Abstract
DNA of 258 patients with treatment-resistant depression was collected in three 8-10 week, two-arm, prospective clinical trials. Forty-four allelic variations were measured in genes for the cytochrome P450 (CYP) enzymes CYP2D6, CYPC19, and CYP1A2, the serotonin transporter (SLC6A4), and the 5-HT2A receptor (HTR2A). The combinatorial pharmacogenomic (CPGx™) GeneSight test results were provided to clinicians to support medication changes from baseline (guided arm), or they were provided at the end of each study to clinicians of unguided patients who were treated as usual (TAU). TAU subjects who at baseline were prescribed medications genetically discordant for them showed only a 12% symptom improvement, far less than the 32.5% or 28.5% improvements of the TAU subjects on yellow-category (‘use with caution’; p = 0.002) or green-category medications (‘use as recommended’; p = 0.02), respectively. The odds of a clinical response were increased 2.3-fold among all GeneSight-guided compared to all TAU subjects (p = 0.004), and overall, the guided group had a 53% greater improvement in depressive symptoms (p = 0.0002), a 1.7-fold relative improvement in response (p = 0.01), and a number needed to treat for one clinical response above that seen in the TAU group of 6.07.
Key Words: Depression, Anxiety, Translational medicine, Genomics, Polymorphism, Allele, Psychiatry, Combinatorial pharmacogenomics
Introduction
Antidepressants are among the most widely prescribed medications, yet only about half of patients achieve a response and even fewer remit following an initial antidepressant trial [1]. It is well established that these treatment-resistant patients become progressively less likely to improve with subsequent antidepressant trials [1,2,3,4]. As a result, treatment resistance among depressed patients generates significant personal, financial, and societal burdens, as measured by higher treatment and health care utilization costs, disability, missed work, and decreased productivity [5].
The field of psychiatric pharmacogenomics has identified single nucleotide polymorphisms (SNPs) within genes that impact the metabolism of and response to antidepressant medications [6,7]. A composite multigenic approach, combinatorial pharmacogenomics (CPGx™), analyzes polymorphisms in genes for cytochrome P450 (CYP) liver enzymes that metabolize antidepressant and antipsychotic drugs, and genes that encode brain response proteins that contribute to their efficacy [7,8,9]. These SNPs have been combined in a proprietary medication decision support tool, GeneSight Psychotropic. Based on the composite phenotype measured for each patient, the GeneSight test generates a report that provides prescribing options for 99% of all FDA-approved antidepressant and antipsychotic drugs used to treat depression. The test has predicted health care utilizations [10] and antidepressant outcomes [11,12,13] of patients with major depressive disorder, and does so with a precision that exceeds traditional pharmacogenomic associations based on single allelic variants [14].
A pharmacogenomic tool with clinical validity might also be expected to increase clinical utility, as measured by greater antidepressant response or remission, by helping clinicians select genetically concordant antidepressants [9,15,16,17]. When provided to clinicians in an actionable report, this information might be expected to augment treatment responsiveness or decrease health care utilizations that accompany failed medication trials [5,18,19].
Three prospective, two-arm studies have evaluated the clinical validity and utility of the GeneSight test in outpatient psychiatric clinics. Subjects were enrolled if their clinicians were considering a medication change for patients who had not responded to prior medication(s) and still had moderate or more severe depressive symptoms. These 8- to 10-week-long studies were the double-blind, randomized Pine Rest study [10] (n = 49), the naturalistic studies from Franciscan Skemp Hospital (La Crosse, Wis., USA) [12] (n = 165), and the Hamm Clinic study (St. Paul, Minn., USA) [11] (n = 44). Each study compared improvements in depression scores between subjects whose clinician received the GeneSight report to select medications (guided arm) with subjects who did not and instead received treatment-as-usual (TAU), standard-of-care prescribing to select medications (TAU arm).
Clinical utility, as defined in the Laboratory-Developed Test - SynFRAME (LDT-SynFRAME), refers to the ability to add value and clinical benefit beyond traditional or previously established practices [20]. This framework can be expected to apply to a test like the GeneSight Psychotropic decision support tool. In the present study, we combined the results of the three clinical studies and analyzed the extent to which the combinatorial pharmacogenomic GeneSight test improved medication response or remission of patients with treatment-resistant depression, compared with unguided subjects that received standard care. This evaluation included the four major LDT-SynFRAME criteria, which determined whether the test was (1) associated with improved outcomes and provided benefits beyond established measures, (2) influenced clinical decision making, (3) reduced uncertainty and increased confidence in decision making for pharmacotherapy, and (4) was generalizable outside of the research setting.
Methods
The following data were extracted from each study: treatment group, sample size, study duration, baseline and 8-week 17-item Hamilton Rating Scale for Depression (HAM-D17) scores, allele frequency and/or genotype frequency, mean age of subjects, and sex ratio of subjects. All participants were outpatients with major depressive disorder or depressive disorder not otherwise specified, and consent was provided if their HAM-D17 score was 14 or higher and they had failed to respond to at least 1 antidepressant medication. At baseline, the average HAM-D17 score ± SEM across all three studies was 21.2 ± 0.8, and in none of the three studies, nor in the pool of all three studies, did the mean score differ between the TAU and GeneSight-guided groups, as determined by an independent t test (table 1). Patients with a serious medical illness, bipolar I disorder, schizophrenia, or schizoaffective disorder were excluded, but patients with other comorbid mental health conditions (e.g., anxiety and axis II disorders) were allowed to participate. Following subject consent, DNA was collected using a buccal swab by a trained health care professional at each site and shipped to the Assurex Health CLIA (Clinical Laboratory Improvement Amendments)- and CAP (College of American Pathologists)-accredited laboratory in Mason, Ohio, USA.
Table 1.
