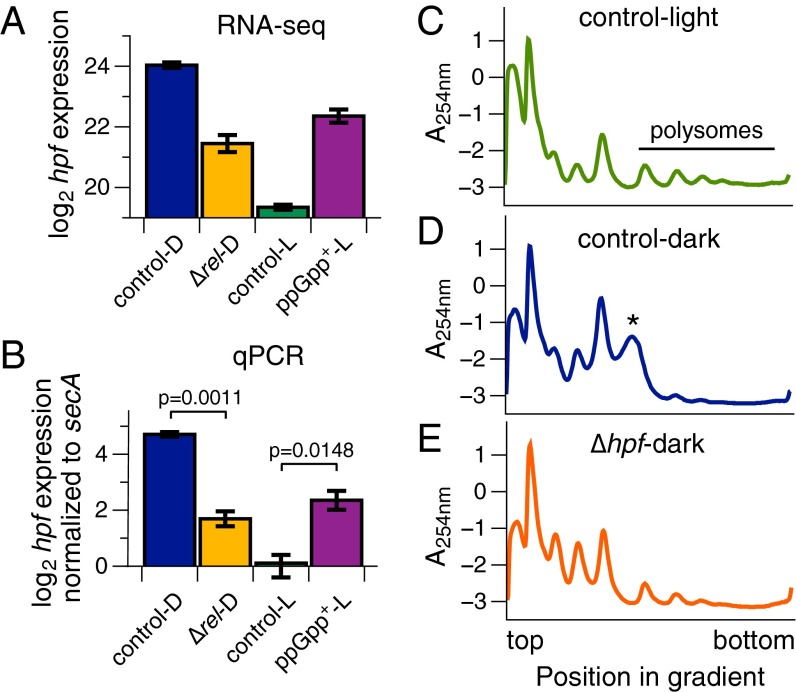
We confirmed (p)ppGpp-dependent gene expression for several genes using quantitative reverse transcriptase PCR (qPCR) and observed trends similar to RNA-seq results (Fig. 5 A and B and SI Appendix, Fig. S6). Furthermore, these trends were consistent after using two or three different reference genes for qPCR normalization (Fig. 5B and SI Appendix, Fig. S6). Fig. 5 A and B shows expression of a highly (p)ppGpp–up-regulated gene, hpf, which encodes ribosomal hibernation promoting factor (see the following section). SI Appendix, Fig. S6 contains verification of expression patterns for gifA and sbtA.
Fig. 5.
(p)ppGpp regulates translation through hibernation promoting factor. (A) RNA-seq expression data reveal striking regulation of hpf by (p)ppGpp. Rockhopper-normalized expression values plotted on a log2 scale. Data are presented as mean ± SEM (for all conditions except ppGpp+, n = 4; for ppGpp+, n = 3). (B) Verification of hpf gene regulation by quantitative reverse transcriptase PCR (qPCR). hpf expression was normalized to secA expression and plotted on a log2 scale relative to control-L. A two-tailed t test between the indicated conditions was performed, and P values are shown. Data are presented as mean ± SEM (n = 4 biological replicates). (C–E) Polysome profiles from Synechococcus lysates analyzed by sucrose density gradient centrifugation. Cultures were grown to midlog phase and shifted into the dark for 2 h where appropriate. Two minutes before harvesting, all cultures were treated with chloramphenicol to arrest translation elongation. Cell lysates were separated on 10 to 40% sucrose gradients by ultracentrifugation. Abundance of RNA species was monitored by absorbance at 254 nm (A254nm; arbitrary units). All traces are representative of two independent biological replicates. (C) Control (WT-KanR) cells in the light were actively translating, as revealed by their abundant polysomes. (D) After a 2-h dark pulse, control cells exhibited decreased translation and instead contained dimerized ribosomes, as indicated by the asterisk (*). (E) hpf is required for ribosome dimerization and decreased translation in the dark. After a 2-h dark pulse, lysate from a ∆hpf mutant contained abundant polysomes and completely lacked dimerized ribosomes.
In other bacteria, the stringent response dramatically decreases production of ribosomal RNAs (rRNAs) (26). We performed qPCR experiments to measure total levels of 16S rRNA as well as rRNA precursor transcripts from cultures grown in light and dark and after induction of ppGpp+ strains. During rRNA maturation in bacteria, the 5′ leader sequence upstream of the 16S rRNA transcript is processed by RNase III, which continues even when rRNA transcription slows or stops (27). By measuring amounts of the 16S 5′ leader sequence (pre-16S rRNA transcripts), one can infer the rate of change of rRNA levels. Total levels of 16S rRNA did not change under any condition in our experiments (SI Appendix, Fig. S7 A and B). Levels of pre-16S transcripts relative to total 16S transcripts decreased ∼10-fold after cells had been in the dark for 12 h (SI Appendix, Fig. S7C), but inducing high (p)ppGpp levels in the light did not affect pre-16S rRNA levels (SI Appendix, Fig. S7D). We conclude that rRNA production rate decreases in the dark but that this decrease requires other factors besides (p)ppGpp.
In sum, (p)ppGpp regulates expression of a number of genes in Synechococcus, some of which seem to be controlled solely through (p)ppGpp, and some of which require additional factors for their expression under light/dark conditions. Although the functions of some (p)ppGpp-regulated genes are known, the majority of genes lack a predicted function.
(p)ppGpp Regulates Ribosomal Status Through Hibernation-Promoting Factor.
One of the most strikingly (p)ppGpp–up-regulated genes encodes ribosomal hibernation promoting factor (hpf), a protein that dimerizes ribosomes into a less active state (Fig. 4) (28, 29). In E. coli, HPF and ribosome modulation factor act together to dimerize ribosomes and block binding of mRNAs, tRNAs, and translation initiation factors to the ribosome (30). HPF is widely conserved across bacterial phyla, including cyanobacteria, as well as in plant plastids, and a long form of HPF from several bacterial clades is sufficient to dimerize ribosomes on its own (28, 29).
