Abstract
Inflammatory bowel disease (IBD) is associated with risk variants in the human genome and dysbiosis of the gut microbiome, though unifying principles for these findings remain largely undescribed. The human commensal Bacteroides fragilis delivers immunomodulatory molecules to immune cells via secretion of outer membrane vesicles (OMVs). We reveal that OMVs require IBD-associated genes, ATG16L1 and NOD2, to activate a non-canonical autophagy pathway during protection from colitis. ATG16L1-deficient dendritic cells do not induce regulatory T cells (Treg) to suppress mucosal inflammation. Immune cells from human subjects with a major risk variant in ATG16L1 are defective in Treg responses to OMVs. We propose that polymorphisms in susceptibility genes promote disease through defects in ‘sensing’ protective signals from the microbiome, defining a potentially critical gene-environment etiology for IBD.
Intestinal microbiota modulate development and function of the immune system, and play a critical role in inflammatory bowel disease (IBD), a family of idiopathic intestinal disorders including Crohn’s disease (CD) and ulcerative colitis (UC) (1-6). Concordance rates of 40-50% between monozygotic twins implicate gene-environment interactions contribute to CD (7-10), albeit in ways that are poorly understood. Advances in DNA sequencing technologies have empowered unprecedented insights into the human genome and the gut microbiome in IBD, enabling detailed genomic characterization of patients (11) and chronicling alterations in the composition and gene content of the gut microbiome (dysbiosis) (12).
Close to 200 risk loci have been proposed for CD, with several susceptibility genes linked to the regulation of autophagy (e.g., ATG16L1) (13-15) or to microbial sensors that activate autophagy (e.g., NOD2) (16-18). While previous studies have shown that disruption of ATG16L1 and NOD2 impacts CD susceptibility through defects in microbial clearance (19-23), recent reports reveal that immune cells impaired in autophagy are hyper-inflammatory (24-29). This suggests that deficiencies in ATG16L1 or NOD2 may contribute to CD risk through impaired anti-inflammatory responses, a hypothesis not mutually exclusive with microbial clearance functions.
The microbiome of CD patients is altered, with emerging evidence for cause and effects relationships to disease. Among other recent examples of host-microbe interactions (3, 5, 6), the human commensal Bacteroides fragilis has evolved beneficial immunomodulatory properties. During colonization of mice, B. fragilis capsular Polysaccharide A (PSA) is packaged in outer membrane vesicles (OMVs) and delivered to intestinal dendritic cells to induce interleukin-10 (IL-10) production from CD4+Foxp3+ regulatory T cells (Tregs), which protect from experimental colitis (30-32). To explore gene-environment interactions during host-microbiota symbiosis, we tested if genetic pathways linked to CD are involved in the immune response to B. fragilis OMVs.
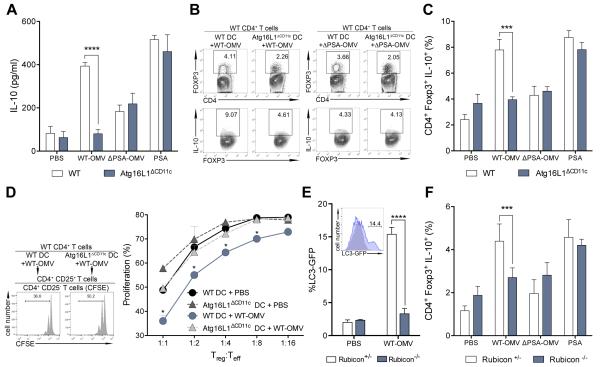
Bone marrow-derived DCs (BMDCs) differentiated from wild-type (WT) and ATG16L1-deficient (Atg16l1fl/fl Cd11cCre; Atg16L1ΔCD11c) mice were pulsed with OMVs harvested from wild-type B. fragilis (WT-OMV) or an isogenic mutant lacking PSA (ΔPSA-OMV), and co-cultured with CD4+ T cells. As previously reported (33), WT-OMVs, but not vehicle or ΔPSA-OMVs, promote IL-10 production (Fig. 1, A to C, figs. S1 and S2). Conversely, ATG16L1-deficient DCs do not support IL-10 production in response to WT-OMVs (Fig. 1, A to C). We observe similar results using Atg16l1fl/fl LysMCre mice (fig. S3). Purified PSA does not require ATG16L1 for its activity (Fig. 1, A and C, fig. S2). Next, we tested functional outcomes using in vitro T cell suppression assays. Tregs isolated from co-cultures with Atg16L1ΔCD11c BMDCs treated with B. fragilis OMVs exhibit impaired suppressive activity (Fig. 1D and fig. S2A). Neither WT-OMVs nor pure PSA have any effect on IL-10 production among CD4+Foxp3− type 1 regulatory T cells (fig. S4). ATG16L1, ATG5 and ATG7 are components of the autophagy elongation complex; BMDCs deleted in these genes likewise do not induce IL-10 production from Tregs (fig. S5). Further, recent reports reveal a role for autophagy components in Treg homeostasis (34, 35). Our findings indicate that ATG16L1-deficient DCs fail to respond to B. fragilis OMVs, demonstrating that autophagy components in DCs are required for commensal-driven Treg induction and function.