Average HAM-D17 scores in the TAU and GeneSightguided groups at baseline in the three clinical studies
| Study | Group | Mean baseline HAM-D17 score | SEM | p value |
|---|---|---|---|---|
| Pine Rest | TAU | 21.1 | 0.83 | 0.70 |
| GeneSight | 21.6 | 0.82 | ||
| Hamm | TAU | 18.4 | 1.10 | 0.20 |
| GeneSight | 20.6 | 1.22 | ||
| La Crosse | TAU | 22.5 | 0.49 | 0.45 |
| GeneSight | 23.0 | 0.49 | ||
| Pooled | TAU | 21.5 | 0.41 | 0.13 |
| GeneSight | 22.4 | 0.42 |
In none of the studies, nor in the pool of the three studies, did these baseline values differ between the two groups.
Following DNA extraction and amplification, polymorphisms in 44 alleles were measured among 5 genes that influence antidepressant and antipsychotic drug metabolism or responses [7,8]: the CYP genes CYP2D6, CYP2C19, and CYP1A2; the long and short 5HTTLPR variants of the serotonin transporter gene (SLC6A4), and the serotonin 2A receptor gene (HTR2A). The CYP2D6, CYP2C19, and CYP1A2 alleles were genotyped using the Luminex xTAG system [11]. Relevant regions were amplified using polymerase chain reaction (PCR) and clarified using Exonuclease I and shrimp alkaline phosphatase. Individual mutations were identified using allele-specific primer extension primers tagged for hybridization to Luminex xTAG beads.
The following 44 alleles or variants were tested for: CYP2D6: *1, *2, *2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *41 and duplications; CYP2C19: *1, *2, *3, *4, *5, *6, *7, and *8; CYP1A2: −3860G>A, −2467delT, −739T>G, −729C>T, −163C>A, 125C>G, 558C>A, 2116G>A, 2473G>A, 2499A>T, 3497G>A, 3533G>A, 5090C>T, 5166G>A, and 5347C>T. *1 alleles were identified by exclusion when no variants were detected. The 5HTTLPR-relevant regions within SLC6A4 and the 102T>C allele of HTR2A were amplified using PCR. The HTR2A PCR product was then digested with the restriction enzyme MspI. The PCR product and digested HTR2A (102T>C) PCR product were then run on a 2% agarose gel to determine the genotype. The short and long forms of the SLC6A4 gene and the HTR2A (102T>C) SNP were identified.
The GeneSight Psychotropic test is a proprietary combination of genotype, phenotype, and drug metabolism information for each subject with respect to CYP2D6, CYP2C19, CYP1A2, SLC6A4, and HTR2A. This was done to reduce the dimensionality inherent in data for 44 alleles and to arrive at a single composite phenotype for each subject. The first level of the composite phenotype was assigned to each subject based on the most severely categorized medication prescribed at baseline. A value of 1 was assigned and scored as ‘green’ (e.g., ‘use as directed’); the second level was assigned a value of 2 and scored as ‘yellow’ (e.g., ‘use with caution’), and the third level was assigned a 3 and scored as ‘red’ (e.g., ‘use with caution and with more frequent monitoring’) [11]. The test report, including the color categorization for each of the antidepressant and antipsychotic medications [12,14] (see online supplement; www.karger.com/doi/10.1159/000430915) as well as the genotypes and corresponding phenotypes for each of the 5 genes (online supplement) was sent within 48 h of sample collection to the clinician of each subject in the GeneSight-guided group to use at his or her discretion in guiding treatment. The report was not available to clinicians of subjects in the TAU (i.e., unguided) group until after completion of each study.
Outcome Variables
Dichotomous Outcomes
Depressive symptoms were measured using the HAM-D17 at baseline and at weeks 2, 4, and 8 for the La Crosse and Hamm studies, and at baseline and at weeks 4, 6, and 10 for the Pine Rest study. Clinical response was defined by a 50% or greater decrease from baseline in depressive symptoms as measured using the HAM-D17. Clinical remission was defined by achieving a score of 7 or less on the HAM-D17.
The number needed to treat (NNT) for response and remission was defined as the number of subjects required to receive GeneSight testing in order for 1 more subject to achieve the clinical outcome of interest above that seen in the TAU group. All dichotomous outcomes were modeled as a function of group status (i.e., GeneSight vs. TAU) when comparisons were extended across groups.
Continuous Outcomes
In order to determine if subjects in the GeneSight arm experienced a greater reduction in depressive symptoms (i.e., compared to TAU), the HAM-D17 percent improvement for each subject was calculated using the following formula: (8- or 10-week score minus baseline score)/baseline score × 100. The mean HAM-D17 percent improvement was then derived from the pooled data for each treatment group. In the current investigation, continuous outcomes were modeled as a function of group status (i.e., GeneSight vs. TAU) when comparisons were extended across groups. Comparisons were based on the ability of composite phenotypes at baseline to predict mean HAM-D17 percent improvements by the end of each study. These comparisons were restricted to the pooled TAU arm due to the heterogeneity in study designs for the GeneSight-guided subjects (e.g., subjects were blinded in the Pine Rest study and unblinded in the Hamm Clinic and La Crosse studies). All TAU subjects across the three clinical studies, however, were fully blinded and treated in exactly the same manner.
Statistics
Dichotomous Outcomes
Odds ratios (ORs), relative benefits (RBs), and their respective 95% confidence intervals (CIs) were calculated from the clinical response and remission data. ORs were derived by taking the ratio of response or remission events in the GeneSight group relative to the TAU group and dividing this ratio by the ratio of nonresponse or nonremission in the GeneSight group relative to the TAU group. The RB of the GeneSight test for improving response or remission was derived as the proportion of responders or remitters per treatment group, where the TAU arm served as the referent group. The derived effect sizes and their associated p values (calculated via Pearson's χ2 tests) served as inputs for the current meta-analysis of response and remission. All pooled effect sizes for each clinical endpoint were calculated using random effects models, and the significance of each pooled effect size was determined using z tests. The NNT was derived using the following equation 1/(CER - EER), where CER is the event rate (i.e., number of responders, number of remitters) in the TAU arm and EER is the event rate in the GeneSight arm.