To determine whether HPF acts in a similar manner in Synechococcus in the dark, we monitored ribosomal status using sucrose density gradient centrifugation. In actively translating cells, ribosomes exist in multiple forms: small (30S) and large (50S) subunits, assembled 70S monosomes, and polysomes, multiple ribosomes translating the same mRNA. Polysomes can be resolved on sucrose gradients as distinct peaks corresponding to two, three, four, etc. ribosomes bound to one mRNA. The presence of polysome peaks in cell lysates can be used to indicate translational status (31).
We determined polysome profiles from Synechococcus and found that hpf is necessary for ribosome dimerization in the dark. Because there have been no reports of polysome analyses in Synechococcus in the literature, we confirmed peak identities by comparison with E. coli lysates (31). Synechococcus ribosomes sedimented similarly to those of E. coli, with characteristic peaks corresponding to small and large ribosomal subunits, monosomes, and polysomes (SI Appendix, Fig. S8A). Treatment of cell lysates with RNase A to cleave mRNA, which should remove polysomes, verified the position of polysome peaks (SI Appendix, Fig. S8B).
We tested Synechococcus lysates from control (WT) cells in the light, and control and ∆hpf cells after a 2-h dark pulse. In the light, control cell lysates contained several polysome peaks, indicating active translation (Fig. 5C). In control cells in the dark, however, a significant fraction of ribosomes exist in a state that sediments between monosomes and the first polysome peak, likely corresponding to dimerized ribosomes (asterisk in Fig. 5D). Furthermore, few polysomes were observed, consistent with the observation that translation rates are lower after 2 h in the dark in Synechococcus (Fig. 2H). In the ∆hpf mutant, ribosomes did not dimerize and instead could be found as polysomes (Fig. 5E). We conclude from these results that ribosomal status is altered in the ∆hpf mutant, so that the ribosomal pool of the ∆hpf mutant resembles that of WT cells in the light. Overall, these data suggest that hpf could be one mechanism by which (p)ppGpp controls translation, tuning protein synthesis in response to environmental cues.
Discussion
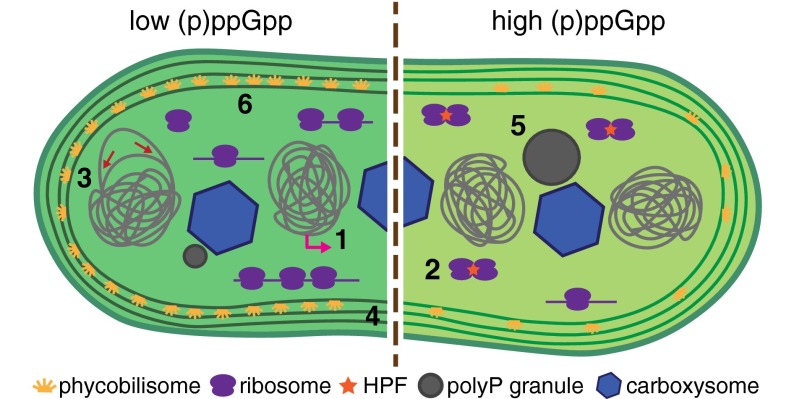
Here, we have shown that the stringent response is an important mechanism by which Synechococcus adapts to darkness. Levels of (p)ppGpp rise in response to a light-to-dark shift, causing dramatic changes in gene expression and regulating ribosomal populations through HPF. By inducing high (p)ppGpp levels, we find that (p)ppGpp can control many fundamental cellular processes in Synechococcus (Fig. 6).
Fig. 6.
Model of (p)ppGpp regulation in Synechococcus. Our results indicate that many fundamental processes are regulated by (p)ppGpp in Synechococcus. These processes are schematized here and include (site 1) transcription, (site 2) translation, (site 3) DNA replication, (site 4) cell growth and division, (site 5) polyP granule formation, and (site 6) photosynthesis.
The Stringent Response as a Coordinator of Light/Dark Physiology in Synechococcus.
It has been known for many years that cyanobacteria respond and adapt to both high-light and low-light conditions. Although there is greater understanding of the photoprotective mechanisms that allow cells to adapt to high light (2), relatively little is known about how cells coordinate their response to darkness.
Our findings provide evidence that the stringent response helps Synechococcus adapt to darkness and help explain previous observations about cyanobacterial physiology. In 1975, Singer and Doolittle reported that translation rates fall dramatically after a light/dark shift in Synechococcus (22). It is also known that treating Synechococcus with inhibitors of photosynthetic electron transport generally suppresses translation (32). We propose that loss of photosynthetic activity leads to increased (p)ppGpp levels, triggering increased HPF production. Higher levels of HPF dimerize ribosomes and likely work with additional cellular factors to decrease translation rates. High (p)ppGpp levels also suppress transcription of a subset of genes, slow or stop DNA replication, and prevent cell division in the dark (Fig. 2).
Circadian rhythm is an important global regulator in Synechococcus and is known to improve cellular fitness in oscillating light/dark cycles (33). It is interesting, therefore, that the capacity to stimulate ppGpp production seems to be independent of circadian rhythm. The experiments presented in Fig. 1 were performed using replicate Synechococcus cultures harvested at different times during the circadian cycle, yet the relative ppGpp levels and kinetics of production are remarkably consistent across replicates. We have shown that the stringent response is important for responding to regular light/dark cycles (Fig. 3B), but it is also activated during unexpected periods of darkness. It is likely that, in its native freshwater environment, Synechococcus could experience intermittent shading, a situation in which an adaptive response to darkness would be of significant benefit.
Conservation and Adaptation of Stringent Response Mechanisms in Diverse Bacteria.
