Abstract
Many studies suggest the presence of aberrations in cellular metabolism in bipolar disorder. We studied the metabolome in bipolar disorder to gain insight into cellular pathways that may be dysregulated in bipolar disorder and to discover evidence of novel biomarkers. We measured polar and nonpolar metabolites in fibroblasts from subjects with bipolar I disorder and matched healthy control subjects, under normal conditions and with two physiologic perturbations: low-glucose media and exposure to the stress-mediating hormone dexamethasone. Metabolites that were significantly different between bipolar and control subjects showed distinct separation by principal components analysis methods. The most statistically significant findings were observed in the perturbation experiments. The metabolite with the lowest p value in both the low-glucose and dexamethasone experiments was α-aminoadipate, whose intracellular level was consistently lower in bipolar subjects. Our study implicates α-aminoadipate as a possible biomarker in bipolar disorder that manifests under cellular stress. This is an intriguing finding given the known role of α-aminoadipate in the modulation of kynurenic acid in the brain, especially as abnormal kynurenic acid levels have been implicated in bipolar disorder.
Key Words: Metabolic profiling, Bipolar disorder, α-Aminoadipate, 2-Aminoadipic acid, 2-Aminoadipate, Kynurenic acid
Introduction
Multiple lines of study suggest a variety of aberrations in cellular metabolism in bipolar disorder [1,2,3]. Altered cerebral metabolism has been reported in bipolar disorder patients, including in medication-naïve patients, using many different techniques [4,5]. Positron emission tomography studies show resting prefrontal hypometabolism and simultaneous resting limbic/paralimbic hypermetabolism in patients with bipolar disorder [6,7]. Nuclear magnetic resonance studies have revealed significantly elevated concentrations of sorbitol and fructose in the cerebrospinal fluid [8] and lower levels of N-acetylaspartate in frontal lobe structures and hippocampi [9,10]. Moreover, treatment with lithium increased levels of N-acetylaspartate in patients with bipolar disorder [11,12]. Gene expression studies in postmortem brains of patients with bipolar disorder showed significant decreases in the expression of genes involved in mitochondrial oxidative phosphorylation and ATP-dependent proteasome degradation [13].
There is evidence for the presence of metabolic dysregulation and gene expression differences not only between bipolar disorder patients and healthy controls but also between different mood states in bipolar disorder patients [14]. Bipolar subjects in a manic state had lactate levels similar to controls in longitudinal magnetic resonance spectroscopy studies, but these lactate levels were lower compared to controls during euthymic phases [15]. Untreated bipolar disorder subjects during depressive episodes had an increased ratio of phosphocreatine/inorganic phosphate in the frontal lobe in a phosphoros-31 magnetic resonance spectroscopy study in comparison to controls as well as to subjects treated with medications [16].
One difficulty in studying the biology of psychiatric illnesses is our inability to directly study live neuronal tissue from patients - brain biopsies of live tissue for research purposes are not feasible for technical and ethical reasons. Given the strong genetic determination of risk, cellular abnormalities that reflect disease biology may also be reflected in peripheral tissue, as noted above, and researchers have often studied cellular abnormalities in accessible surrogate tissue [17,18,19]. Studies in fibroblasts and lymphocytes suggest that there may indeed be disease-related cellular phenotypes that can be observed in peripheral cells. Examination of mitochondrial structure in bipolar disorder showed that patients had distinct differences in mitochondrial size and distribution not only in postmortem brains but also in fibroblasts and lymphocytes [20]. Similarly, gene expression studies in lymphocytes from patients with bipolar disorder and normal controls showed marked differences in the expression patterns of genes coding for mitochondrial electron transfer chain proteins in response to low-glucose conditions [21]. Patients with bipolar disorder, including medication-naïve patients, have an increased risk for insulin resistance, impaired glucose tolerance and diabetes [22,23,24,25,26]. Taken together, these studies indicate that cellular metabolic dysregulation may be an innate and crucial feature of bipolar disorder. Crucially, the presence of similar abnormalities of metabolism in peripheral cells, in vivo brain studies and in postmortem brains suggests that the peripheral abnormalities are specific to the disease biology.
Given the evidence for metabolic dysregulation in bipolar disorder and studies showing that some disease-related cellular features can be observed in fibroblasts, we undertook a study to profile the metabolome of fibroblasts from subjects with bipolar I disorder and age- and sex-matched healthy controls. We measured metabolites under normal growth conditions and under two stressful perturbations - growth in low-glucose media and exposure to dexamethasone. We identified metabolites that were differentially regulated in the bipolar disorder and healthy control groups under these three different conditions. We report an especially intriguing finding of lower levels of α-aminoadipate in fibroblasts from bipolar disorder subjects when they are ‘perturbed’ with cellular stress, which is consistent with a proposed role for the kynurenic acid pathway in the biology of bipolar disorder [27,28,29,30,31].
Methods
Subjects
Subjects with bipolar I disorder with psychotic features were recruited from the Schizophrenia and Bipolar Disorder Program at the McLean Hospital with Institutional Review Board (IRB) approval. The initial subject recruitment referrals were based on diagnoses by treating psychiatrists. The subject enrollment process included a semi-structured interview using the Structured Clinical Interview for DSM Disorders to ascertain the diagnoses. For healthy controls, subjects were chosen who had no previous psychiatric diagnoses, treatments or first-degree relatives with a major psychotic or affective disorder. Exclusion criteria included subjects who had any comorbid neurological disorder.
Isolation of Fibroblasts
Fibroblasts were obtained by informed consent through punch biopsies performed by a physician, under a protocol approved by the IRB. The biopsy specimen was minced into 0.5-mm pieces and placed in the center of a 6-well plate with 3 ml of fibroblast media (DMEM Gibco 11995-065 containing 25 mM glucose, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin). After a week of incubation in 37°C and 5% CO2, dense fibroblast outgrowths were treated with 0.05% trypsin and passed through a 70-mm strainer after addition of media to remove large pieces of tissue. Cells were passaged at 1:3 ratios every 5-7 days until the cells reached 80% confluency and then were frozen and stored in liquid nitrogen until ready for use.