Continuous Outcomes
Parametric modeling assumptions were tested and verified prior to statistical modeling. Raw HAM-D17 scores for subjects from each study were first pooled, and the mean percent improvement in HAM-D17 scores from baseline was compared across groups via an independent t test. Within the TAU arm only, the ability of a given subject's composite phenotype to predict the percent improvement in HAM-D17 score was evaluated by employing a one-way ANOVA model. Four statistical comparisons were made: (1) an omnibus ANOVA; (2) a planned comparison between subjects stratified into the green composite phenotype category and those subjects stratified into the red category; (3) a planned comparison between those stratified into the yellow category and those subjects stratified into the red category, and (4) a planned comparison of subjects stratified into the green and yellow phenotype categories to those subjects stratified into the red category. To depict the medication category level changes within each treatment group, descriptive data for the distribution of subjects in each group within each medication category level at study entry and again at study conclusion were calculated for subjects who were prescribed at least 1 panel medication at baseline and by the end of the study. Due to the above-mentioned heterogeneity in study designs, no statistical tests were conducted during this analysis.
The Šidák correction was employed using the formula 1 - (1 - α)1/n, where n is the number of independent tests and α is the nominal level (i.e., 0.05) of statistical significance, and all reported p values were adjusted for multiple testing. All analyses were conducted using SAS 9.3 [21] and CMA software [22].
Results
Odds of Response and Remission
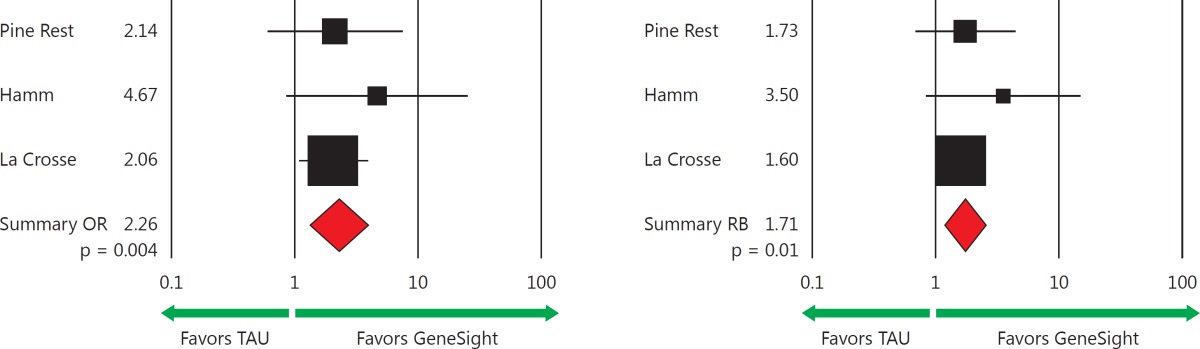
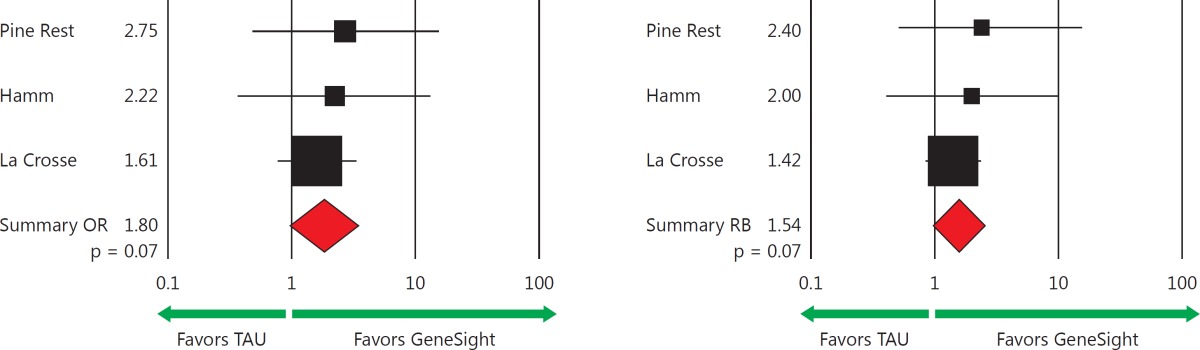
The odds (OR) of clinical response for the GeneSight-guided versus the TAU group in the Pine Rest, Hamm, and La Crosse clinical studies were 2.14, 4.67, and 2.06, respectively (table 2). The odds of clinical remission of 2.75, 2.22, and 1.61, respectively, in the same three studies also favored the GeneSight-guided subjects (table 3). A meta-analysis of these three studies using random effects models revealed an average of a 2.26 greater odds of achieving a clinical response for GeneSight-guided as compared to TAU subjects (95% CI: 1.30-3.92; p = 0.004) (table 2) and a nonsignificant increase in odds of remission of 1.80 for the GeneSight group (95% CI: 0.95-3.40; p = 0.07) (table 3). Forest plots of the 258 subjects who had complete clinical follow-up data across the three studies show the significantly increased OR for response (table 2, bottom left) and the trend for an increased OR for remission (table 3, bottom left).
Table 2.
Odds of clinical response
| Pine Rest | Hamm | La Crosse | Pooled | |
|---|---|---|---|---|
| Achieving clinical response | ||||
| Subjects, % | 49 | 44 | 165 | 258 |
| GeneSight, % (n) | 36 (9/25) | 31.8 (7/22) | 43.1 (31/72) | 39.5 (47/119) |
| TAU, % (n) | 20.8 (5/24) | 9.1 (2/22) | 26.9 (25/93) | 23.0 (32/139) |
| Difference, % | 15.2 | 22.7 | 16.2 | ~17 |
| Individual and pooled effect sizes for clinical response | ||||
| OR 2.14 | 4.67 | 2.06 | 2.26 | |
| 95% CI | 0.56–7.69 | 0.85–25.75 | 1.07–3.95 | 1.30–3.92 |
| p value | 0.24 | 0.06 | 0.03 | 0.004 |
| z value | – | – | – | 2.90 |
| Weight, % | 18.6 | 10.4 | 71.0 | 100 |
| RB | 1.73 | 3.50 | 1.60 | 1.71 |
| 95% CI | 0.68–4.42 | 0.82–15.01 | 1.04–2.46 | 1.17–2.49 |
| p value | 0.25 | 0.09 | 0.03 | 0.01 |
| z value | – | – | – | 2.79 |
| Weight, % | 16.0 | 6.7 | 77.3 | 100 |
Numbers and percentages of responding subjects in the three prospective studies, where response is defined by a 50% or greater decrease from baseline in HAM-D17 score. The forest plots reveal an increased OR of 2.26 for achieving a clinical response (p = 0.004) and a greater RB of 1.71-fold (p = 0.01) for subjects in the GeneSight groups as compared to those in the TAU groups. ORs and RBs are reported as the odds and likelihood, respectively, of subjects in the GeneSight group achieving a clinical response as compared to the TAU group. Each pooled OR, RB, and 95% CI, p value, and z value was obtained from random effects models via meta-analysis.
