Abstract
Aim:
To develop an improved delivery system for nucleic acids.
Materials & methods:
We designed, synthesized and characterized a new polymer of lactic-co-glycolic acid-modified polyethylenimine (LGA-PEI). Functions of LGA-PEI polymer were determined.
Results:
The new LGA-PEI polymer spontaneously formed nanoparticles (NPs) with DNA or RNA, and showed higher DNA or RNA loading efficiency, higher or comparable transfection efficacy, and lower cytotoxicity in several cell types including PANC-1, Jurkat and HEK293 cells, when compared with lipofectamine 2000, branched or linear PEI (25 kDa). In nude mouse models, LGA-PEI showed higher delivery efficiency of plasmid DNA or miRNA mimic into pancreatic and ovarian xenograft tumors. LGA-PEI/DNA NPs showed much lower toxicity than control PEI NPs in mouse models.
Conclusion:
The new LGA-PEI polymer is a safer and more effective system to deliver DNA or RNA than PEI.
Keywords: : lactic-co-glycolic acid, LGA-PEI polymer, nanoparticle, nucleic acid delivery, polyethylenimine, xenograft tumors
Current gene therapy relies on two systems, viral and nonviral vectors [1]. Although nonviral vectors, such as liposome, in general have more advantages, such as lower immunogenicity and more versatility than viral vectors, they may have unfavorable pharmacokinetics and lower transfection efficiency in clinical gene therapy [2–4]. One of the most frequently studied cationic polymers for DNA delivery is polyethylenimine (PEI) [3]. In fact, the PEI delivery system has been approved for use in clinical trials of gene therapy for bladder cancer [5] and HIV therapy [6,7]. PEI in both linear and branched forms delivers nucleic acids into cells through endocytosis [8–11]. Positively charged PEI amine groups can electrostatically interact with negatively charged nucleic acid phosphate groups, condense nucleic acids to form nanoparticles (NPs), which can protect the nucleic acids from degradation during the delivery path and facilitate cellular uptake of the nucleic acids. In addition, the proton-sponge effect of PEI enhances endosomal escape leading to an efficient release of DNA complexes into the cytoplasm [10]. Low-molecular-weight PEIs have fewer amine groups per molecule, having lower overall transfection efficiencies. High-molecular-weight PEIs can condense DNA more efficiently, but they may have less effective release and high cytotoxicity to cells. Currently, tissue distribution, degradation and removal process of PEI polymers from the body as well as the mechanisms of PEI-induced cytotoxicity are not fully understood. High-molecular-weight PEI lacks degradable linkages and is not easy to be broken down and excreted from the body. Due to its high positive charges, PEI could disrupt cell membranes [12,13]; and it has the tendency to aggregate red blood cells and bind complement components [14,15]. PEI can induce cell death through multiple mechanisms including necrosis, apoptosis and autophagy with damaging mitochondrial functions [13,16–21]. Mid-range-molecular-weight PEIs such as branched PEI (B-PEI; 25 kDa) and linear PEI (L-PEI; 22–25 kDa) are the most popular PEIs as transfection agents [10,15,22–24]. B-PEI may have all types of primary, secondary and tertiary amino groups; while L-PEI contains only secondary and limited primary amino groups. B-PEI has a stronger electrostatic interaction with DNA than L-PEI, accounting for greater compaction, higher zeta potentials and smaller nanoparticle sizes at equivalent DNA concentrations [10,15,23,25]. For example, the particle size formed from B-PEI (25 kDa) and DNA is 70 nm; while the particle size formed from L-PEI (25 kDa) and DNA was 782 nm [26]. B-PEI can enhance extracellular binding by fivefold as compared with L-PEI [27]. In addition, B-PEI is internalized by cholesterol dependent pathways; while L-PEI is independent to clathrin and cholesterol pathways for its internalization [27]. A number of studies have shown that B-PEI generates stronger transgene expression than L-PEI [26,28–31]. In contrast, other studies have shown that L-PEI generates stronger transgene expression in comparison to B-PEI [22–23,25,32–36]. However, both B-PEI and L-PEI polymers have certain degrees of cytotoxicity that has impeded their broad clinical applications although L-PEI is generally less toxic than B-PEI [32,37–40]. Currently, chemical and structure modifications of PEI have undergone extensive research, aiming to reduce their cytotoxicity and enhance transfection efficiency or targeted delivery. B-PEI, but not L-PEI, is often chosen for these investigations mainly because of its structure and properties [4,41–48]. B-PEI has much more primary amine groups than L-PEI has. As primary amines generally are more reactive than secondary amines, B-PEI is easier to modify and retain its nucleic acid condensation capability and intrinsic endosomal activity [49–51]. For these reasons, we have chosen B-PEI (25 kDa) for covalent linkage of LGA single units in the current study.