Sample Preparation for Polar Metabolites
Fibroblasts were controlled for passage number between the different lines to ensure that variations observed are not due to the differences in passage number seen in cellular senescence. Fibroblasts were thawed and grown in 6-well cell culture plates (Corning, CLS3506) to 80% confluency. Cells were then incubated for 6 h in fresh high-glucose-, low-glucose-, or dexamethasone-containing media. For the low-glucose condition experiments, cells were grown in media containing only 1 mM glucose for 6 h, compared to 25 mM under normal growth conditions. For the dexamethasone experiments, cells were cultured in the presence of 1 μM dexamethasone for 6 h in normal fibroblast media. At the end of the 6-hour incubation with normal media, low-glucose media, or media with dexamethasone, cells were washed with cold PBS (Mg2+-/Ca2+-) to initiate metabolite extraction. After aspirating the PBS, 800 μl of cold 80% methanol (−80°C) was immediately added to the plates and incubated at −80°C for 15 min. Cells were then lifted with cell scrapers and the cell lysate/methanol mixture was transferred to 1.5-ml centrifuge tubes on dry ice. Tubes containing the lysate/methanol mixture were centrifuged (9,000 g, 4°C, 10 min) to pellet cell debris and proteins. The supernatants were transferred to new 1.5-ml centrifuge tubes on dry ice, while keeping the old 1.5-ml tubes containing pellets for further extraction. Pellets were resuspended in 100 μl 80% methanol (−80°C) and centrifuged (9,000 g, 4°C, 5 min), and the supernatants were pooled in tubes containing the earlier supernatants. The collected supernatants were stored at −80°C.
Sample Preparation for Lipids
At the end of the 6-hour incubation with normal media, low-glucose media, or media with dexamethasone, cells were washed once with cold PBS (Mg2+-/Ca2+-) to initiate metabolite extraction. 800 μl of cold isopropranol (4°C) was added to the plate and cells were immediately lifted with cell scrapers. The cell lysate/isopropranol mixtures were transferred to 1.5-ml centrifuge tubes on ice, and incubated at 4°C for 1 h, while covered with aluminum foil to avoid exposure to light. Tubes containing the lysate mixtures were then vortexed and centrifuged to remove cell debris and proteins (9,000 g, 4°C, 10 min). Supernatants were collected and stored at −80°C.
Quantitation of Metabolites
The endogenous metabolites in the fibroblasts were identified and quantified using liquid chromatography-tandem mass spectrometry (LC-MS/MS) [32]. Liquid chromatography, through the application of a number of distinct stationary phase chemistries, affords reproducible separation of metabolites in complex mixtures on the basis of their physical properties [33]. Mass spectrometry enables further resolution of metabolites on the basis of mass-to-charge ratio and quantitation over a wide linear dynamic range [33]. This LC-MS/MS methodology uses three chromatographic stationary phases and analyzed sugars, sugar phosphates, nucleotides, amino acids, biogenic amines, organic acids, bile acids, and lipids in a targeted manner. Metabolites were identified by the parent ion mass (MS) and dominant product ion mass (MS/MS) on a sensitive mass spectrometer in combination with the retention time on an appropriate chromatography column. While none of these three parameters is individually sufficient to uniquely identify a metabolite in a biological sample, the three in combination provide a ‘tag’ that marks the metabolite to permit identification and quantitation. We collected data on 11,993 metabolites, of which 347 were annotated, known metabolites. Differences in individual metabolites, as well as groups of metabolites that are part of definable cellular functions, were identified using the software MultiQuant (v2.1, AB SCIEX) and Progenesis CoMet software (v2.0, Nonlinear Dynamics) [34]. MultiQuant (v 2.1, AB SCIEX) rapidly integrates large numbers of peaks in the samples and visualizes the results for inspection of quality [34]. Targeted data were processed using MultiQuant software, and compound identities were confirmed using reference standards and reference samples. Nontargeted data were processed using Progenesis CoMet software (v2.0, Nonlinear Dynamics) to detect peaks, perform chromatographic retention time alignment, and integrate peak areas [35]. Nontargeted metabolite LC-MS peaks were initially identified by matching measured retention times and masses to a database of >500 characterized compounds, and secondarily by matching exact masses only to a database of >40,000 metabolites (Human Metabolome Database v3) [36]. Bioinformatics analyses were conducted using MetaboAnalyst 3.0 [37]. Univariate and multivariate analyses were undertaken to identify group differences among annotated metabolites between the two groups that each contained 9 biological replicates.
Results
Fibroblasts were cultured from 9 subjects who met criteria for the DSM-IV-TR bipolar I disorder as well as 9 healthy controls matched for sex and age. The average age at the time of sample collection was 35.8 years for the bipolar disorder subjects and 37 years for healthy controls. All the bipolar subjects were in treatment - 2 subjects were treated with lithium, 7 with antipsychotic medications, 5 with benzodiazepines and 2 with antidepressants. The two groups consisted of 6 pairs of male subjects and 3 pairs of female subjects that were matched for age. None of the subjects had any diagnoses of diabetes or any cardiovascular disease.