Table 3.
Odds of remission
| Pine Rest | Hamm | La Crosse | Pooled | |
|---|---|---|---|---|
| Achieving clinical remission | ||||
| Subjects, % | 49 | 44 | 165 | 258 |
| GeneSight, % (n) | 20 (5/25) | 18.2 (4/22) | 30.5 (22/72) | 26.1 (31/119) |
| TAU, % (n) | 8.3 (2/24) | 9.1 (2/22) | 21.5 (20/93) | 17.3 (24/139) |
| Difference, % | 11.7 | 9.1 | 9 | ~9 |
| Individual and pooled effect sizes for clinical remission | ||||
| OR 2.75 | 2.22 | 1.61 | 1.80 | |
| 95% CI | 0.48–15.79 | 0.36–13.61 | 0.77–3.37 | 0.95–3.40 |
| p value | 0.26 | 0.39 | 0.21 | 0.07 |
| z value | – | – | – | 1.81 |
| Weight, % | 13.3 | 12.4 | 74.3 | 100 |
| RB | 2.40 | 2.00 | 1.42 | 1.54 |
| 95% CI | 0.51–11.21 | 0.41–9.82 | 0.84–2.39 | 0.96–2.47 |
| p value | 0.27 | 0.39 | 0.19 | 0.07 |
| z value | – | – | – | 12.79 |
| Weight, % | 9.4 | 8.8 | 81.8 | 100 |
Remission, defined as achieving a HAM-D17 score of 7 or less for the same patients reported on in table 2, who had medication changes guided by GeneSight or guided by standard-of-care empirical prescribing (TAU) in the three studies. The forest plots reveal a nonsignificant increased odds of remission (1.80; p = 0.07) and RB (1.54; p = 0.07) with psychiatric medications for subjects in the GeneSight groups as compared to those in the TAU groups. ORs and RBs are reported as the odds and likelihood, respectively, of subjects in the GeneSight group achieving clinical remission as compared to the TAU group. Each pooled OR, RB, and 95% CI, p value, and z value was obtained from random effects models via meta-analysis.
RBs of Response and Remission
The RB of response by the end of the three studies was also greater for the 119 GeneSight-guided than for the 139 TAU subjects (table 2). The average rate of those attaining an antidepressant response was 39.5% for the GeneSight-guided group, as compared with 23% for the TAU subjects. A meta-analysis of these data revealed a significant 1.71-fold increase in the likelihood of response of guided subjects above that of the TAU group (95% CI: 1.17-2.49; p = 0.01) (table 2) using random effects models. The average remission rate was 26.1% among the guided subjects, and this exceeded the 17.3% remission rate of the TAU subjects. A 1.54-fold, but nonsignificant, increase was found in the likelihood of remission of the pooled guided subjects compared with that of the TAU group (95% CI: 0.96-2.47; p = 0.07) (table 3).
Number Needed to Treat
Response
The number of subjects requiring GeneSight testing for 1 more subject to achieve a clinical response above that of TAU subjects in the Pine Rest study was 6.59; it was 4.40 in the Hamm Clinic study, and 6.18 in the La Crosse study. The unweighted, pooled NNT estimate for response across these three studies was 6.07.
Remission
The number of subjects requiring GeneSight testing to achieve clinical remission above that for TAU subjects was 8.57 in the Pine Rest study; it was 11.00 in the Hamm Clinic study, and 11.05 in the La Crosse study. The unweighted, pooled NNT estimate for remission across these three studies was 11.38.
Medication Changes
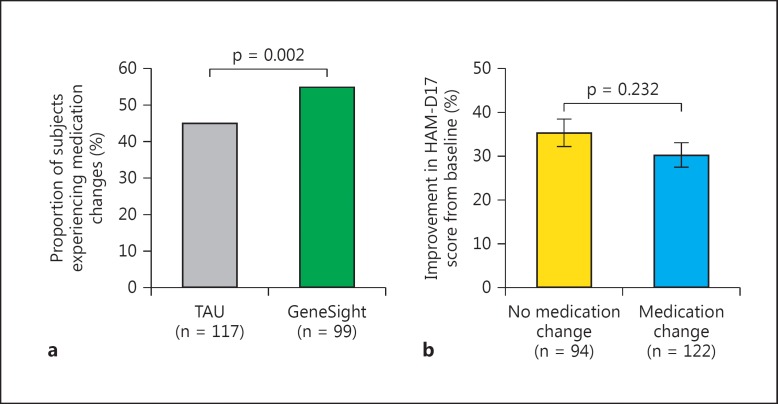
We assessed the clinical influence of the test during the 8- to 10-week-long studies through changes in medication prescriptions in the 216 subjects among the 258 who were prescribed at least 1 GeneSight panel medication at baseline and by the study end, and who also had complete clinical outcome data. Health care providers changed medications for 54.9% of those in the GeneSight-guided group, which was 21.7% more often than for the 45.1% of those in the TAU group (p = 0.002) (fig. 1a). Importantly, when these subjects were pooled and then segregated by the presence or absence of any change in panel medication dose or type, the 35.3% average decrease in HAM-D17 score for the 94 patients with a medication change did not differ (p > 0.05) from the 30.2% change of the 122 patients without a medication change (fig. 1b), indicating that the act of changing medications per se did not influence clinical improvement.