Fig. 1. ATG16L1 signals via a non-canonical autophagy pathway during OMV-mediated Treg induction.
(A) ELISA for IL-10 production during DC–T cell co-cultures with WT or Atg16L1ΔCD11c BMDCs treated with PBS, B. fragilis WT-OMV, ΔPSA-OMV or purified PSA. (B and C) Representative flow cytometry plots (B) and frequency (C) of CD4+Foxp3+IL-10+ Tregs from DC–T cell co-cultures with WT or Atg16L1ΔCD11c DCs treated with PBS, B. fragilis WT-OMV, ΔPSA-OMV or purified PSA. (D) T cell suppression assay analyzing in vitro generated Tregs from WT or Atg16L1ΔCD11c DCs treated with WT-OMVs. (E) Quantification of LC3-GFP accumulation by B. fragilis WT-OMV treatment of Rubicon+/− or Rubicon−/− DCs. Representative flow cytometry histogram plot (inset). PBS, grey; WT-OMV, blue. (F) Frequency of CD4+Foxp3+IL-10+ Tregs from Rubicon+/− or Rubicon−/− DC–T cell co-cultures treated with PBS, B. fragilis WT-OMV, ΔPSA-OMV or purified PSA. Error bars represent S.E.M. * p < 0.05, *** p < 0.001, **** p < 0.0001. Two-way ANOVA, followed by Tukey’s post-hoc analysis. Data are representative of at least 2 independent experiments.
ATG16L1, ATG5 and ATG7 participate in both canonical and non-canonical autophagy pathways (36). Interestingly, the classical autophagy-specific genes Ulk1, Fip200 or Atg14 are not required for CD4+Foxp3+IL-10+ Treg induction upon WT-OMV treatment (fig. S6). We hypothesized that OMVs utilize the non-canonical autophagy pathway, LC3-associated phagocytosis (LAP), which is specifically activated by microbial ligands delivered as particles rather than soluble molecules. LAP activation requires RUBICON, which represses canonical autophagy (36). Rubicon+/− but not Rubicon−/− BMDCs display increased accumulation of lipidated, membrane-bound LC3-GFP (LC3-II) upon B. fragilis WT-OMV treatment (Fig. 1E). As expected, neither ΔPSA-OMVs nor purified PSA are able to activate LAP (fig. S7). Moreover, treatment of Rubicon−/− DCs fails to induce Treg responses (Fig. 1F). As RUBICON is upstream of ATG16L1 signaling, OMVs preferentially utilize the non-canonical autophagy pathway LAP to mediate tolerogenic responses to B. fragilis. Further, these data suggest a reconsideration of previous literature assigning the role of ATG16L1 in IBD to defects exclusively in autophagy.