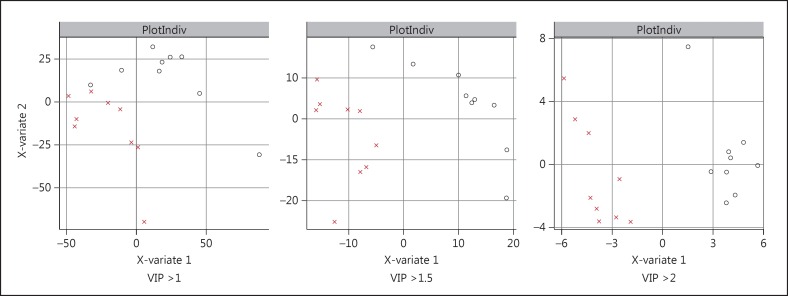
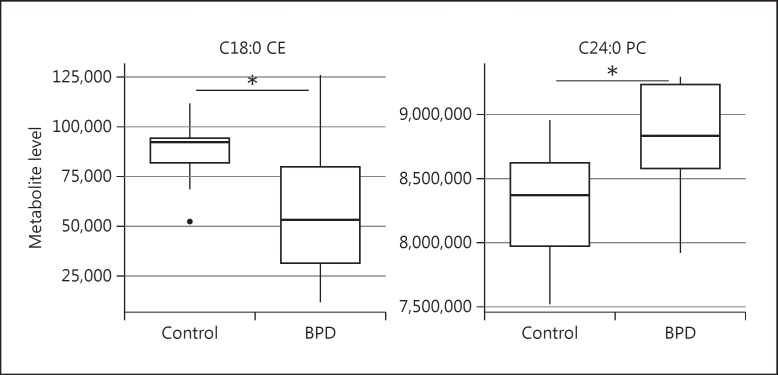
Measurement values for 11,993 total metabolites, including 347 annotated metabolites, were obtained as described above. Raw data for each metabolite were normalized to the total metabolites for that particular experimental well before analysis. In analyzing the metabolite data collected under normal growth conditions, we carried out univariate analyses to identify group differences among the metabolites between bipolar disorder cells and healthy control cells. We used these data to generate a metabolite signature and employed a multivariate method with a partial least squares discriminant analysis (PLS-DA) approach to model the separation of the two groups. We used metabolite profiles obtained with different VIP thresholds and found that the profiles with higher VIP thresholds were in much better separation of the two groups (fig. 1). Under normal growth conditions, the analysis revealed two annotated metabolites that were different at statistically significantly different levels between the two groups, at a significance threshold of p < 0.05 by univariate analysis (table 1). The range of values for the metabolites that were statistically significantly different between the two groups are shown in figure 2.
Fig. 1.
Separation of bipolar disorder patients and controls in a PLS-DA model with a panel of metabolites that were significantly different between the two groups. PLS-DA plots are shown for metabolite profiles with different VIP thresholds. O refers to bipolar subjects and X to healthy control subjects.
Table 1.
Annotated metabolites that significantly distinguish (p < 0.05, unpaired t test) bipolar I disorder patients and healthy control subjects under normal growth conditions
| Metabolite | p value |
|---|---|
| C18:0 CE | 0.043672 |
| C24:0 PC | 0.047946 |
Fig. 2.
Distribution of normalized metabolite levels in bipolar disorder patients (BDP) and control subjects for the metabolites that were significantly different between the two groups when grown under normal conditions. * Denotes significance at p < 0.05 (unpaired t test).
Peturbational Profiling
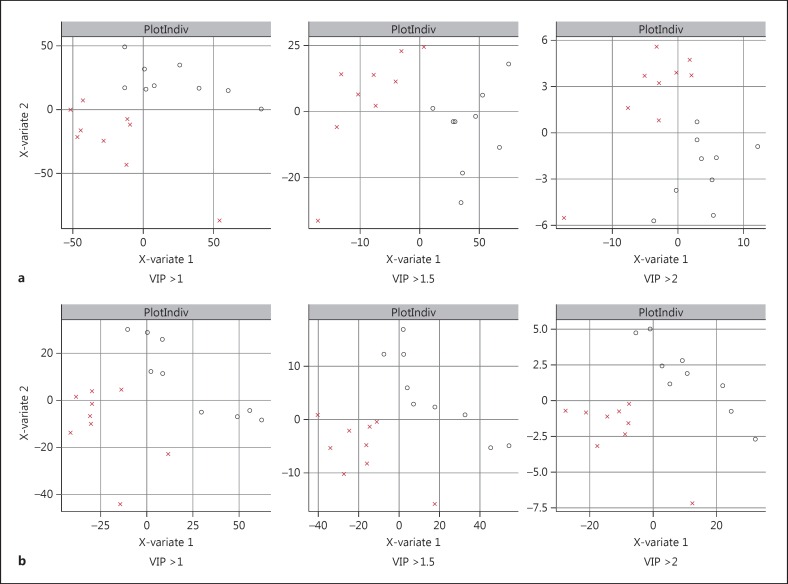
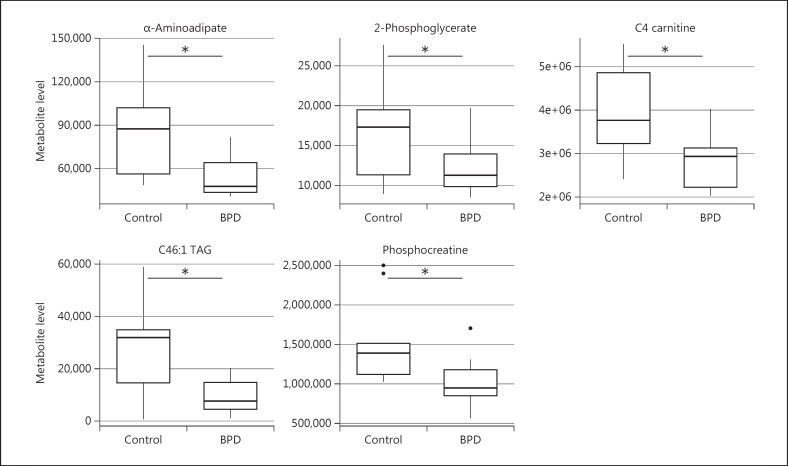
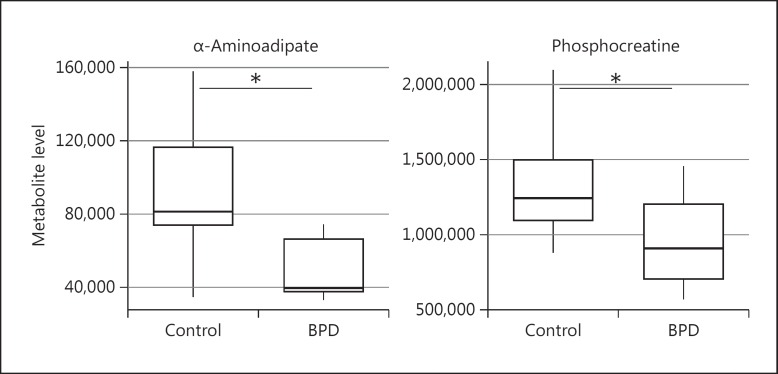
We analyzed the data similarly for metabolites collected for the experiments where fibroblasts were exposed to two different stress-inducing perturbations for 6 h - growth under low-glucose conditions and exposure to 1 μM dexamethasone. PLS-DA models generated with these metabolite signatures again showed a clear separation of the bipolar disorder samples from healthy controls under both perturbation conditions, with clearer separation between the groups at higher VIP thresholds (fig. 3). The list of annotated metabolites was different than the ones under normal growth conditions and the highest-ranked metabolites had smaller p values in the perturbation experiments. We identified five metabolites that were statistically significantly different between the two groups (p < 0.05) under low-glucose conditions, while there were two metabolites that were significantly different between the two groups (p < 0.05) in the dexamethasone experiment (table 2). We compared the levels of these individual metabolite hits between bipolar disorder and healthy controls. We found that the range of values of these metabolites in the bipolar disorder group was generally tighter than in the healthy control group under the perturbed conditions (fig. 4, 5).
Fig. 3.
Separation of bipolar disorder patients and controls with perturbational profiling. Separation of bipolar disorder patients and control subjects in a PLS-DA model is shown for metabolites using different VIP thresholds for the low-glucose experiment (a) and the dexamethasone experiment (b). O refers to bipolar subjects and X to healthy control subjects.
Table 2.
Annotated metabolites that significantly distinguish (p < 0.05, unpaired t test) bipolar I disorder patients and healthy control subjects under low-glucose conditions and with dexamethasone treatment
| Metabolite | p value |
|---|---|
| Low-glucose conditions | |
| α-Aminoadipate | 0.011848 |