Poly(lactic-co-glycolic acid (PLGA) is a US FDA-approved polymer that is used in therapeutic devices and drug delivery systems because it is biodegradable, biocompatible, and has low toxicity and excellent pharmacokinetic parameters [52]. Although some studies have used PLGA to deliver plasmid DNA into cells, this use has limited applications in gene therapy because PLGA's chemical characteristics are not favorable for DNA loading, stability and release [53]. After analyzing the chemical structures of both B-PEI (25 kDa) and PLGA (12∼16 kDa), we hypothesized that the primary amines of PEI would readily react with PLGA by attacking its ester bonds; this would result in the formation of amides between PEI and lactide-co-glycolide acid (LGA). During this reaction, PLGA molecules at certain concentrations would be completely fragmented into LGA single units by PEI, while PEI remained intact. Covalent linkage of LGA units to PEI would form a new polymer that can simultaneously form NPs with nucleic acids, improve DNA or RNA delivery, and decrease toxicity in vitro and in vivo as compared with unmodified B-PEI. This new LGA-PEI polymer is fundamentally different from other reported PEI-modified PLGA gene delivery systems [44,54]. Our hypothesis was tested and confirmed in the current study; LGA-PEI polymer may have significant applications as an improved gene delivery system for the development of new clinical therapies.
Materials & methods
Chemicals & reagents
PLGA (50:50) (12 kDa–16 kDa, lactide:glycolide 50:50 mol/mol, iv. 0.50–0.65), branched and linear PEIs with average MW approximately 25 kDa were obtained from Polysciences Inc. (PA, USA). Ethidium bromide and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were obtained from Sigma-Aldrich (MO, USA). Green fluorescence protein (GFP)- or red fluorescence protein (RFP)-containing DNA plasmids (5 kb and 8 kb, respectively) were prepared by Aldevron (ND, USA). Custom miRIDIAN mimic (Thermo Fisher Scientific Biosciences) for miR-520h (double stranded) was synthesized with DY-547 fluorescent tag on the passenger strand. The mature sequence of miR-520h is 22 bp: ACAAAGUGCUUCCCUUUAGAGU. The TurboRFP labeled pLemiR lentiviral vector (Thermo Scientific Open Biosystems) with miR-520h insert is approximately 11.7 kb. Artificially modified 1070-base GFP mRNA (mmRNA) was provided by Dr. Eduard Yakubov (Houston Methodist Research Institute). Oligonucleotides (DNA primer, 24 bases) were obtained from Sigma-Aldrich.
Preparation of LGA-PEI polymer
The LGA-PEI polymer was prepared by directly mixing PLGA (12 kDa–16 kDa) and B-PEI (25 kDa) in organic solvent. Typically, 250 mg B-PEI and 120 mg PLGA were dissolved separately in 10 ml tetrahydrofuran (THF) each, combined, and then moderately stirred at room temperature for 48 h. The soft precipitate was separated from the THF solution and washed with THF solvent two-times. The solid was then dried in vacuum at room temperature overnight. Four types of LGA-PEI polymers were prepared at the PLGA/PEI weight ratios of 0.5:1, 1:1, 2.5:1 and 5:1. The yield of each new LGA-PEI polymer was determined by dry weight analysis. PEI amount and primary amine content were determined with the Cu(II) method [55] and the trinitrobenzene sulfonate assay, respectively [56].