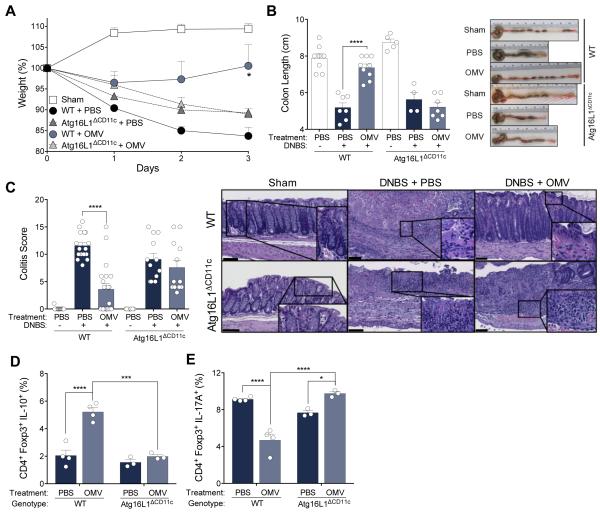
As a CD-risk gene, we investigated the in vivo requirement for ATG16L1 in CD11c+ DCs during OMV-mediated protection from experimental colitis. Indeed, WT mice treated by oral gavage with WT-OMVs are protected from 2,4-dinitrobenzenesulfonic acid (DNBS) colitis (33), whereas Atg16L1ΔCD11c mice exhibit acute weight loss and increased mortality similar to untreated mice (Fig. 2A and fig. S8A). WT, but not Atg16L1ΔCD11c mice, orally administered OMVs are protected from shortening of the colon, a hallmark of colitis models (Fig 2B), with colitis scoring and cytokine profiles verifying protection from disease (Fig. 2C and fig. S8B). Prevention of colitis is not due to an overall defect in Treg development in Atg16L1ΔCD11c mice (fig. S9). Further, while proportions of CD4+Foxp3+ cells are comparable in all groups of mice during colitis (fig. S10), Atg16L1ΔCD11c mice produce significantly less IL-10 from gut Foxp3+ Tregs compared to WT mice following WT-OMV treatment (Fig. 2D and fig. S8C). Thus, WT-OMVs require ATG16L1 within DCs to induce IL-10 expression from Foxp3+ Tregs and to suppress intestinal inflammation in a colitis model.
Fig. 2. B. fragilis OMVs require ATG16L1 in CD11c+ DCs for protection from colitis.
(A and B) Weight loss (A), colon length and gross pathology (B) of WT and Atg16L1ΔCD11c mice orally treated with PBS or B. fragilis WT-OMV during DNBS colitis. Sham groups were treated with ethanol. (C) Colitis scores by a blinded pathologist using a standard scoring system, and representative H & E images. Scale bar represents 100 μm. (D and E) Mesenteric lymph node (MLN) lymphocytes isolated post-DNBS analyzed for IL-10 (D) and IL-17A (E) production among CD4+Foxp3+ Tregs, as assessed by flow cytometry. Error bars represent S.E.M. * p < 0.05, *** p < 0.001, **** p < 0.0001. Two-way ANOVA, followed by Tukey’s post-hoc analysis. Data are representative of at least 3 independent experiments, with 3-9 mice/group.
In addition to impaired IL-10 production in response to OMV treatment, Atg16L1ΔCD11c mice display an increase in IL-17A expression (Fig. 2E), but not IFN-γ (fig. S11), among mucosal CD4+Foxp3+ T cells during colitis. Further, in vitro co-cultures of OMV-pulsed Atg16L1ΔCD11c BMDCs result in impaired IL-10 expression among Tregs (Fig. 1C), and increased IL-17A production in CD4+Foxp3+ T cells (fig. S12). Interestingly, while OMVs from other enteric bacteria each elicited a unique ATG16L1-dependent immune profile, only B. fragilis OMVs exclusively induce an anti-inflammatory response (fig. S13). Together, these data suggest ATG16L1-deficiency in DCs alters the quality of the T cell response to OMVs.
As DCs coordinate adaptive immunity, we sought to determine how Atg16L1ΔCD11c DCs are impaired in promoting tolerogenic responses. Following OMV stimulation, we observe no differences by WT or Atg16L1ΔCD11c DCs in internalizing OMVs, or in surface expression of MHC II, CD80 and CD86 (fig. S14) (27). However, stimulation with OMVs results in an increase transcription of multiple pro-inflammatory cytokines in Atg16L1ΔCD11c DCs compared to WT cells (fig. S15). These data are consistent with previous reports of a hyper-inflammatory response in ATG16L1-deficient macrophages and DCs stimulated with other microbial ligands (24, 26). Abrogation of Treg responses by ATG16L1-deficient DCs is likely due to increased pro-inflammatory cytokine production, which may impair DC-T cell interactions. Atg16L1ΔCD11c mice do not display more severe colitis than WT mice in the absence of OMV treatment (Fig. 2), suggesting that lack of protection is not due to more fulminant inflammation, but rather an inability to induce Tregs in mice deficient in ATG16L1 among CD11c+ DCs.