| C4 carnitine | 0.026489 |
| 2-Phosphoglycerate | 0.029856 |
| Phosphocreatine | 0.037766 |
| C46:1 TAG | 0.041136 |
| Dexamethasone treatment | |
| α-Aminoadipate | 0.013575 |
| Phosphocreatine | 0.045586 |
Fig. 4.
Metabolite patterns under low-glucose conditions. Differences in metabolite levels between bipolar disorder patients (BPD) and healthy controls subjects for metabolites significantly different between the two groups. * Denotes significance at p < 0.05 (unpaired t test).
Fig. 5.
Metabolite patterns with exposure to dexamethasone. Differences in metabolite levels between bipolar disorder patients (BDP) and control subjects for metabolites significantly different between the two groups. * Denotes significance at p < 0.05 (unpaired t test).
There were two metabolites that were shared in the lists of significantly different metabolites under both perturbation conditions. Interestingly, the top-ranked metabolite with the lowest p value was the same in both perturbation experiments - α-aminoadipate (table 2). Under both conditions, α-aminoadipate was significantly less abundant in bipolar disorder compared to the healthy control group (fig. 4, 5). The other metabolite that was present at significantly different levels in the bipolar disorder and healthy control group in both perturbation experiments was phosphocreatine. In addition, levels of C4 carnitine, 2-phosphoglycerate and C46:1 TAG were significantly different between the two groups under low-glucose conditions, but not under high-glucose or dexamethasone conditions.
Discussion
Metabolic profiling of patient tissue has been used successfully to investigate the pathophysiology of various human diseases [38,39,40]. Studies applying metabolomic profiling of patient serum are also starting to be used to explore the biology of psychiatric disorders [41,42,43,44,45,46]. Such studies have suggested the possibility of identifying biomarkers for schizophrenia [47,48,49,50,51,52], major depressive disorder [53,54] and posttraumatic stress disorder [55], and have also been used to study medication-induced metabolic changes [56,57]. In bipolar disorder, metabolomic studies have been undertaken in urine samples to identify potential biomarkers [58,59,60,61]. We report our findings from unbiased metabolic profiling of fibroblasts from patients with bipolar disorder and healthy controls, under normal growth conditions as well as under physiologic perturbations. We report on a number of metabolites that were present at significantly different levels between bipolar disorder patients and healthy control subjects under normal conditions as well as metabolites that were significantly different between the two groups under perturbation. We found that these metabolite profiles led to a clear separation of bipolar subjects from control subjects (fig. 1, 3).
One metabolite stood out as the most significant separator under both perturbation conditions, with p values that were much smaller than other metabolites in the three lists. α-Aminoadipate was significantly less abundant in the cells from bipolar disorder subjects compared to the healthy control subjects in the presence of stressful perturbations but not under normal growth conditions. α-Aminoadipate is a component of the lysine metabolism pathway and a marker of oxidative stress [62,63]. A recent metabolomic study of diabetes in human plasma samples suggested that α-aminoadipate may be a modulator of glucose homeostasis and diabetes risk [64]. While little is known about its biology in the human brain, α-aminoadipate has been reported to induce neurogenesis in the visual system in adult mice [65]. Studies in rodents have also shown that α-aminoadipate modulates kynurenic acid levels in the brain. In experiments with rat brain tissue slices, α-aminoadipate exposure resulted in a substantial decrease in levels of kynurenic acid [66]. Similarly, in vivo studies in free-moving rats exposed to α-aminoadipate through microdialysis in the hippocampus resulted in a robust decrease in kynurenic acid levels [67]. Another study has shown that α-aminoadipate inhibited the synthesis of kynurenic acid in tissue culture as well as in the rat hippocampus [68].