The stringent response was first identified in E. coli (34), and more recent studies have uncovered targets and mechanisms of this pathway in B. subtilis, C. crescentus, and several bacterial pathogens (35–37). Although the stringent response works to restore metabolic homeostasis in both E. coli and B. subtilis, the specific targets of (p)ppGpp are distinct in these organisms. Although (p)ppGpp binds directly to RNA polymerase and works with the transcription factor DksA in E. coli to control gene expression, (p)ppGpp regulates GTP biosynthesis in B. subtilis by directly inhibiting enzymes in this pathway, including guanylate kinase (GMK) (38). Altered GTP levels then affect gene expression in B. subtilis and other Firmicutes through the GTP-sensing transcription factor CodY (39). A recent phylogenetic analysis suggests that regulation of RNA polymerase by (p)ppGpp is conserved throughout the Proteobacteria whereas regulation of GTP biosynthesis is conserved throughout the Firmicutes (40). This analysis does not predict a mechanism for the stringent response in cyanobacteria, but several observations suggest that neither of these strategies fully accounts for the global regulation enacted by (p)ppGpp in Synechococcus. In vitro assays with Synechococcus GMK have shown that it is insensitive to (p)ppGpp (41), and its genome lacks CodY and DksA homologs as well as the (p)ppGpp-interacting motif on RNA polymerase (42). Although it is likely that alteration of GTP pools as a result of (p)ppGpp synthesis could affect cell physiology and metabolic processes, we did not observe dramatic decreases in rRNA precursor levels when (p)ppGpp levels were high (SI Appendix, Fig. S7), even though GTP is the initiating nucleotide for rRNA transcription in Synechococcus (43).
As a photoautotroph, Synechococcus lives a very different metabolic lifestyle than the bacterial model systems in which the stringent response has been studied. Because photosynthesis forms the foundation of cyanobacterial growth, it is logical that (p)ppGpp would be synthesized in conditions that are unfavorable for photosynthesis, like darkness, and that this pathway could feed back to regulate levels of light-harvesting pigments. A study in the facultative phototroph Rhodobacter capsulatus suggested a link between the stringent response and regulation of photosynthetic gene expression/genome structure by the nucleoid protein HvrA (44), but this α-proteobacterium is only distantly related to cyanobacteria and has greater metabolic flexibility. It is likely that the strategies used by cyanobacteria to respond to darkness differ from those used by facultative phototrophs. We are beginning to appreciate the mechanisms behind the stringent response in Synechococcus, but much work remains to determine how (p)ppGpp levels can lead to altered physiology in phototrophic organisms.
HPF and Translational Regulation by (p)ppGpp.
Regulation of translation is a classic mechanism of the stringent response that can be accomplished by immediate action of (p)ppGpp on the translation apparatus, or by longer term changes in gene expression of rRNAs, tRNAs, and ribosomal and ribosome-associated proteins. These adaptations are important during starvation, because production of ribosomes can consume a significant fraction of the cell’s energy supply (45). Immediately after encountering a stress, the cell contains its full complement of ribosomes and uses posttranslational mechanisms to decrease the activity of these preexisting ribosomes. One way bacteria can decrease translation rate is through direct inhibition of ribosomal GTPases by (p)ppGpp, as has been shown for the translation initiation factor IF2 (46), as well as for GTPases of Staphylococcus aureus (47). Alternatively, expression of factors such as HPF can modulate ribosome activity over longer timescales (48).
HPF is known to dimerize ribosomes in Listeria monocytogenes, B. subtilis, and E. coli in response to conditions that increase (p)ppGpp levels, such as stationary phase (49, 50). We have shown that hpf expression is (p)ppGpp-regulated and that hpf affects ribosomal status under conditions that decrease translation rates in Synechococcus, suggesting that it may contribute to this phenotype. Twenty years ago, HPF was identified as a protein made at high levels in the dark in Synechococcus sp. PCC 7002, and a mutation in this gene seems to alter patterns of protein synthesis after dark adaptation (51). This result is an intriguing one, but little is known about the extent to which HPF affects translation rates in vivo in any organism or whether HPF might alter translational specificity. Synechococcus synthesizes a distinct set of “dark-specific” polypeptides after a shift to darkness (8)—which is consistent with the transcriptional changes seen under these conditions—but the identities of these proteins and how production of specific proteins is controlled remain unknown.
Signals That Trigger the Stringent Response Are Different in Photosynthetic Organisms.
The range of stresses sensed by the stringent response in other organisms encompasses carbon sources, nitrogen sources, and inorganic nutrients. Light represents an equally—if not more—important signal of nutritional status in Synechococcus. A cyanobacterial cell experiences dramatic intracellular changes in its redox state and pH, for example, depending on whether it is actively photosynthesizing (52). Redox state regulates centrally important metabolic enzymes in the cyanobacterium Synechocystis through thioredoxins, which mediate reduction of disulfides (53). Similar signals could trigger (p)ppGpp synthesis once photosynthesis stops. Inhibition of photosynthesis is known to reduce translation in Synechococcus (32), an effect we also observed after inducing high (p)ppGpp levels. It would be interesting to determine whether inhibitors of photosynthesis mimic the effects of darkness and induce other physiological effects of the stringent response. Several observational studies have shown that ppGpp levels in Synechococcus rise in response to stresses, such as elevated temperature (8) and nitrogen starvation (14, 54), but these studies did not investigate the mechanisms upstream of ppGpp synthesis or the physiological responses of the organism under these conditions.
Plants also contain (p)ppGpp metabolic enzymes (55). In Arabidopsis thaliana, all of these proteins localize to the chloroplast (19, 56), and ppGpp levels rise in plants in response to darkness and other stresses, including wounding (57). Two recent studies have also shown that ppGpp levels affect photosynthetic capacity and chloroplast development in Arabidopsis (19, 58). Thus, it is likely that aspects of the stringent response pathway are conserved between cyanobacteria and chloroplasts.
Diverse bacterial taxa have different configurations of (p)ppGpp synthetases and hydrolases, but the fact that they are present in all but obligate pathogens suggests that they have been adapted to incredibly diverse lifestyles. Cyanobacteria represent an understudied group of organisms that could provide clues to the core principles and flexible components of this conserved bacterial stress response, while at the same time telling us about the unique stresses faced by phototrophs.
Materials and Methods
Bacterial Strains and Culture Conditions.