Fig. 1.
a The proportion of the 99 subjects in the GeneSight-guided group exceeded that of the 117 subjects in the TAU group who experienced an addition, removal, or change in dose of any panel medication during the clinical trial (p = 0.002, d.f. = 1, χ2 = 9.32). b No difference in antidepressant efficacy, as measured by change in HAM-D17 score, was found between the 122 subjects with and the 94 subjects without a change in a panel drug or dose during each study.
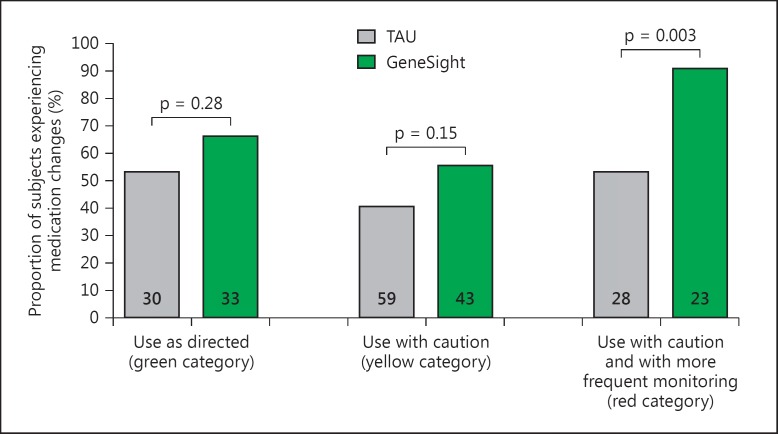
Among subjects who were prescribed a red-category medication at baseline, 21 out of 23 GeneSight-guided subjects (91.3%) experienced a medication change or dose adjustment during the study, as compared to only 15 of the 28 unguided, TAU subjects (53.6%; p = 0.003) (fig. 2, far right). Among subjects prescribed 1 or more yellow-category medications at baseline, and no red-category medications, 24 of 43 GeneSight-guided subjects (55.8%) and 24 of 59 TAU subjects (40.7%) experienced a medication or dose change during the study (p = 0.13). Among subjects prescribed 1 or more green-category medications at baseline, and no red- or yellow-category medications, 22 of 33 GeneSight-guided subjects (66.7%) and 16 of 30 TAU subjects (53.3%) experienced a medication or dose change during the study (p = 0.28) (fig. 2).
Fig. 2.
Proportion of all subjects in the TAU and GeneSight-guided groups (numbers of subjects at the bottom of each bar) who experienced an addition, removal, or change in dose of any panel medication during the clinical trial, as a function of the most severe category (green, yellow, or red) of the medications they were prescribed at baseline. The proportion of subjects on 1 or more red-category medications who experienced a change was higher for the guided subjects than the TAU subjects (p = 0.003, d.f. = 1, χ2 = 8.66).
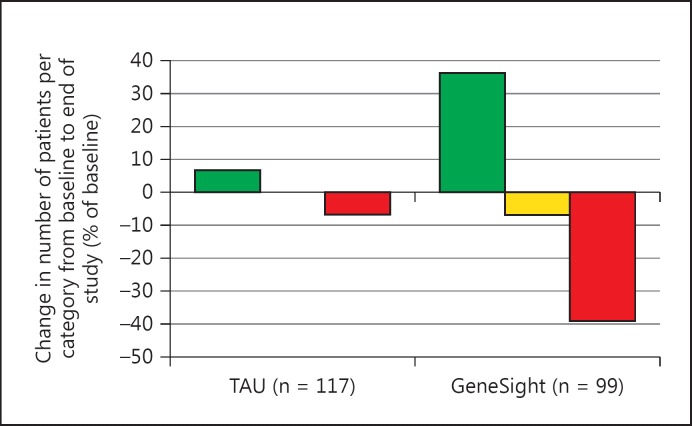
Relatively few clinicians of TAU subjects made prescription changes from baseline to the study end that changed the status of their patients' most genetically discordant medication. This produced a modest (6.7%) increase in those moved to green-category medications and even fewer away from red- or yellow-category medications (fig. 3). An amplification of this pattern was found for GeneSight-guided subjects, since 36.4% more were prescribed green-category medications, and 7.0 and 39.1% fewer were prescribed yellow- or red-category medications, respectively, by the study end (fig. 3). By the end of the study, among subjects who remained on a panel medication, only 14.1% of the subjects in the GeneSight-guided arm were prescribed 1 or more medications from the red category, which was fewer than the 22.2% of subjects in the unguided, TAU group prescribed red-category medications (data not shown; p = 0.02).
Fig. 3.
GeneSight guidance amplifies the tendency of clinicians of unguided patients to shift patients to genetically appropriate medications. From baseline to the end of the 8- or 10-week studies, 36.4% of the 99 GeneSight-guided patients switched to medications whose most severe category status was green, while 39.1% were switched off of medications whose most severe category status was red. Far smaller proportions (6.7 and 7.1%, respectively) of the 117 TAU subjects were switched in the same directions.
Percent Improvement in HAM-D17 Score
Between-Group Comparison
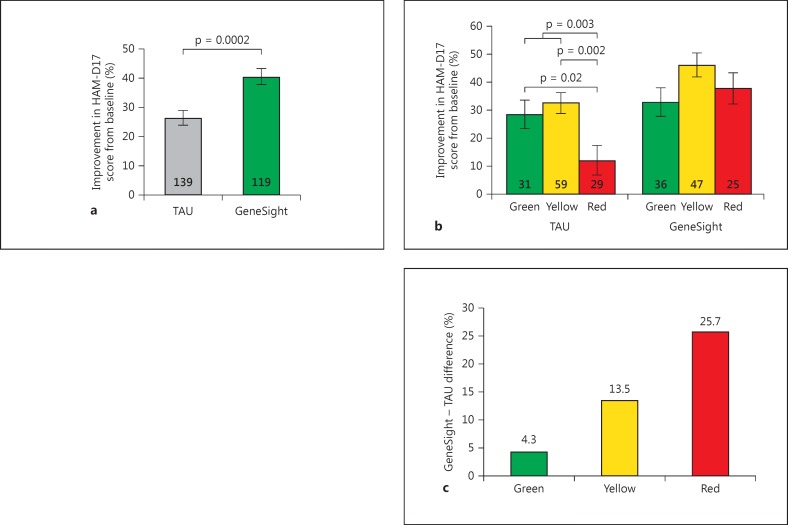
The percent improvement in HAM-D17 depression scores from baseline to week 8 or 10 was 53% greater for subjects guided by the GeneSight test (n = 119; 40.5% improvement) relative to TAU subjects, who were not guided by the pharmacogenomic report (n = 139; 26.5% improvement; p < 0.0002) (fig. 4a).