There is consistent evidence that kynurenic acid plays a role in the disease biology of bipolar disorder. Elevated levels of kynurenic acid have been reported in patients with bipolar disorder [28,29]. Moreover, it was shown that the cerebrospinal fluid levels of kynurenic acid were positively correlated with lifetime history of mania and psychosis in bipolar disorder patients [28]. A recent study examining the kynurenine 3-monoxygenase (KMO) found that bipolar disorder patients with psychotic features had decreased expression of KMO mRNA in the prefrontal cortex [27]. The same study also reported an association between the KMO Arg452 allele and the presence of psychosis during manic episodes [27]. A recent genome-wide association study linked abnormal metabolism of kynurenic acid to a specific gene associated with bipolar disorder [69]. We are intrigued by the identification of α-aminoadipate, a key regulator of kynurenic acid in the brain, as the highest-ranked metabolite in an unbiased profiling of bipolar disorder and control subjects.
The interplay of α-aminoadipate and kynurenic acid reflects interesting biology. α-Aminoadipate is a substrate of the enzyme α-aminoadipate aminotransferase II, which has been shown to be the same enzyme as kynurenine aminotransferase II, and is responsible for the transamination of L-kynurenine to kynurenic acid [70,71]. α-Aminoadipate levels dictate the availability of kynurenine aminotransferase II for the transamination of L-kynurenine to kynurenic acid [72]. Hence, high levels of α-aminoadipate result in decreased kynurenic acid while low levels of α-aminoadipate lead to increased kynurenic acid [67,68,72]. We hypothesize that the genetic background that lends itself to lower levels of α-aminoadipate in fibroblasts may also give rise to similar biology in the central nervous system, where kynurenic acid plays a major role [30]. As observed in fibroblasts, there may be lower levels of α-aminoadipate in the brain in bipolar disorder, which would result in increased levels of kynurenic acid, as has been reported in the cerebrospinal fluid of patients with bipolar disorder [27,28,29].
The metabolite with the second lowest p value after α-aminoadipate was C4 carnitine, the levels of which were lower in fibroblasts from bipolar disorder patients when they were grown under low-glucose conditions. Carnitine is necessary for the transport of long-chain fatty acids into the mitochondrial matrix for generating metabolic energy from lipid breakdown [73]. While there are no studies specifically looking at C4 carnitine in bipolar disorder, carnitine biology in general has been studied in the context of new treatment approaches for bipolar disorder [74,75]. Polymorphisms in a carnitine transporter SLC22A16 are strongly associated with bipolar I disorder [76]. C30:1 PC, a lecithin, was also lower both under normal conditions and in the presence of dexamethasone. This is of some interest since lecithins have also been explored as putative treatment agents in bipolar disorder [77,78].
In our study, we undertook perturbational studies to explore whether stressful conditions may expose disease-related vulnerabilities in patient cells. This approach attempts to recreate a gene-environment interaction in vitro. Our model was guided by previous studies comparing mitochondrial gene expression in bipolar disorder which had shown that mitochondrial gene expression patterns were abnormal in bipolar disorder cells only when grown under stressful low-glucose conditions [21]. In our study, we found that metabolites that were significantly different between the two groups in the perturbation experiments had lower, more statistically significant p values than metabolites that were different under normal growth conditions (tables 1, 2). Interestingly, α-aminoadipate was the most significantly different metabolite under both perturbation (stress) conditions.
While these findings are intriguing, they are preliminary. There are caveats and limitations to our study. One limitation is the sample size, a consequence of the cost of the metabolic profiling experiments. We identified a number of metabolites that were different between the groups at a significance threshold of p < 0.05 but these were not significant after adjusting for multiple testing. Our results should be followed by a targeted study to validate the top metabolites identified in these profiling experiments. Another caveat relates to the patient population studied. Our subjects had bipolar disorder with psychotic features and it will need to be seen if these results are specific for subjects with bipolar disorder with psychotic features or whether they are applicable to all subjects with bipolar disorder. Since the bipolar subjects were not medication-naïve, it is possible that the results could have been confounded by the medications that the patients were taking. Such a scenario is not very likely since the fibroblasts had been grown and passaged in fresh media for multiple passages before the metabolic profiling experiments. Another possible limitation is that we used peripheral cells in a profiling study of a brain disorder [79]. As one of our goals is to find reliable biomarkers, we chose to work with tissue that was easily accessible. In addition, given the strong genetic determination of risk for bipolar disorder, we hypothesized that dysregulation in metabolic pathways associated with disease in the central nervous system may also be discoverable in peripheral tissues such as fibroblasts [18]. Analysis of whole-genome biomarker expression in blood and brain samples shows that about 22% of total transcriptome in postmortem brain is expressed at a similar level and pattern in blood elements [80]. Studies comparing gene-expression patterns, epigenetic differences and subcellular organelles in bipolar disorder and schizophrenia have found disease-related changes that are present in both the brain and in peripheral cells [13,20,21,81,82,83,84]. Nonetheless, it is important to follow up these results in human neuronal cells, derived from patient-induced pluripotent stem cells to see which group-related metabolite differences observed in the fibroblasts are present in neuronal cells as well.
Statement of Ethics
The fibroblasts used in the study were obtained from human subjects after informed consent through a protocol approved by the Institutional Review Board.