S. elongatus PCC 7942 was grown in BG-11 media (59) at 30 °C with shaking (185 rpm) under white fluorescent lights at 60–100 μE. Cultures grown in diurnal (12-h light/12-h dark) cycles were incubated in a programmable photosynthetic incubator (Percival Scientific); otherwise, cultures grown in constant light were incubated in an environmental (30 °C) room. All processing of samples under dark conditions was performed in a dark room with a minimal amount of light provided by a green LED. Unless otherwise indicated, Synechococcus cultures were induced with 50 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) when appropriate. Single antibiotics (chloramphenicol, kanamycin) were used at 10 μg/mL, and double antibiotics were used at 2 μg/mL each. E. coli was grown in LB media at 37 °C with shaking (250 rpm), unless otherwise noted. When appropriate, chloramphenicol was used at 25 μg/mL, and kanamycin was used at 60 μg/mL for E. coli. Measurements of culture optical density (OD600 or OD750, subscript indicating wavelength in nm) were performed using a Thermo Scientific Genesys 20 spectrophotometer.
Plasmid and Strain Construction.
All plasmids were constructed using a Golden Gate cloning strategy (60) and were propagated in E. coli DH5α. Primers were designed to amplify genes such that a BsaI restriction enzyme site would be added onto both ends, and overhangs generated by digestion with BsaI would be complementary to those present in Golden Gate destination plasmids. Golden Gate reactions (incorporating cycles of restriction enzyme digestion and ligation) were incubated for 50 cycles of 45 °C for 5 min and 16 °C for 2 min, followed by incubations at 50 °C for 10 min and 80 °C for 10 min to inactivate the enzymes. Standard methods were used for PCR, gel purification of PCR products, E. coli transformation, and DNA sequencing to verify cloned constructs. SI Appendix, Table S4 lists the plasmids and primers used in this study.
Synechococcus was transformed by growing cultures to log phase (OD750 0.2–0.6), harvesting cells by centrifugation (16,000 × g, 2 min, 25 °C), washing cells once in 0.5× volume of 10 mM NaCl, and resuspending cells in 0.1× volume of BG-11 media. Approximately 200 ng of the plasmid to be transformed was added, and cultures were wrapped in foil and incubated overnight at 30 °C. The next day, transformations were plated on BG-11 plates containing the appropriate selective antibiotic. All deletion strains were verified by colony PCR, using primers to detect both the native locus and the deletion construct, and were fully penetrant. SI Appendix, Table S5 lists and describes the strains used in this study.
ppGpp Measurements by HPLC and Estimation of Intracellular Concentrations.
Synechococcus cultures grown under the conditions specified in the text were harvested by filtration. For the light/dark shift experiment shown in Fig. 1A, a WT culture was grown to log phase (OD750 ∼0.25) and split into seven flasks with 57 mL each (corresponding to ∼13 OD750 units), and each culture was harvested at the appropriate time point. For the experiment shown in Fig. 1B, cultures of the appropriate strains were grown to log phase (OD750 ∼0.4), and strains in the light (control and ppGpp+) were induced with IPTG for 17 h before harvesting 20 OD750 units per culture.
Cells were isolated by filtration, resuspended, and lysed in 400 μL of 13 M formic acid, subsequently flash-frozen in liquid nitrogen, and stored at −80 °C until further processing. Lysates were thawed at room temperature, were subjected to three freeze/thaw cycles alternating between a dry ice/ethanol bath and room temperature, and were clarified by centrifugation (16,000 × g, 5 min, 4 °C). Extracts were filtered using PES syringe filters and were flash-frozen in liquid nitrogen and stored at −80 °C until HPLC analysis.
Levels of ppGpp were measured using anion-exchange chromatography and a Phenomenex Luna NH2 column, 50 × 4.60 mm. Using a protocol modified from ref. 61, an isocratic method was developed to isolate the ppGpp peak on an Agilent Technologies 1200 series HPLC. The buffer consisted of 0.85 M ammonium phosphate, pH 2.1. Samples were thawed on ice before transfer to a prechilled HPLC vial; all samples were kept on ice until just before injection. Then, 25 μL was injected at time 0, and the method was run at 1 mL/min for 8–10 min. The column temperature was maintained at 30 °C, and absorbance was monitored at 254 nm. A small ppGpp peak was found to elute at ∼3.5 min. Concentrations were estimated by manually integrating peaks using Agilent Chemstation software and comparing with a ppGpp standard added to the sample. The ppGpp standard was obtained from TriLink Biotechnologies. Using this method, the limit of detection for ppGpp was ∼1 μM. We did not measure pppGpp by HPLC because we were unable to resolve any candidate peaks eluting after ppGpp.
To estimate intracellular ppGpp concentrations, we used the equation
where each parameter was determined as follows: mAU⋅s, value determined for each sample by integrating ppGpp peaks from HPLC traces; moles/(mAU⋅s) conversion, determined by running a standard curve with known molar amounts of the ppGpp standard vs. measured mAU⋅s and taking the slope of a linear regression (R2 = 0.998) [value = 2.4 × 10−12 moles/(mAU⋅s)]; fraction of total lysate measured, 25 μL was analyzed by HPLC from an original volume of 400 μL (value = 16); efficiency of ppGpp extraction, determined by spiking a known amount of ppGpp into the 400 μL of formic acid used to lyse cells and measuring how much was recovered compared with that measured after spiking in ppGpp at the end of the extract preparation (∼37% efficiency; value = 2.69); number of cells, determined by plating dilutions from cultures harvested for ppGpp extraction and averaging the number of colony-forming units across replicates (value = 4.69 × 109 cells); and estimated cell volume, determined by analyzing microscopy images of WT Synechococcus using MicrobeTracker software (62) and converting pixels3 into μm3 into L (value = 3 fL = 3 × 10−15 L).
ppGpp Measurements by 32P-TLC.