Characterization of the LGA-PEI (0.5:1 w/w) polymer
Fourier Transform infrared spectroscopy measurements were performed with a Nicolet iS10 FT-IR Spectrometer (Thermoscientific). Scans were taken from 650 cm-1 to 4000 cm-1 at a resolution of 0.48 cm-1. PLGA, B-PEI or LGA-PEI samples were loaded on the probe of the spectrometer and measured. NMR spectroscopy was performed with a GE QE 300 MHz spectrometer; the polymers were dissolved in CDCl3 or DMSO-d6 at a concentration of 10 mg/ml. The molecular mass of LGA-PEI polymer (0.5:1 w/w) was determined with Zetasizer Nano ZS90 instrument and Debye plot analysis. The commercial B-PEI (25 kDa) or LGA-PEI polymer samples were prepared in D.I. water at concentrations from 1 to 50 mg/ml. After the materials were dissolved, the solutions were filtered through 0.2 μm membrane before the measurement. The molecular weight of LGA-PEI polymer was measured on a Zetasizer Nano ZS90 instrument. The principle of this measurement is based on the static light scattering using Debye plot. In cuvette mode, measurements from a few different concentrations are combined to draw a Debye plot. The intercept of the Debye plot is used to determine the molecular weight, and the slope is used to calculate the second virial coefficient [57].
LGA-PEI polymer & nucleic acid interaction
NPs of LGA-PEI (0.5:1 w/w) polymer and nucleic acids, including plasmid DNA, mmRNA, miRNA mimic and DNA oligonucleotides, were prepared at different polymer to nucleic acid weight ratios by adding the nucleic acid solution to the water solution of LGA-PEI polymer, and vortexing for 5 s. For example, 10 μg plasmid DNA in 50 μl of water was added to 25 μg of LGA-PEI polymer in 50 μl water. These NP suspensions were kept at room temperature for 30 min before use without any further treatment. Similarly, PEI/DNA NPs were prepared as controls.
NP nucleic acid loading efficiency was measured with spectrophotometry and the gel retardation assay. For example, LGA-PEI/DNA NPs were prepared by mixing 10 μg plasmid DNA in 50 μl of water with 0 to 30 μg of LGA-PEI in 50 μl water to make a final 100 μl solution. After 30 min, 50 μl of each suspension was aliquoted and centrifuged at 15 krpm (Eppendorf, centrifuge 5424) for 10 min. The absorption of the supernatant at 260 nm was measured with an Agilent 8453 spectrophotometer. Gel electrophoresis was performed on 0.8% agarose gels containing 25 nM ethidium bromide. Each lane was loaded with 10 μl of the above suspension mixed with 5 μl of negatively charged dye. The samples were run at 80 mV for 45 min and imaged on a VersaDoc Imaging System with software ‘Quantity One 4.6.7’ (Bio-Rad, USA).
Particle size & zeta potential measurements
The sizes of LGA-PEI/nucleic acid NPs were determined with dynamic light scattering using ZetaPlus Particle Sizing (Brookhaven Instruments Corp). The operation parameters were: wavelength 658 nm; angle 90 degree; reference index fluid 1.33; and viscosity 0.89 cp. The zeta potential of the NPs was measured with a PALs Zeta Potential Analyzer (Brookhaven Instruments Corp). The operation parameters were: conductance 39 µS; current 0.22 mA; electric field 10.22 V/cm; reference count rate 1251 kcps; and temperature 25°C.
Scanning electronic microscopy & atomic force microscopy
LGA-PEI/DNA NPs were analyzed with scanning electronic microscopy (SEM) and atomic force microscopy (AFM). The samples were prepared by mixing 10 μg DNA and 15, 25 or 35 μg LGA-PEI polymer and then following the procedure described for the size distribution studies. For SEM imaging, 50 μl of the NP suspension was dropped onto a carbon tape attached to an aluminum mount. After air drying at room temperature, the samples were coated with gold-palladium at 15 mA for 40 s to minimize surface charging. SEM imaging was conducted with a Nova NanoSEM 230 (FEI) instrument at an accelerating voltage of 10 kV. For AFM imaging, LGA-PEI/nuclei acid NPs in water were placed on a mica surface and air dried. AFM imaging was conducted with a MultiMode® 8 atomic force microscope (Bruker), and the images were analyzed with ‘NanoScope Analysis’ software.