Disclosure Statement
The authors of this paper do not have any commercial associations that might pose a conflict of interest in connection with this paper.
Acknowledgement
This study was carried out with funding from the Ryan Licht Sang Bipolar Foundation.
References
- 1.Kaidanovich-Beilin O, Cha DS, McIntyre RS. Crosstalk between metabolic and neuropsychiatric disorders. F1000 Biol Rep. 2012;4:14. doi: 10.3410/B4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calkin CV, et al. The relationship between bipolar disorder and type 2 diabetes: more than just co-morbid disorders. Ann Med. 2013;45:171–181. doi: 10.3109/07853890.2012.687835. [DOI] [PubMed] [Google Scholar]
- 3.Kato T. Mitochondrial dysfunction and bipolar disorder. Curr Top Behav Neurosci. 2011;5:187–200. doi: 10.1007/7854_2010_52. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz-Yesiloglu A, Ankerst DP. Neurochemical alterations of the brain in bipolar disorder and their implications for pathophysiology: a systematic review of the in vivo proton magnetic resonance spectroscopy findings. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:969–995. doi: 10.1016/j.pnpbp.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Frey BN, et al. Abnormal cellular energy and phospholipid metabolism in the left dorsolateral prefrontal cortex of medication-free individuals with bipolar disorder: an in vivo 1H MRS study. Bipolar Disord. 2007;9((suppl 1)):119–127. doi: 10.1111/j.1399-5618.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 6.Brooks JO, et al. Prefrontal and paralimbic metabolic dysregulation related to sustained attention in euthymic older adults with bipolar disorder. Bipolar Disord. 2010;12:866–874. doi: 10.1111/j.1399-5618.2010.00881.x. [DOI] [PubMed] [Google Scholar]
- 7.Brooks JO, Hoblyn JC, Ketter TA. Metabolic evidence of corticolimbic dysregulation in bipolar mania. Psychiatry Res. 2010;181:136–140. doi: 10.1016/j.pscychresns.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Regenold WT, et al. Relationship of cerebrospinal fluid glucose metabolites to MRI deep white matter hyperintensities and treatment resistance in bipolar disorder patients. Bipolar Disord. 2008;10:753–764. doi: 10.1111/j.1399-5618.2008.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bearden CE, et al. Greater cortical gray matter density in lithium-treated patients with bipolar disorder. Biol Psychiatry. 2007;62:7–16. doi: 10.1016/j.biopsych.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cecil KM, et al. Frontal lobe differences in bipolar disorder as determined by proton MR spectroscopy. Bipolar Disord. 2002;4:357–365. doi: 10.1034/j.1399-5618.2002.02235.x. [DOI] [PubMed] [Google Scholar]
- 11.Hajek T, et al. Large positive effect of lithium on prefrontal cortex N-acetylaspartate in patients with bipolar disorder: 2-centre study. J Psychiatry Neurosci. 2012;37:185–192. doi: 10.1503/jpn.110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forester BP, et al. Brain lithium, N-acetyl aspartate and myo-inositol levels in older adults with bipolar disorder treated with lithium: a lithium-7 and proton magnetic resonance spectroscopy study. Bipolar Disord. 2008;10:691–700. doi: 10.1111/j.1399-5618.2008.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konradi C, et al. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 14.Le-Niculescu H, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- 15.Brady RO, et al. A longitudinal pilot proton MRS investigation of the manic and euthymic states of bipolar disorder. Transl Psychiatry. 2012;2:e160. doi: 10.1038/tp.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi XF, et al. Frontal lobe bioenergetic metabolism in depressed adolescents with bipolar disorder: a phosphorus-31 magnetic resonance spectroscopy study. Bipolar Disord. 2012;14:607–617. doi: 10.1111/j.1399-5618.2012.01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viswanath B, et al. Cellular models to study bipolar disorder: a systematic review. J Affect Disord. 2015;184:36–50. doi: 10.1016/j.jad.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 18.Munkholm K, et al. A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl Psychiatry. 2015;5:e614. doi: 10.1038/tp.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy MJ, et al. Genetic and clinical factors predict lithium's effects on PER2 gene expression rhythms in cells from bipolar disorder patients. Transl Psychiatry. 2013;3:e318. doi: 10.1038/tp.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cataldo AM, et al. Abnormalities in mitochondrial structure in cells from patients with bipolar disorder. Am J Pathol. 2010;177:575–585. doi: 10.2353/ajpath.2010.081068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naydenov AV, et al. Differences in lymphocyte electron transport gene expression levels between subjects with bipolar disorder and normal controls in response to glucose deprivation stress. Arch Gen Psychiatry. 2007;64:555–564. doi: 10.1001/archpsyc.64.5.555. [DOI] [PubMed] [Google Scholar]
- 22.McIntyre RS, et al. Bipolar disorder and diabetes mellitus: epidemiology, etiology, and treatment implications. Ann Clin Psychiatry. 2005;17:83–93. doi: 10.1080/10401230590932380. [DOI] [PubMed] [Google Scholar]
- 23.Russell JD, Johnson GF. Affective disorders, diabetes mellitus and lithium. Aust NZ J Psychiatry. 1981;15:349–353. doi: 10.3109/00048678109159459. [DOI] [PubMed] [Google Scholar]
- 24.Regenold WT, et al. Increased prevalence of type 2 diabetes mellitus among psychiatric inpatients with bipolar I affective and schizoaffective disorders independent of psychotropic drug use. J Affect Disord. 2002;70:19–26. doi: 10.1016/s0165-0327(01)00456-6. [DOI] [PubMed] [Google Scholar]
- 25.Taylor V, MacQueen G. Associations between bipolar disorder and metabolic syndrome: a review. J Clin Psychiatry. 2006;67:1034–1041. doi: 10.4088/jcp.v67n0704. [DOI] [PubMed] [Google Scholar]