Synechococcus strains were grown in low-phosphate BG-11 media (as in ref. 59, except containing 44 μM rather than 175 μM K2HPO4) to log phase (OD750 ∼0.5). E. coli strains were grown in low-phosphate minimal media (50 mM KCl, 10 mM NH4Cl, 0.5 mM MgSO4, 0.2 mM KH2PO4, 50 mM Hepes, pH 7.5, 100 μM FeCl3, 0.4% glucose, 0.2% casamino acids) at 30 °C to midlog phase (OD600 ∼0.5). H332PO4 (Perkin-Elmer) was added to the appropriate low-phosphate medium at 20 μCi/mL, along with 50 μM IPTG for Synechococcus cultures where appropriate, and cell pellets (normalized to 0.5 OD units) were resuspended in low-phosphate media containing H332PO4. Cultures were incubated without shaking in a 30 °C incubator for at least one generation time (30 min for E. coli; 27 h for Synechococcus) to allow for 32P uptake and incorporation. E. coli cultures were treated with serine hydroxamate (Sigma-Aldrich) at 1 mg/mL to mimic amino acid starvation for 10 min before harvesting.
Cells were harvested and excess H332PO4 was removed by centrifugation (10,000 × g, 2 min, 25 °C), and pellets were resuspended in the appropriate 32P-free low-phosphate medium. An equal volume of 13 M formic acid was added to cell suspensions, which were then subjected to three freeze–thaw cycles alternating between a dry ice/ethanol bath and room temperature. Lysates were clarified by centrifugation (10,000 × g, 2 min, 25 °C) and were spotted onto PEI-cellulose TLC plates (EMD Millipore) along with a [γ-32P]-GTP standard (Perkin-Elmer). TLC plates were run in a chamber containing 1.5 M KH2PO4, pH 3.4, for 30 min and exposed to a phosphorscreen for ∼24 h. Imaging and quantitation were performed using a GE Healthcare Typhoon phosphorimager. Spot intensities were quantified using ImageQuant software.
Microscopy and Image Analysis.
Synechococcus cultures were harvested at the appropriate time point (26 h after IPTG induction for the experiment shown in Fig. 2) and were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) at 1 mg/mL for 15 min in the dark at room temperature. Stained cells were washed two times in PBS and resuspended in BG-11 medium. Cells were immobilized by spotting cultures onto agarose pads (2% agarose in BG-11 medium). Imaging was performed on a Zeiss AXIO Observer.Z1 inverted microscope equipped with a 100× phase-contrast oil objective (NA 1.4) and an Excelitas Technologies X-Cite 120Q fluorescent light source. Images were acquired using a Hamamatsu Photonics ORCA-Flash 4.0 scientific complementary metal oxide semiconductor (sCMOS) camera and Zeiss ZEN 2012 software.
Cyanobacteria naturally fluoresce in the red portion of the visible spectrum due to both chlorophyll-containing photosystems and the light-harvesting pigments present in phycobilisomes, both of which are found in the thylakoid membranes (63). Pigment fluorescence was assayed using a standard red fluorescent protein (RFP) filter set (excitation 572 nm; emission 629 nm). Polyphosphate (polyP) was imaged using a standard cyan fluorescent protein (CFP) filter set (excitation 436 nm; emission 480 nm). Upon binding to negatively charged polyP, the positively charged DAPI molecule exhibits a spectral Stokes shift that permits DAPI-polyP fluorescence to be distinguished from DAPI-DNA fluorescence using these wavelengths (64). At the same time, a standard DAPI filter set was used to assay DAPI-DNA fluorescence (excitation 365 nm; emission 445 nm). Microscopy images were analyzed using MicrobeTracker and SpotFinder, bacterial image analysis programs written in Matlab (62). Segmentation of cells was manually verified and corrected when necessary.
Flow Cytometry.
Synechococcus cultures were diluted to OD750 ∼0.03 and were stained with Vybrant DyeCycle Green (Thermo Fisher) at 26 h after IPTG induction. The final dilution of the stain was 1:5,000. Cultures were stained for 45 min in the dark at 30 °C. Stained cells were analyzed on an EMD Millipore Guava EasyCyte HT flow cytometer; 10,000 events were analyzed for each condition.
14C-Leu Protein Incorporation Measurements.
Synechococcus cultures were grown in constant light to log phase (OD750 ∼0.4), and ppGpp+ cultures were induced with 50 μM IPTG for 12 h. Aliquots (0.5 mL) of cultures were incubated in the dark for 2 h where appropriate, and all cultures were then labeled with 0.2 μCi 14C-Leu (Perkin-Elmer) for 1 h. Cells were lysed by adding SDS to a final concentration of 1% and heating at 80 °C for 15 min, followed by cooling on ice. To precipitate proteins, ice-cold trichloroacetic acid (TCA) was added to cell lysates at a final concentration of 5%, and lysates were incubated on ice for 30 min. Lysates were centrifuged (21,000 × g, 10 min, 25 °C), and pellets were washed once in ice-cold 5% TCA to remove unincorporated 14C-Leu and pellets were centrifuged again. The entire protein pellet was resuspended in 8 M urea, and 14C counts were determined using a Beckman LS6500 scintillation counter.
Absorbance Scans.
Absorbance scans of Synechococcus cultures were performed using a Tecan Infinite M1000 Pro plate reader, measuring absorbance between 350 and 800 nm at 5-nm intervals. An identical absorbance scan was performed using sterile media, and these values were subtracted from culture readings.
RNA-seq: RNA Library Preparation, Sequencing, and Data Analysis.
RNA was isolated from four different conditions as follows: control-L, WT cultures (WT-CmR) in the light (+50 μM IPTG for 18 h); ppGpp+-L, ppGpp+ cultures in the light (induced with 50 μM IPTG for 18 h); control-D, WT cultures (WT-CmR) at the end of a 2-h dark pulse; and ∆rel-D, ∆rel cultures at the end of a 2-h dark pulse. All cultures were synchronized by two 12-h light/12-h dark cycles and were harvested at the same time during the subjective day (L7) to minimize effects of circadian rhythm on gene expression. Cultures (100 mL; OD750 0.2–0.4) were harvested by filtration; cell material was scraped off filters and resuspended in 400 μL of AE buffer (50 mM sodium acetate, pH 5.5, 10 mM EDTA, pH 8.0). Cell suspensions were flash-frozen in liquid nitrogen and stored at −80 °C until further processing.