Fig. 4.
a The percent decrease in HAM-D17 depression scores (positive y-axis values) from baseline to the study end of GeneSight-guided subjects (green bar, n = 119) exceeded that of TAU subjects (gray bar, n = 139; p = 0.0002). b The greater decreases over 8 weeks in HAM-D17 depression score for the GeneSight as compared to the TAU group are a function of medication category status. The y-axis shows the greater improvement for the GeneSight-guided subjects by subtracting the mean value for that group from the value for the TAU subjects. Results are plotted for groups whose subjects' most cautionary drug status at baseline was in the green (n = 31, TAU; n = 36, GeneSight), yellow (n = 59; n = 47), and red (n = 29; n = 25) category. c Clinical improvement as a function of the most severely categorized baseline medication for each subject in the TAU and GeneSight-guided groups. p values are derived via independent t tests for all subgroup comparisons within the TAU group.
Within-Group Comparison (Composite Phenotype)
Of the 258 subjects who had complete clinical outcome data, 227 (87.9%) were prescribed a GeneSight Psychotropic panel medication at the time of study entry (fig. 4b). Of these, 47.6% were in the GeneSight arm, and a similar proportion (52.4%) was in the TAU arm. Among these 227 TAU and GeneSight subjects, 54 (23.8%) were prescribed 1 or more medications in the most severe, red category at baseline, while 106 subjects (46.7%) were prescribed yellow-category medications and 67 subjects (29.5%) were prescribed green-category medications.
The 119 fully blinded TAU subjects differed in their HAM-D17 improvement as a function of their most severe medication category level at baseline (F = 5.05, d.f. = 2, p = 0.0079). Among subjects who remained on a panel medication by the end of the study, those classified by the green category at baseline experienced a mean 28.5% improvement in HAM-D17 depression scores, and those in the yellow category experienced a similar 32.5% improvement in HAM-D17 depression scores (p = 0.52) (fig. 4b). In contrast, TAU subjects in the red category experienced only a 12% improvement in HAM-D17 depression scores, which was significantly lower than that for patients on green- (t = 2.22, p = 0.02), yellow- (t = 3.15, p = 0.002), or yellow- and green-category medications (t = 2.97, p = 0.003) (fig. 4b).
The GeneSight-guided subjects whose highest cautionary medication level at baseline was green, yellow, or red experienced mean improvements in HAM-D17 depression scores of 32.9, 46.1, and 37%, respectively, during the study. As mentioned previously, it was not feasible to statistically test for pooled medication category differences within the GeneSight arm because, unlike subjects in the TAU group, the guided subjects who were combined for this analysis had received heterogeneous treatments (i.e., blinding in the Pine Rest study). However, for descriptive purposes, the mean improvements in depressive symptoms for TAU and guided subjects in the green, yellow, or red categories were subtracted. Improvements for the GeneSight-guided subjects were 4.3, 13.5, and 25.7% higher than for the TAU patients prescribed green-, yellow-, or red-category medications, respectively, at baseline (fig. 4c).
Discussion
The three prospective, two-arm studies La Crosse [12], Hamm [11], and Pine Rest [10] measured subjects' improvements in depression outcomes with GeneSight guidance compared to TAU and evaluated the feasibility of introducing GeneSight into current psychiatric practice. Our present analysis of data from these combined studies demonstrates that GeneSight-guided treatment is associated with a greater reduction in overall depression symptoms and increases in response rates compared to standard-of-care empiric prescribing.
Providing clinicians with the GeneSight interpretive report improved the proportion of antidepressant responders by 71% as compared with unguided patients. A 2.26-fold increase in the odds of clinical response was also found for the guided patients as compared with the unguided patients. These improvements paralleled changes in drug dosing or selection, in that a greater proportion of guided patients experienced medication changes. These changes resulted in 40% of the guided patients initially on red-category medications being shifted to yellow- or green-category medications, and 35% more patients prescribed green-category medications, by the study end. Medication change per se did not explain the greater antidepressant efficacy in the guided subjects, because no difference in clinical improvements occurred between subjects who experienced a medication change and those who did not. The influence of pharmacogenomic information was even greater for guided individuals initially prescribed red-category medications, of whom 91.3% experienced a medication change and obtained the greatest clinical benefit from the test. In contrast, only 53.6% of the unguided subjects on red-category medications had a prescription change, and their improvement was significantly smaller.
These results show that GeneSight-guided physicians were more likely to adjust medication dosing or selection than were physicians who did not have access to test information, and that the improved outcomes of guided subjects derived from the use of the GeneSight results, rather than from a medication change per se. In a similar kind of evaluation, Jürgens et al. [23] studied the relationship between medication prescribing and the information clinicians received regarding the poor metabolizer or ultrarapid metabolizer phenotypes for specific CYP enzymes. In that approach, physicians took a clinical action for only 52% of the subjects with a ‘deviant’ genotype-drug interaction [23], which was far less than the 91.3% of the GeneSight-guided subjects initially taking a red-category medication who had a medication change by the study end. This indicates that medication decision support in the form of a comprehensive, combinatorial pharmacogenomic report such as GeneSight improves adherence by clinicians to its recommendations more than providing information about single, and aberrant, gene phenotypes.