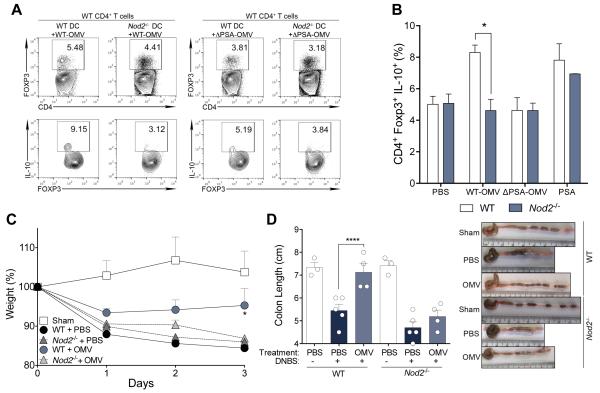
NOD2 encodes for an intracellular sensor of bacterial peptidoglycan, and polymorphisms in this gene contribute to the largest fraction of genetic risk for CD (13). NOD2 has been shown to physically recruit ATG16L1 (20, 21), a process that is impaired in human cells homozygous for a NOD2 frameshift mutation (20). Accordingly, Nod2−/− BMDCs pulsed with WT-OMVs are unable to support IL-10 production from Foxp3+ Tregs during in vitro co-cultures (Fig. 3, A and B), revealing a crucial role for NOD2 signaling in microbiome-mediated immune tolerance. This notion is supported with in vivo studies showing that Nod2−/− mice are not protected from colitis by WT-OMV treatment (Fig. 3, C and D). Similar to Atg16L1ΔCD11c animals, Nod2−/− mice produce significantly less IL-10 from Foxp3+ Tregs of the MLN following WT-OMV treatment (fig. S16A), while proportions of Tregs remain unchanged during DNBS colitis (fig. S16B). Previous studies have shown that Toll-like receptor 2 (TLR2) is required for the PSA response (33, 37). While the role of NOD2 in inducing LAP is currently unknown, signaling through TLR2 potently activates LAP (36, 38). B. fragilis OMVs induce reactive oxygen species (ROS) from WT DCs, a known product of LAP activation (36), but at significantly reduced levels in Nod2−/− or Tlr2−/− DCs (fig. S17). Though further studies are needed to define the mechanism of LAP activation by OMVs, these data reveal that NOD2 and ATG16L1 may cooperate as part of a common pathway to promote anti-inflammatory immune responses to the microbiome.
Fig. 3. NOD2 is required for OMV-mediated Tregs induction and protection from colitis.
(A and B) Representative flow cytometry plots (A) from WT-OMV (left) and ΔPSA-OMV (right) treated BMDCs co-cultured with CD4+ T cells, and frequency (B) of CD4+Foxp3+IL-10+ Tregs from DC–T cell co-cultures. (C and D) Weight loss (C), colon length and gross pathology (D) of WT or Nod2−/− mice treated with PBS or B. fragilis WT-OMV during DNBS colitis. Error bars represent S.E.M. * p < 0.05, ****p < 0.0001. Two-way ANOVA, followed by Tukey’s post-hoc analysis. Data are representative of at least 3 independent experiments, with 3-5 mice/group.
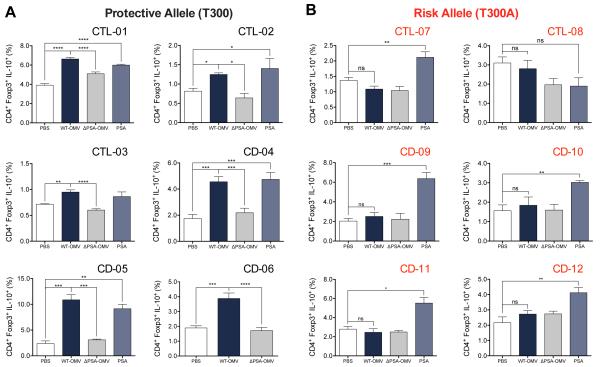
To extend and validate gene deletion approaches, we tested responses to OMVs by immune cells carrying the CD-associated variant of ATG16L1 (13, 14, 39). The ATG16L1 T300A variant leads to protein instability and altered cellular responses (23). BMDCs from transgenic mice expressing the T300A allele are also unable to promote IL-10 expression from Foxp3+ Tregs in response to WT-OMVs (fig. S18A). Further, ATG16L1 T300A transgenic mice are not protected from DNBS colitis and do not mount a potent Treg response when administered WT-OMV compared to WT mice (fig. S18, B to F). These findings prompted us to investigate if human immune cells from CD patients with the ATG16L1 T300A risk variant (table S1) are also defective in promoting Foxp3+ Treg development by B. fragilis OMVs. Monocyte-derived dendritic cells (MoDC) from CD patients and healthy controls harboring either the protective allele (T300) or the risk allele (T300A) were pulsed with OMVs or PSA and co-cultured with syngeneic CD4+ T cells. Consistent with our mouse data, human cells homozygous for the risk allele are unable to support induction of IL-10 from Foxp3+ Tregs by WT-OMVs compared to MoDCs carrying the protective allele (Fig. 4). Remarkably, all samples tested display the predicted outcome based on genotype, and not disease status. However, cells from most subjects, regardless of genotype, respond to purified PSA (Fig. 4). Collectively, we conclude that mouse and human DCs require functional ATG16L1 for induction of CD4+Foxp3+IL-10+ Tregs in response to B. fragilis OMVs.