RNA was isolated using a method based on ref. 4 in which cells were lysed using hot acid phenol/SDS and phenol/chloroform extraction. Cell suspensions were thawed at room temperature. Then, 0.1× volume (40 μL) of 10% SDS was added to each tube, followed by an equal volume (440 μL) of phenol (equilibrated with AE buffer). Tubes were incubated at 65 °C for 4 min, frozen in a dry ice/ethanol bath for ∼1 min, and centrifuged to separate aqueous and phenol phases (16,000 × g, 5 min, 25 °C). The aqueous phase was transferred to a phase lock gel heavy tube (5 Prime), to which an equal volume (∼350 μL) of acid phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5) was added. These tubes were centrifuged to separate aqueous and phenol phases (16,000 × g, 5 min, 25 °C). Then, 200 μL of the aqueous phase was further purified using a Qiagen RNeasy Mini kit according to the manufacturer’s protocol. RNA concentrations were determined using a Thermo Scientific Nanodrop 2000 spectrophotometer. Total RNA (5 μg) was treated with 2 units of DNase I (Thermo Scientific) to remove genomic DNA contamination according to the manufacturer’s recommendations. Twenty units of RNase inhibitor (SUPERaseIn; Thermo Fisher) were added to reactions, which were incubated at 37 °C for 75 min. EDTA was added to reactions at a final concentration of 2.5 mM, and reactions were incubated at 65 °C for 10 min to stop DNase I activity. DNase-treated RNA was further purified using a Zymo Research RNA Clean and Concentrator-5 kit according to the manufacturer’s protocol. RNA integrity was verified at multiple steps using the Agilent Technologies RNA 6000 Nano kit and an Agilent 2100 Bioanalyzer according to the manufacturer’s protocol.
Total RNA (∼1.5 μg) was depleted of ribosomal RNA using an Illumina RiboZero rRNA depletion kit (Bacteria), and RNA-seq libraries were constructed using the BIOO Scientific, NEXTflex Rapid Illumina Directional RNA-Seq Library Prep Kit (cat no. 5138-08) using 15 cycles of PCR. Samples were barcoded, pooled in equimolar ratios, and sequenced in one lane on an Illumina HiSeq2500, generating 50-bp single-end reads. Each condition sequenced had four biological replicates, except for ppGpp+, which had three.
RNA-seq library adapter contamination was removed using Scythe (bioinformatics.ucdavis.edu/research-computing/software/). The resulting data were aligned to the Synechococcus genome, normalized, and analyzed using Rockhopper, a bacterial RNA-seq analysis platform (65). Rockhopper performs upper-quartile normalization to generate RNA-seq expression values for each condition. RNA-seq expression values were plotted in DataGraph (Visual Data Tools, Inc.), and a power regression analysis was performed. Genes with expression values at least one SD higher or lower than the values predicted by the regression line were considered differentially expressed.
Quantitative RT-PCR.
For qPCR experiments to verify RNA-seq results, RNA isolation, DNase treatment, and RNA cleanup were performed as described for RNA-seq experiments. RNA concentrations were determined using a Thermo Scientific Nanodrop 2000 spectrophotometer and were standardized to the same amount of input RNA (∼1 μg) for reverse transcription reactions, which were performed using Clontech RNA to cDNA EcoDry Premix with random hexamers according to the manufacturer’s protocol. The resulting cDNA was diluted 1:10 before amplification in qPCR reactions.
For qPCR experiments to measure pre-16S and 16S rRNA levels, cultures were synchronized by one 12-h light/12-h dark cycle, and 50 mL at an OD750 of 0.2–0.4 (maintained over the course of the experiment by manual back-dilution) was harvested at the appropriate time point by filtration. Cell material was scraped off filters, resuspended in 1 mL of TRIzol reagent (Invitrogen), flash-frozen in liquid nitrogen, and stored at −80 °C until further processing. RNA isolation was performed according to the manufacturer’s protocol, with one additional incubation at 90 °C for 5 min immediately after thawing frozen samples. DNase treatment and RNA cleanup were performed as described for RNA-seq experiments. Reverse transcription reactions were performed with the same amount of input RNA (1 μg), using Invitrogen SuperScript III reverse transcriptase with random primers according to the manufacturer’s protocol. The resulting cDNA was diluted 1:100,000 before amplification in qPCR reactions.
All qPCR reactions were performed using the Thermo Fisher DyNAmo HS SYBR Green qPCR kit according to the manufacturer’s instructions with 0.5 μM of each primer (see SI Appendix, Table S4 for primer sequences). Reactions were run on an Applied Biosystems StepOnePlus real-time PCR machine, using fluorescence of the ROX reference dye for normalization. Cycling conditions were as follows: 95 °C for 15 min; 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s; and melt curve analysis from 60 °C to 95 °C. Amplicons ranged from 120 to 140 base pairs, and product specificity was verified by melt curve analysis and agarose gel electrophoresis. No template controls were performed and showed minimal primer–dimer formation. Three candidate reference genes, secA (Synpcc7942_0289), rimM (Synpcc7942_2259), and rnpA (Synpcc7942_1615), were selected for normalization based on a previous analysis performed with Synechococcus sp. PCC 7002 (66) and after confirming similar expression levels across conditions from our RNA-seq data. CT values for hpf (Synpcc7942_2352), gifA (Synpcc7942_0900), and sbtA (Synpcc7942_1475) were normalized to CT values for the appropriate reference gene, and expression values were determined relative to the control-L condition (using the ∆∆ CT analysis method).
Polysome Analysis by Sucrose Density Gradient Centrifugation.