The greater influence and effectiveness of GeneSight versus traditional guidance may be due to the predictive validity of the gene variants used in the combinatorial approach [24]. We evaluated the clinical validity of the CPGx test by how well it predicted clinical outcomes in the 119 TAU subjects or health care utilizations in another 96 subjects [14]. Using the multigenic composite CPGx phenotype, both types of outcomes were predicted for subgroups of patients prescribed medications metabolized by CYP2D6, CYP2C19, or CYP1A2, and to a lesser and statistically nonsignificant extent by SSRIs whose mechanism depends upon the product of the SLC6A4 PD gene, the high-affinity serotonin transporter. In contrast, phenotypes ascribed to any of the single genes failed to predict clinical outcomes and health care utilizations, demonstrating the superiority of combinatorial pharmacogenomics for producing clinical validity. When added to the combinatorial method, other gene variants may be able to predict differential outcomes between medication classes, such as selective norepinephrine or combined norepinephrine-serotonin reuptake blockers, versus SSRIs.
Greater effectiveness may also benefit from the simple interpretative guidance provided by the GeneSight report. As a result, the NNT to show an antidepressant response for guided subjects above that for TAU subjects prescribed antidepressants according to the standard of care was 6.07 (≈7). Importantly, the NNT to generate a response to SSRI antidepressants over that seen in placebo-treated patient groups is approximately 8.7 (≈9), as reported in 8 studies [25,26]. Thus, the use of CPGx decision support to guide medication selection generates an antidepressant effect that is about equal in magnitude to that of the antidepressants themselves. This is consistent with the doubling of response rates by GeneSight guidance as compared to the ‘one-size-fits-all’, standard-of-care approach that has been used to evaluate antidepressant medications versus placebo response in FDA approval studies.
The GeneSight test also identified subjects on red-category medications for whom clinical improvements were suboptimal over the following 8-10 weeks, as compared to those on yellow- or green-category medications. In another study [10], subjects prescribed red-category medications were found to have 69% more health care visits, nearly 3 times the number of days off work, and nearly 4 times the number of disability claims, and they spent USD 5,188 more on medical care over 1 year. The problematic nature of drugs in the red category derives from their more aberrant metabolism by CYP enzymes, diminished brain response, or both due to individual gene variant-dependent pharmacokinetic and pharmacodynamic phenotypic alterations [7,8,27].
Several observations from the results of the combined studies also indicate that improvements in the GeneSight-guided groups are not due to placebo effects. The improvements in antidepressant efficacy in the guided subjects were greatest for the longer, 8- to 10-week periods in these studies [10,12]; yet, the majority of improvements in responses to antidepressants found in 47 studies, including those in placebo groups, occurred in the first 2-3 weeks [28]. Also, the fully blinded, unguided TAU subjects who were prescribed red-category medications showed the poorest responses, and the red-category subjects who were guided by the test results benefitted the most.
It is interesting to see in figure 4 that 23.8% (54 of the 227) of the treatment-resistant subjects in the combined studies were prescribed red-category medications, as compared with only 10% of a general psychiatric population of 96 subjects [10]. This difference suggests that treatment resistance in major depressive disorder may in part result from patients being prescribed one or more medications that are mismatched with their genetic propensity for optimal response. A combinatorial pharmacogenomic test that identifies those who are at risk for poor response, and medications more likely to generate an improved clinical response, should enhance treatment responsiveness.
One of the goals of pharmacogenomic-based personalized medicine is to provide information that can better define treatments for patients and increase the rate or amount of their therapeutic improvement. The results of the three prospective clinical trials provided here are consistent with these goals. Each of the criteria from the Centers for Disease Control and Prevention [29] and the LDT-SynFRAME for the clinical utility of biomarker tests [20] was satisfied by the present results. The GeneSight test: (1) improved patient outcomes beyond those obtained by established (TAU) methods; (2) influenced clinical decision making by greatly increasing the prescription of green-category medications and lessening the prescription of those in the red category, and (3) reduced uncertainty in decision making for pharmacotherapy, as evidenced by the increased percentage of clinicians making medication changes compared with the standard of care; finally, (4) the test did all this outside of the research setting, in the real-world clinical treatment of patients with major depression.
This meta-analysis of results with the GeneSight test demonstrates that clinical utility can be provided by an integrated, multigenic analysis of an individual's polymorphisms among carefully measured and incorporated pharmacokinetic and pharmacodynamic genes. The increased clinical utility may be expected to broaden the adoption of combinatorial pharmacogenomics because it lessens the variability, and guesswork, that has traditionally been inherent in prescribing medications for major depressive disorder.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
Funding for the Hamm and La Crosse studies was provided by the Mayo Clinic Discovery Translation Grant, and genetic testing was funded by AssureRx Health, Inc. The Pine Rest study was fully funded by AssureRx Health, Inc. C.A.A., J.C., J.D.A., J.W., and B.D. are employees of AssureRx Health, Inc.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
References