- 26.Leopold K, et al. Type 2 diabetes and pre-diabetic abnormalities in patients with bipolar disorders. J Affect Disord. 2015;189:240–245. doi: 10.1016/j.jad.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Lavebratt C, et al. The KMO allele encoding Arg452 is associated with psychotic features in bipolar disorder type 1, and with increased CSF KYNA level and reduced KMO expression. Mol Psychiatry. 2014;19:334–341. doi: 10.1038/mp.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsson SK, et al. Elevated levels of kynurenic acid in the cerebrospinal fluid of patients with bipolar disorder. J Psychiatry Neurosci. 2010;35:195–199. doi: 10.1503/jpn.090180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsson SK, et al. Cerebrospinal fluid kynurenic acid is associated with manic and psychotic features in patients with bipolar I disorder. Bipolar Disord. 2012;14:719–726. doi: 10.1111/bdi.12009. [DOI] [PubMed] [Google Scholar]
- 30.Myint AM. Kynurenines: from the perspective of major psychiatric disorders. FEBS J. 2012;279:1375–1385. doi: 10.1111/j.1742-4658.2012.08551.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller CL, et al. Upregulation of the initiating step of the kynurenine pathway in postmortem anterior cingulate cortex from individuals with schizophrenia and bipolar disorder. Brain Res. 2006;1073-1074:25–37. doi: 10.1016/j.brainres.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 32.Roberts LD, et al. Targeted metabolomics. Curr Protoc Mol Biol. 2012;Chapter 30: Unit 30.2.1-24 doi: 10.1002/0471142727.mb3002s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogeser M, Parhofer KG. Liquid chromatography tandem-mass spectrometry (LC-MS/MS) - technique and applications in endocrinology. Exp Clin Endocrinol Diabetes. 2007;115:559–570. doi: 10.1055/s-2007-981458. [DOI] [PubMed] [Google Scholar]
- 34.Colangelo CM, et al. Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods. 2013;61:287–298. doi: 10.1016/j.ymeth.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Galarza FF, et al. A critical appraisal of techniques, software packages, and standards for quantitative proteomic analysis. OMICS. 2012;16:431–442. doi: 10.1089/omi.2012.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, et al. HMDB 3.0 - The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J, et al. MetaboAnalyst 3.0 - making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis GD, et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–3512. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaham O, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaham O, et al. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc Natl Acad Sci USA. 2010;107:1571–1575. doi: 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bot M, et al. Serum proteomic profiling of major depressive disorder. Transl Psychiatry. 2015;5:e599. doi: 10.1038/tp.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz E, et al. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry. 2012;17:494–502. doi: 10.1038/mp.2011.42. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz E, et al. Alterations of primary fatty acid amides in serum of patients with severe mental illness. Front Biosci (Elite Ed) 2011;3:308–314. doi: 10.2741/e246. [DOI] [PubMed] [Google Scholar]
- 44.Cai HL, et al. Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naive schizophrenia patients after treatment with risperidone. J Proteome Res. 2012;11:4338–4350. doi: 10.1021/pr300459d. [DOI] [PubMed] [Google Scholar]
- 45.Sethi S, Brietzke E. Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychopharmacol. 2015;19 doi: 10.1093/ijnp/pyv096. pii pyv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turck CW, Miliou MD. What have mass spectrometry-based proteomics and metabolomics (not) taught us about psychiatric disorders? Mol Neuropsychiatry. 2015;1:69–75. doi: 10.1159/000381902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perkins DO, et al. Towards a psychosis risk blood diagnostic for persons experiencing high-risk symptoms: preliminary results from the NAPLS project. Schizophr Bull. 2015;41:419–428. doi: 10.1093/schbul/sbu099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan MK, et al. Development of a blood-based molecular biomarker test for identification of schizophrenia before disease onset. Transl Psychiatry. 2015;5:e601. doi: 10.1038/tp.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomasik J, et al. Blood test for schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2012;262(suppl 2):S79–S83. doi: 10.1007/s00406-012-0354-3. [DOI] [PubMed] [Google Scholar]
- 50.Koike S, et al. A snapshot of plasma metabolites in first-episode schizophrenia: a capillary electrophoresis time-of-flight mass spectrometry study. Transl Psychiatry. 2014;4:e379. doi: 10.1038/tp.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orešič M, et al. Metabolome in schizophrenia and other psychotic disorders: a general population-based study. Genome Med. 2011;3:19. doi: 10.1186/gm233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xuan J, et al. Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. J Proteome Res. 2011;10:5433–5443. doi: 10.1021/pr2006796. [DOI] [PubMed] [Google Scholar]
- 53.Zheng P, et al. A novel urinary metabolite signature for diagnosing major depressive disorder. J Proteome Res. 2013;12:5904–5911. doi: 10.1021/pr400939q. [DOI] [PubMed] [Google Scholar]
- 54.Zheng P, et al. Plasma metabonomics as a novel diagnostic approach for major depressive disorder. J Proteome Res. 2012;11:1741–1748. doi: 10.1021/pr2010082. [DOI] [PubMed] [Google Scholar]
- 55.Karabatsiakis A, et al. Metabolite profiling in posttraumatic stress disorder. J Mol Psychiatry. 2015;3:2. doi: 10.1186/s40303-015-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sussulini A, et al. Metabolic profiling of human blood serum from treated patients with bipolar disorder employing 1H NMR spectroscopy and chemometrics. Anal Chem. 2009;81:9755–9763. doi: 10.1021/ac901502j. [DOI] [PubMed] [Google Scholar]