Synechococcus cultures (500 mL) were synchronized by one 12-h dark period, were grown in constant light to log phase (OD750 0.2–0.5), and then shifted into the dark for 2 h where appropriate. E. coli cultures (50 mL) were grown to log phase (OD600 ∼0.5). All cultures were treated with chloramphenicol at a final concentration of 0.5 mg/mL for 2 min to arrest translation elongation. Cultures were poured over crushed ice in centrifuge bottles (to allow rapid cooling), and cells were harvested by centrifugation (10,000 × g, 10 min, 4 °C). Pellets were washed in 5 mL of ice-cold polysome lysis buffer (10 mM Tris⋅HCl, pH 8.0, 10 mM MgCl2) and centrifuged again (4,000 × g, 10 min, 4 °C), and pellets were resuspended in 1 mL of ice-cold polysome lysis buffer. This cell material was transferred to 2-mL screw-cap tubes containing ∼0.5 mL of 0.1-mm glass beads, which were flash-frozen in liquid nitrogen and stored at −80 °C until further processing.
Cell lysates were prepared on the same day as sucrose gradient analysis to minimize the number of freeze–thaw cycles. Cell material was thawed at room temperature and lysed in a bead beater for 3 × 30-s cycles, with 1 min on ice in between each beating. Tubes were centrifuged quickly (16,000 × g, 30 s, 4 °C) to sediment beads, the supernatant was transferred to a new tube, and the lysate was clarified by centrifugation (16,000 × g, 10 min, 4 °C). This supernatant was transferred to a new tube, and absorbance at 260 nm was measured using a Nanodrop 2000 spectrophotometer. Samples in a particular experiment were normalized to the same A260nm (∼20 A260 units) in a total volume of 300 μL (diluted with polysome lysis buffer where necessary), to which 40 units of RNase inhibitor (SUPERaseIn; Thermo Fisher) was added before loading lysates onto sucrose gradients. For RNase A-treated samples (and negative controls), 33 μL of 3 M NaCl was added to the 300 μL of cell lysate, to which 3 mg of RNase A (Thermo Fisher) was added where appropriate. Lysates were incubated at room temperature for 10 min and were then frozen in liquid nitrogen to stop the reaction (these samples were subjected to one additional freeze–thaw cycle.)
Sucrose gradients (10 to 40%, wt/vol) were prepared in ultracentrifuge tubes by layering 6 mL of 40% sucrose buffer solution (20 mM Tris⋅HCl, pH 7.8, 10 mM MgCl2, 100 mM NH4Cl, 2 mM DTT, 40% sucrose) underneath 6 mL of 10% sucrose buffer solution (20 mM Tris⋅HCl, pH 7.8, 10 mM MgCl2, 100 mM NH4Cl, 2 mM DTT, 10% sucrose). Linear gradients were created using a Gradient Mate and allowed to equilibrate for at least 15 min at 4 °C before loading. Then, 300 μL of cell lysate was loaded on top of the sucrose gradient, and gradient tubes were centrifuged in a Beckman ultracentrifuge (40,000 × g, 105 min, 4 °C). Sucrose gradients were then analyzed by a UV-Vis detector, monitoring absorbance at 254 nm.
Supplementary Material
Acknowledgments
We thank A. Whiteley and the D. Portnoy laboratory for providing E. coli strains and for assistance with ppGpp detection experiments; K. Sogi and the S. Stanley laboratory for assistance with 14C incorporation experiments and use of equipment; the University of California, Davis sequencing facility for constructing RNA-seq libraries, acquiring sequencing data, and initial data analysis; and V. Yu and the J. Cate laboratory for assistance with polysome experiments and use of equipment. We are grateful to R. Yokoo for critical reading of the manuscript, and C. Cassidy-Amstutz for help with statistical analysis. This work was supported by Department of Energy Office of Science Early Career Research Program Grant DE-SC0006394 through the Office of Basic Energy Sciences and by an Alfred P. Sloan Research Fellowship (to D.F.S.). R.D.H. and A.F. were supported by the National Science Foundation Graduate Research Fellowship Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524915113/-/DCSupplemental.
References
- 1.Külheim C, Agren J, Jansson S. Rapid regulation of light harvesting and plant fitness in the field. Science. 2002;297(5578):91–93. doi: 10.1126/science.1072359. [DOI] [PubMed] [Google Scholar]
- 2.Bailey S, Grossman A. Photoprotection in cyanobacteria: Regulation of light harvesting. Photochem Photobiol. 2008;84(6):1410–1420. doi: 10.1111/j.1751-1097.2008.00453.x. [DOI] [PubMed] [Google Scholar]
- 3.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106(33):14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106(52):22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle WF. The cyanobacterial genome, its expression, and the control of that expression. Adv Microb Physiol. 1979;20:1–102. doi: 10.1016/s0065-2911(08)60206-4. [DOI] [PubMed] [Google Scholar]
- 6.Binder BJ, Chisholm SW. Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J Bacteriol. 1990;172(5):2313–2319. doi: 10.1128/jb.172.5.2313-2319.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93(19):10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surányi G, Korcz A, Pálfi Z, Borbély G. Effects of light deprivation on RNA synthesis, accumulation of guanosine 3‘(2’)-diphosphate 5′-diphosphate, and protein synthesis in heat-shocked Synechococcus sp. strain PCC 6301, a cyanobacterium. J Bacteriol. 1987;169(2):632–639. doi: 10.1128/jb.169.2.632-639.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosokawa N, et al. Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc Natl Acad Sci USA. 2011;108(37):15396–15401. doi: 10.1073/pnas.1019612108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takano S, Tomita J, Sonoike K, Iwasaki H. The initiation of nocturnal dormancy in Synechococcus as an active process. BMC Biol. 2015;13(1):36. doi: 10.1186/s12915-015-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68(5):1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272(34):21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 13.Seki Y, Nitta K, Kaneko Y. Observation of polyphosphate bodies and DNA during the cell division cycle of Synechococcus elongatus PCC 7942. Plant Biol (Stuttg) 2014;16(1):258–263. doi: 10.1111/plb.12008. [DOI] [PubMed] [Google Scholar]
- 14.Friga GM, Borbély G, Farkas GL. Accumulation of guanosine tetraphosphate (ppGpp) under nitrogen starvation in Anacystis nidulans, a cyanobacterium. Arch Microbiol. 1981;129(5):341–343. doi: 10.1007/BF00406458. [DOI] [PubMed] [Google Scholar]
- 15.Nanamiya H, et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67(2):291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 16.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117(1):57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 17.Steinchen W, et al. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc Natl Acad Sci USA. 2015;112(43):13348–13353. doi: 10.1073/pnas.1505271112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collier JL, Herbert SK, Fork DC, Grossman AR. Changes in the cyanobacterial photosynthetic apparatus during acclimation to macronutrient deprivation. Photosynth Res. 1994;42(3):173–183. doi: 10.1007/BF00018260. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa M, et al. Impact of the plastidial stringent response in plant growth and stress responses. Nat Plants. 2015;1:15167. doi: 10.1038/nplants.2015.167. [DOI] [PubMed] [Google Scholar]