Cell viability experiments
The cytotoxicity of LGA-PEI/DNA NPs at different polymer to DNA ratios was determined in a pancreatic cancer cell line (PANC-1) and in HEK293 cells with the MTT assay. The experimental conditions of the MTT assays were similar to those of the transfection assays described below. Cells (5000) were initially cultured in the 96-well plate with DMEM medium containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, in a 5% CO2 incubator at 37°C for 24 h. The cell medium was replaced with 100 μl opti-MEMTM medium (Thermo Fisher Scientific) and LGA-PEI/DNA NPs (with a DNA concentration of 0.2 or 1 μg/well and various LGA-PEI polymer to DNA weight ratios) was added into the culture and incubated for 16 h. Then, the opti-MEM medium was replaced with 100 μl fresh medium with 10% FBS and cultured until time for analysis. After that, the medium was replaced with a fresh medium containing 1 mg/ml MTT and 2% FBS, and the cells were incubated for 3 h or more. Finally, 100 μl of 20% SDS in N, N-dimethylformamide (DMF)/H2O (1:1 w/w) were added to the medium and the cells incubated overnight. Cell viability was determined by reading the absorbance of formazan at 570 nm on a microplate reader, and the data were expressed as a percentage of the readings of untreated cells. For separate experiments, cytotoxicity with the same MTT protocol described above was determined for different amounts of plasmid DNA with a fixed LGA-PEI/DNA ratio (2.5:1 w/w), 25 kDa PEI (1.5:1 w/w) or industry-recommended lipofectamine 2000/DNA ratio (4 μl reagent/1 μg DNA).
Cell transfection
Several cell types including pancreatic cancer cell lines (PANC-1 and ASPC-1 cells), human cholangiocarcinoma cell lines (M213, M214, M055 and M139) [58], THP-1 (human monocytic cell line derived from an acute monocytic leukemia patient), human foreskin fibroblast (BJ), human embryonic kidney 293 cells (HEK293), human breast cancer cell line (MCF-7), human umbilical vein endothelial cells (HUVEC), immortalized human T lymphocyte cell line (Jurkat), mouse embryo fibroblasts and mouse embryo stem cells were used in the cell transfection experiments. In general, 30,000 cells were initially cultured in the 12-well plate with a culture medium with 10% FBS for 24 h; the medium was replaced with opti-MEM medium, and LGA-PEI/DNA, RNA, or miRNA NPs, PEI/DNA, or lipofectamine 2000/DNA were added into the culture for 16 h; and, then, the opti-MEM medium was replaced with the fresh medium with 10% FBS for different times until analysis. For example, NPs, prepared by mixing 2 μg DNA with 5 μg LGA-PEI polymers in 100 μl H2O, were kept at room temperature for 30 min before adding them into the 12-well plate culture. Gene expression was measured by observing GFP-positive or RFP-positive cells under a fluorescence microscope. Several experiments were performed in the 96-well plates and 6-well plates with a similar protocol. A few experiments were also carried out with 8-well chamber slides (Lab-Tek II). Transfection efficiency of LGA-PEI polymers of a large modified mouse artificial chromosome plasmid DNA containing a GFP gene (∼200 kb) was tested in PANC-1 cells as described above. Cell density was recorded by phase contrast microscopy.
In vivo toxicity & gene delivery
The following animal work was approved by the Office for Protection from Research Risks and Animal Welfare Act guidelines under an animal protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. In vivo toxicity tests of PEI, LGA-PEI, PEI/DNA and LGA-PEI/DNA NPs were carried out by injecting Balb/c mice with polymer or polymer/DNA NPs via the tail vein. The maximum tolerated dose was defined as 0.5 mg/kg below the dose at which any animal had died within 10 h of iv. injection [59]. Mice received iv. injection with B-PEI (25 kDa) at serial doses (8, 6.5, 6, 5.5 or 5 mg/kg body weight) or with LGA-PEI (0.5:1 w/w) polymer at serial doses (29, 26, 23, 22, 21 and 20.5 mg/kg body weight). Four doses (30, 50, 100 and 200 μg DNA) of PEI/DNA (1.5:1 w/w, n = 3) and LGA-PEI/DNA (2.5:1 w/w, n = 4) were injected in a volume 200 μl. The mice were given food and water before or after the injection and were monitored closely after iv. injection.