- 57.Ma D, Guest PC, Bahn S. Metabonomic studies of schizophrenia and psychotropic medications: focus on alterations in CNS energy homeostasis. Bioanalysis. 2009;1:1615–1626. doi: 10.4155/bio.09.144. [DOI] [PubMed] [Google Scholar]
- 58.Aberg KA, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen JJ, et al. Sex-specific urinary biomarkers for diagnosing bipolar disorder. PLoS One. 2014;9:e115221. doi: 10.1371/journal.pone.0115221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen JJ, et al. Combined application of NMR- and GC-MS-based metabonomics yields a superior urinary biomarker panel for bipolar disorder. Sci Rep. 2014;4:5855. doi: 10.1038/srep05855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu XJ, et al. 2,4-Dihydroxypyrimidine is a potential urinary metabolite biomarker for diagnosing bipolar disorder. Mol Biosyst. 2014;10:813–819. doi: 10.1039/c3mb70614a. [DOI] [PubMed] [Google Scholar]
- 62.Yuan W, et al. Amine metabolomics of hyperglycemic endothelial cells using capillary LC-MS with isobaric tagging. J Proteome Res. 2011;10:5242–5250. doi: 10.1021/pr200815c. [DOI] [PubMed] [Google Scholar]
- 63.Zeitoun-Ghandour S, et al. C. elegans metallothioneins: response to and defence against ROS toxicity. Mol Biosyst. 2011;7:2397–2406. doi: 10.1039/c1mb05114h. [DOI] [PubMed] [Google Scholar]
- 64.Wang TJ, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda M, et al. α-Aminoadipate induces progenitor cell properties of Müller glia in adult mice. Invest Ophthalmol Vis Sci. 2008;49:1142–1150. doi: 10.1167/iovs.07-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gramsbergen JB, et al. Brain-specific modulation of kynurenic acid synthesis in the rat. J Neurochem. 1997;69:290–298. doi: 10.1046/j.1471-4159.1997.69010290.x. [DOI] [PubMed] [Google Scholar]
- 67.Wu HQ, Ungerstedt U, Schwarcz R. L-α-aminoadipic acid as a regulator of kynurenic acid production in the hippocampus: a microdialysis study in freely moving rats. Eur J Pharmacol. 1995;281:55–61. doi: 10.1016/0014-2999(95)00224-9. [DOI] [PubMed] [Google Scholar]
- 68.Chang YF, et al. L-α-aminoadipate inhibits kynurenate synthesis in rat brain hippocampus and tissue culture. Neurochem Res. 1997;22:825–829. doi: 10.1023/a:1022035926832. [DOI] [PubMed] [Google Scholar]
- 69.Sellgren CM, et al. A genome-wide association study of kynurenic acid in cerebrospinal fluid: implications for psychosis and cognitive impairment in bipolar disorder. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.186. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hallen A, Jamie JF, Cooper AJ. Lysine metabolism in mammalian brain: an update on the importance of recent discoveries. Amino Acids. 2013;45:1249–1272. doi: 10.1007/s00726-013-1590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchli R, et al. Cloning and functional expression of a soluble form of kynurenine/alpha-aminoadipate aminotransferase from rat kidney. J Biol Chem. 1995;270:29330–29335. doi: 10.1074/jbc.270.49.29330. [DOI] [PubMed] [Google Scholar]
- 72.Schwarcz R, et al. Kynurenines in the mammalian brain: when physiology meets pathology. Nat Rev Neurosci. 2012;13:465–477. doi: 10.1038/nrn3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kerner J, Hoppel C. Fatty acid import into mitochondria. Biochim Biophys Acta. 2000;1486:1–17. doi: 10.1016/s1388-1981(00)00044-5. [DOI] [PubMed] [Google Scholar]
- 74.Brennan BP, et al. A placebo-controlled trial of acetyl-L-carnitine and α-lipoic acid in the treatment of bipolar depression. J Clin Psychopharmacol. 2013;33:627–635. doi: 10.1097/JCP.0b013e31829a83f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nierenberg AA, et al. Mitochondrial modulators for bipolar disorder: a pathophysiologically informed paradigm for new drug development. Aust NZ J Psychiatry. 2013;47:26–42. doi: 10.1177/0004867412449303. [DOI] [PubMed] [Google Scholar]
- 76.Fan J, et al. Linkage disequilibrium mapping of the chromosome 6q21-22.31 bipolar I disorder susceptibility locus. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:29–37. doi: 10.1002/ajmg.b.30942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen BM, Lipinski JF, Altesman RI. Lecithin in the treatment of mania: double-blind, placebo-controlled trials. Am J Psychiatry. 1982;139:1162–1164. doi: 10.1176/ajp.139.9.1162. [DOI] [PubMed] [Google Scholar]
- 78.Potter M, Moses A, Wozniak J. Alternative treatments in pediatric bipolar disorder. Child Adolesc Psychiatr Clin N Am. 2009;18:483–514. doi: 10.1016/j.chc.2008.11.001. xi. [DOI] [PubMed] [Google Scholar]
- 79.Hayashi-Takagi A, Vawter MP, Iwamoto K. Peripheral biomarkers revisited: integrative profiling of peripheral samples for psychiatric research. Biol Psychiatry. 2014;75:920–928. doi: 10.1016/j.biopsych.2013.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rollins B, et al. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Auta J, et al. DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophr Res. 2013;150:312–318. doi: 10.1016/j.schres.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guidotti A, et al. Toward the identification of peripheral epigenetic biomarkers of schizophrenia. J Neurogenet. 2014;28:41–52. doi: 10.3109/01677063.2014.892485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwamoto K, et al. Expression of HSPF1 and LIM in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. J Hum Genet. 2004;49:227–231. doi: 10.1007/s10038-004-0136-5. [DOI] [PubMed] [Google Scholar]
- 84.Harris LW, et al. Comparison of peripheral and central schizophrenia biomarker profiles. PLoS One. 2012;7:e46368. doi: 10.1371/journal.pone.0046368. [DOI] [PMC free article] [PubMed] [Google Scholar]