Fig. 4. The T300A risk variant of ATG16L1 in human cells is unable to support OMV responses.
(A and B) MoDCs with either the protective (A) or risk (B) allele were treated with PBS, B. fragilis WT-OMV, ΔPSA-OMV or purified PSA, washed and co-cultured with syngeneic CD4+ T cells. IL-10 expression was analyzed by flow cytometry among CD4+Foxp3+ Tregs. Human samples were processed and analyzed in a blinded fashion. CTL, control subjects; CD, Crohn’s Disease subjects. Error bars represent S.E.M. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant. One-way ANOVA, followed by Tukey’s post-hoc analysis.
IBD impacts over 1.5 million people in the US, with rates of diagnosis increasing and treatment options remaining limited (40, 41). The etiology of IBD is complex and incompletely resolved (1). We describe herein that interactions between genetic (ATG16L1/NOD2) and environmental (microbiome) factors cooperate to promote beneficial immune responses. B. fragilis OMVs utilize LAP, an ATG16L1-dependent cellular trafficking and signaling pathway, to induce mucosal tolerance. The hyper-inflammatory responses that occur with mutations in ATG16L1 likely alter antigen-processing pathways and impair signaling by DCs to T cells, and may explain why CD-associated polymorphisms abrogate Treg induction by OMVs. Collectively, discovery of genetic circuits co-opted by the microbiome to engender health provides unprecedented functional insights into gene-environment interaction relevant to the pathogenesis of IBD. We propose an additional role for genes previously implicated in killing bacteria—namely, mutations in genetic pathways linked to IBD result in an inability to sense and/or respond to beneficial microbes. This hypothesis may represent a new perspective for the etiology of microbiome-related diseases.
Supplementary Material
Acknowledgements
We thank L. Hwang, E. Park, and M. Salas for clinical research coordination (Cedars-Sinai); A. Maskell, L. Sandoval, and C. Rumaldo for animal husbandry (Caltech); and members of the Mazmanian laboratory for discussions and critical reading of the manuscript. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by the National Institutes of Health (NIH) under Ruth L. Kirschtein National Research Service Award (DK100109) to H.C.; NIH DK097485 to R.J.X.; NIH PO1DK046763, the Cedars-Sinai F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute Research Funds, The Feintech Family Chair in IBD to S.R.T.; The Lupus Research Institute and NIH AI40646 to D.R.G; NIH U19 AI109725 (H.W.V.); The Lisa Z. Greer Endowed Chair in IBD Genetics, NIH DK062413, NIH AI067068, NIH DE023789-01, grant 305479 from the European Union, The Crohn's and Colitis Foundation of America and The Leona M. and Harry B. Helmsley Charitable Trust to D.P.B.M.; NIH AI109725 to H.W.V.; and NIH DK078938, NIH GM099535, The Crohn's and Colitis Foundation of America and the Heritage Medical Research Institute to S.K.M.
Rubicon and ULK1 knockout mice were obtained from Douglas R. Green and Mondira Kundu, respectively, under a materials transfer agreement with St. Jude Children’s Research Hospital. A provisional patent application entitled “Beneficial Activation of Autophagy Components by the Microbiome” has been filed by H. C., H. W. V. and S. K. M.