- 20.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128(5):865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesley JA, Shapiro L. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol. 2008;190(20):6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer RA, Doolittle WF. Control of gene expression in blue-green algae. Nature. 1975;253(5493):650–651. doi: 10.1038/253650a0. [DOI] [PubMed] [Google Scholar]
- 23.Luque I, Forchhammer K. Nitrogen assimilation and C/N balance sensing. In: Herrero A, Flores E, editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic; Norfolk, UK: 2008. pp. 335–382. [Google Scholar]
- 24.García-Domínguez M, Reyes JC, Florencio FJ. Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci USA. 1999;96(13):7161–7166. doi: 10.1073/pnas.96.13.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muro-Pastor MI, Reyes JC, Florencio FJ. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem. 2001;276(41):38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- 26.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 27.Cangelosi GA, Brabant WH. Depletion of pre-16S rRNA in starved Escherichia coli cells. J Bacteriol. 1997;179(14):4457–4463. doi: 10.1128/jb.179.14.4457-4463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueta M, et al. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem. 2008;143(3):425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 29.Ueta M, et al. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells. 2013;18(7):554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 30.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336(6083):915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin D, Fredrick K. Analysis of polysomes from bacteria. Methods Enzymol. 2013;530:159–172. doi: 10.1016/B978-0-12-420037-1.00008-7. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz O, Tsinoremas NF, Schaefer MR, Anandan S, Golden SS. General effect of photosynthetic electron transport inhibitors on translation precludes their use for investigating regulation of D1 biosynthesis in Synechococcus sp. strain PCC 7942. Photosynth Res. 1999;62(2):261–271. [Google Scholar]
- 33.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr Biol. 2004;14(16):1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 34.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221(5183):838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 35.Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: Starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80(3):695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalebroux ZD, Swanson MS. ppGpp: Magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10(3):203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 37.Kriel A, et al. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol Cell. 2012;48(2):231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197(7):1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8(2):203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Liu K, et al. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015;57(4):735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nomura Y, et al. Diversity in guanosine 3′,5′-bisdiphosphate (ppGpp) sensitivity among guanylate kinases of bacteria and plants. J Biol Chem. 2014;289(22):15631–15641. doi: 10.1074/jbc.M113.534768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumano M, Tomioka N, Shinozaki K, Sugiura M. Analysis of the promoter region in the rrnA operon from a blue-green alga, Anacystis nidulans 6301. Mol Gen Genet. 1986;202(2):173–178. [Google Scholar]
- 44.Masuda S, Bauer CE. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J Bacteriol. 2004;186(1):235–239. doi: 10.1128/JB.186.1.235-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12(5):327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 46.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci USA. 2006;103(38):13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corrigan RM, Bellows LE, Wood A, Gründling A. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci USA. 2016;113(12):E1710–E1719. doi: 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starosta AL, Lassak J, Jung K, Wilson DN. The bacterial translation stress response. FEMS Microbiol Rev. 2014;38(6):1172–1201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wada A, Mikkola R, Kurland CG, Ishihama A. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J Bacteriol. 2000;182(10):2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tagami K, et al. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. MicrobiologyOpen. 2012;1(2):115–134. doi: 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan X, Varughese M, Widger WR. A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J Biol Chem. 1994;269(33):20905–20912. [PubMed] [Google Scholar]
- 52.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42(4):504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 53.Lindahl M, Florencio FJ. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci USA. 2003;100(26):16107–16112. doi: 10.1073/pnas.2534397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borbély G, Kaki C, Gulyás A, Farkas GL. Bacteriophage infection interferes with guanosine 3′-diphosphate-5′-diphosphate accumulation induced by energy and nitrogen starvation in the cyanobacterium Anacystis nidulans. J Bacteriol. 1980;144(3):859–864. doi: 10.1128/jb.144.3.859-864.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6(8):e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masuda S, et al. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 2008;49(2):135–141. doi: 10.1093/pcp/pcm177. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc Natl Acad Sci USA. 2004;101(12):4320–4324. doi: 10.1073/pnas.0308555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugliani M, et al. An ancient bacterial signaling pathway regulates chloroplast function to influence growth and development in Arabidopsis. Plant Cell. 2016;28(3):661–679. doi: 10.1105/tpc.16.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen MM. Simple conditions for growth of unicellular blue-green algae on plates. J Phycol. 1968;4(1):1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x. [DOI] [PubMed] [Google Scholar]
- 60.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3(11):e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bokinsky G, et al. HipA-triggered growth arrest and β-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J Bacteriol. 2013;195(14):3173–3182. doi: 10.1128/JB.02210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80(3):612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yokoo R, Hood RD, Savage DF. Live-cell imaging of cyanobacteria. Photosynth Res. 2015;126(1):33–46. doi: 10.1007/s11120-014-0049-x. [DOI] [PubMed] [Google Scholar]
- 64.Aschar-Sobbi R, et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18(5):859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- 65.McClure R, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41(14):e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szekeres E, Sicora C, Dragoş N, Drugă B. Selection of proper reference genes for the cyanobacterium Synechococcus PCC 7002 using real-time quantitative PCR. FEMS Microbiol Lett. 2014;359(1):102–109. doi: 10.1111/1574-6968.12574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.