- 1.Rush AJ, Trivedi MH, Wisniewski SR, Stewart JW, Nierenberg AA, Thase ME, Ritz L, Biggs MM, Warden D, Luther JF, Shores-Wilson K, Niederehe G, Fava M; STAR*D Study Team. Bupropion-SR, sertraline, or venlafaxine-XR after failure of SSRIs for depression. N Engl J Med. 2006;354:1231–1242. doi: 10.1056/NEJMoa052963. [DOI] [PubMed] [Google Scholar]
- 2.Trivedi MH, Rush J, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Lebowitz B, McGrath PJ, Shores-Wilson K, Biggs MM, Balasubramani GK, Fava M; STAR*D Study Team. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 3.Warden D, Rush AJ, Trivedi MH, Fava M, Wisniewski SR. The STAR*D project results: a comprehensive review of findings. Curr Psychiatry Rep. 2007;9:449–459. doi: 10.1007/s11920-007-0061-3. [DOI] [PubMed] [Google Scholar]
- 4.Gaynes RN, Rush AJ, Trivedi MH, Wisniewski SR, Spencer D, Fava M. The STAR*D study: treating depression in the real word. Cleve Clin J Med. 2008;75:57–66. doi: 10.3949/ccjm.75.1.57. [DOI] [PubMed] [Google Scholar]
- 5.Mrazek DA, Hornberger JA, Altar CA, Degtiar I. A review of the clinical, economic, and societal burden of treatment-resistant depression: 1996-2013. Psychiatr Serv. 2014;65:977–987. doi: 10.1176/appi.ps.201300059. [DOI] [PubMed] [Google Scholar]
- 6.Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- 7.Altar CA, Hornberger J, Shewade A, Cruz V, Garrison J, Mrazek D. Clinical validity of cytochrome P450 metabolism and serotonin gene variants in psychiatric pharmacotherapy. Int Rev Psychiatry. 2013;25:509–533. doi: 10.3109/09540261.2013.825579. [DOI] [PubMed] [Google Scholar]
- 8.Mrazek DA. Psychiatric pharmacogenomic testing in clinical practice. Dialogues Clin Neurosci. 2010;12:69–76. doi: 10.31887/DCNS.2010.12.1/dmrazek. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung S, Xiaofan L. The clinical use of pharmacogenomic testing in treatment-resistant depression. Prim Psychiatry. 2010;17:46–51. [Google Scholar]
- 10.Winner J, Allen J, Altar CA, Mihajlovic A. Genotype predicts health resource utilization of outpatients with anxiety and depression in a staff model health maintenance organization. Transl Psychiatry. 2013;3:e300. doi: 10.1038/tp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall-Flavin DK, Winner JG, Allen JD, Jordan JJ, Nesheim RS, Snyder KA, Drews MS, Eisterhold LL, Biernacka JM, Mrazek DA. Using a pharmacogenomic test to guide the treatment of depression. Transl Psychiatry. 2012;2:e172. doi: 10.1038/tp.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Procter B, Snyder KA, Drews MS, Eisterhold LL, Geske J, Mrazek DA. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23:535–548. doi: 10.1097/FPC.0b013e3283649b9a. [DOI] [PubMed] [Google Scholar]
- 13.Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo B. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16:219–227. [PubMed] [Google Scholar]
- 14.Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical validity: combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2014.85. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, Brockmöller J. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9:442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 16.Licinio J, Wong ML. Pharmacogenomics of antidepressant treatment effects. Dialogues Clin Neurosci. 2011;13:63–71. doi: 10.31887/DCNS.2011.13.1/jlicinio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rundell JR, Harmandayan M, Staab JP. Pharmacogenomic testing and outcome among depressed patients in a tertiary care outpatient psychiatric consultation practice. Transl Psychiatry. 2011;1:e6. doi: 10.1038/tp.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Empaire I, Guico-Pabia CJ, Preskorn SH. Antidepressant treatment and altered CYP2D6 activity: are pharmacokinetic variations clinically relevant? J Psychiatr Pract. 2011;17:330–339. doi: 10.1097/01.pra.0000405363.95881.01. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova JI, Birnbaum HG, Kidolezi Y, Subramanian G, Khan SA, Stensland MD. Direct and indirect costs of employees with treatment-resistant and non-treatment-resistant major depressive disorder. Curr Med Res Opin. 2010;26:2475–2484. doi: 10.1185/03007995.2010.517716. [DOI] [PubMed] [Google Scholar]
- 20.Hornberger J, Doberne J, Chien R. Laboratory-developed test - SynFRAME: an approach for assessing laboratory-developed tests synthesized from prior appraisal frameworks. Genet Test Mol Biomarkers. 2012;16:605–614. doi: 10.1089/gtmb.2011.0177. [DOI] [PubMed] [Google Scholar]
- 21.SAS Institute . SAS 9.3 Software. Cary: SAS Institute; 2011. [Google Scholar]
- 22.Borenstein M, Hedges L, Higgins J, Rothstein H. Comprehensive Meta-Analysis Version 2. Englewood: Biostat; 2005. [Google Scholar]
- 23.Jürgens G, Jacobsen CB, Rasmussen HB, Werge T, Nordentoft M, Andersen SE. Utility and adoption of CYP2D6 and CYP2C19 genotyping and its translation into psychiatric clinical practice. Acta Psychiatr Scand. 2012;125:228–237. doi: 10.1111/j.1600-0447.2011.01802.x. [DOI] [PubMed] [Google Scholar]
- 24.Mrazek DA, O'Kane DJ, Black JL.Methods for selecting medications US Patent No. 2007/0003931 A1. 2007
- 25.Cohn C, Shrivastava R, Mendels J, Cohn J, Fabre L, Claghorn J, Dessain E, Itil T, Lautin A. Double-blind, multicenter comparison of sertraline and amitriptyline in elderly depressed patients. J Clin Psychiatry. 1990;51:28–33. [PubMed] [Google Scholar]
- 26.Bridge J, Iyengar S, Salary C, Barbe R, Birmaher B, Pincus H, Ren L, Brent D. Clinical response and risk for reported suicidal ideation and suicide attempts in pediatric antidepressant treatment: a meta-analysis of randomized controlled trials. JAMA. 2007;297:1683–1696. doi: 10.1001/jama.297.15.1683. [DOI] [PubMed] [Google Scholar]
- 27.Mrazek DA, Biernacka JM, O'Kane DJ, Black JL, Cunningham JM, Drews MS, Snyder KA, Stevens SR, Rush AJ, Weinshilboum RM. CYP2C19 variation and citalopram response. Pharmacogenet Genomics. 2011;21:1–9. doi: 10.1097/fpc.0b013e328340bc5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posternak MA, Zimmerman M. Is there a delay in the antidepressant effect? A meta-analysis. J Clin Psychiatry. 2005;66:148–158. doi: 10.4088/jcp.v66n0201. [DOI] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention . Genomic Testing: ACCE Model Process for Evaluating Genetic Tests. Atlanta: Centers for Disease Control and Prevention; 2010. http://www.cdc.gov/genomics/getsting/ACCE/index.htm. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data