References and Notes
- 1.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. published online EpubJun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. published online EpubJul. [DOI] [PubMed] [Google Scholar]
- 5.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. published online EpubMar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. published online EpubJun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011;17:1–5. doi: 10.1002/ibd.21385. published online EpubJan. [DOI] [PubMed] [Google Scholar]
- 8.Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scandinavian journal of gastroenterology. 2000;35:1075–1081. doi: 10.1080/003655200451207. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 9.Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–1773. doi: 10.1016/s0016-5085(03)00385-8. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 10.Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577–591. doi: 10.1016/j.chom.2015.04.008. published online EpubMay 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. published online EpubFeb (ng1954 [pii]10.1038/ng1954) [DOI] [PubMed] [Google Scholar]
- 14.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. published online EpubMay (ng2032 [pii]10.1038/ng2032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. published online EpubJul (ng2061 [pii]10.1038/ng2061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardet A, Xavier RJ. Common alleles that influence autophagy and the risk for inflammatory bowel disease. Curr Opin Immunol. 2012;24:522–529. doi: 10.1016/j.coi.2012.08.001. published online EpubOct (S0952-7915(12)00124-0 [pii]10.1016/j.coi.2012.08.001) [DOI] [PubMed] [Google Scholar]
- 17.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/3507910735079107. published online EpubMay 31. [pii] [DOI] [PubMed] [Google Scholar]
- 18.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/3507911435079114. published online EpubMay 31. [pii] [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. published online EpubFeb 4. [DOI] [PubMed] [Google Scholar]
- 20.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. published online EpubJan (ni.1823 [pii]10.1038/ni.1823) [DOI] [PubMed] [Google Scholar]
- 21.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. published online EpubJan (nm.2069 [pii]10.1038/nm.2069) [DOI] [PubMed] [Google Scholar]
- 22.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. published online EpubFeb 27. [DOI] [PubMed] [Google Scholar]
- 23.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. published online EpubMay 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. published online EpubNov 13 (nature07383 [pii]10.1038/nature07383) [DOI] [PubMed] [Google Scholar]
- 25.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. published online EpubJun 25 (S0092-8674(10)00545-3 [pii]10.1016/j.cell.2010.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard-Lucey VM, Shono Y, Maurer K, West ML, Singer NV, Ziegler CG, et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity. 2014;41:579–591. doi: 10.1016/j.immuni.2014.09.011. published online EpubOct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S, Buck MD, Desai C, Zhang X, Loginicheva E, Martinez J, et al. Autophagy Genes Enhance Murine Gammaherpesvirus 68 Reactivation from Latency by Preventing Virus-Induced Systemic Inflammation. Cell Host Microbe. 2016;19:91–101. doi: 10.1016/j.chom.2015.12.010. published online EpubJan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q, Yokoyama CC, Williams JW, Baldridge MT, Jin X, DesRochers B, et al. Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe. 2016;19:102–113. doi: 10.1016/j.chom.2015.12.011. published online EpubJan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–569. doi: 10.1038/nature16451. published online EpubDec 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. published online EpubMay 29 (nature07008 [pii]10.1038/nature07008) [DOI] [PubMed] [Google Scholar]
- 31.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. published online EpubJul 6 (0909122107 [pii]10.1073/pnas.0909122107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allan SE, Broady R, Gregori S, Himmel ME, Locke N, Roncarolo MG, et al. CD4+ T-regulatory cells: toward therapy for human diseases. Immunol Rev. 2008;223:391–421. doi: 10.1111/j.1600-065X.2008.00634.x. published online EpubJun (IMR634 [pii]10.1111/j.1600-065X.2008.00634.x) [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Torchia M. L. Giardino, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. published online EpubOct 18 (S1931-3128(12)00275-2 [pii]10.1016/j.chom.2012.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. published online EpubMar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabat AM, Harrison OJ, Riffelmacher T, Moghaddam AE, Pearson CF, Laing A, et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. eLife. 2016;5 doi: 10.7554/eLife.12444. published online EpubFeb 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature cell biology. 2015;17:893–906. doi: 10.1038/ncb3192. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. published online EpubMay 20 (science.1206095 [pii]10.1126/science.1206095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. published online EpubDec 20 (nature06421 [pii]10.1038/nature06421) [DOI] [PubMed] [Google Scholar]
- 39.C. Wellcome Trust Case Control Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. published online EpubJun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. published online EpubJan (S0016-5085(11)01378-3 [pii]10.1053/j.gastro.2011.10.001) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.