For in vivo gene delivery, NPs prepared with 75 μg LGA-PEI polymer and 30 μg RFP DNA (2.5:1 w/w; 10 N/P), or 45 μg PEI and 30 μg DNA (1.5:1 w/w; 9.4 N/P) were mixed in water and then injected into wild-type Balb/c mice (20 to 30 g in weight; about 200 μl/mice, 3 mice/group) through the tail vein. Control mice (n = 3) were injected with water only. The mice were injected the same NP suspension in 200 μl, three-times in 5 days, and sacrificed on day 8 (n = 3) and day 19 (n = 3). Mouse organs were collected for histological analysis, and RFP expression was determined with fluorescence microscopy. To determine the DNA delivery efficiency to subcutaneous xenograft tumors, nude mice (n = 3) were injected subcutaneously with another human pancreatic cancer cell line (AsPC-1 cells). Briefly, 2 × 106 cells (50 μl) were injected into the right flank of 6–8 week old female nude mice. The tumors grew for 2 weeks. NPs prepared with 25 μg LGA-PEI polymer and 10 μg RFP DNA (2.5:1 w/w; 10 N/P) were diluted to 30 μl sterile H2O and injected directly into the tumor. The mice were injected once and sacrificed 3 days later. Mouse tumor tissues were collected for histological analysis, and RFP expression was determined with fluorescence microscopy. In addition, the human ovarian cancer cell line (SKOV3, 1 × 105 cells) was orthotopically injected into the bilateral ovaries of female nude mice. After xenograft tumors were developed in 5 weeks, LGA-PEI (75 μg)/RFP DNA (30 μg) NPs were injected via tail vein into the nude mice three-times in 5 days (n = 5); 5 days later, the mice were sacrificed and organ cryosections were analyzed under a fluorescence microscope. In a separate experiment, orthotopical SKOV3 tumors were similarly established in 5 weeks. LGA-PEI/miR-520h mimic (DY547 tag, 15 μg) NPs were injected into the mice via tail vein once (n = 3); 5 days later, the mice were sacrificed and organ cryosections were analyzed under the fluorescence microscope.
Statistical analysis
All data are presented as mean ± SEM. Differences among three or more groups were analyzed with one way analysis of variance (ANOVA). Student t-test was used to compare two groups. A p-value < 0.05 was regarded as statistically significant.
Results
LGA-PEI polymer synthesis & characterization
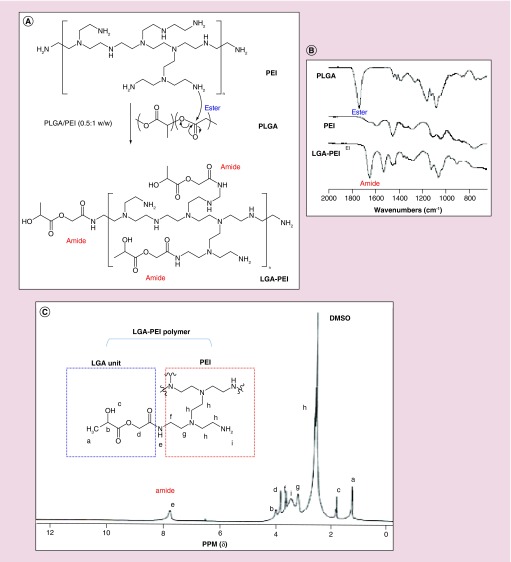
Initially, we synthesized four LGA-PEI polymers based on PLGA and PEI weight ratios (0.5:1, 1:1, 2:1 and 5:1). PLGA (12∼16 kDa) and branched PEI (25 kDa) dissolved well in THF. After stirring the reaction mixture for 48 h, we separated the soft precipitate from the solution by filtration. The yield of each new LGA-PEI polymer is >95% by dry weight analysis. The w/w ratios of PLGA to PEI ranged from 0.5:1 to 5:1, corresponding to molar ratios (n/n) that ranged from 1:1 to 10:1, respectively. For LGA-PEI (0.5:1 w/w) polymer, the PEI content in the polymer was 64.7.0 ± 1.6% according to the results of the copper sulfate assay [55], and the primary amine number was 49 ± 3% of that of PEI according to the trinitrobenzene sulfonate assay [56]. For LGA-PEI (1:1 w/w) polymer, the PEI content in the polymer was 48.0 ± 0.8%, and the primary amine number was 9.5 ± 2.1% of that of PEI. For LGA-PEI (2:1 w/w) and LGA-PEI (5:1 w/w) polymers, the primary amine numbers were 11.3 and 8.2% of that of PEI, respectively (Table 1). Therefore, about 90% of primary amines of PEI were accessible for the reaction with LGA. Only 10% of primary amines of PEI were not able to react with LGA. DNA readily interacts with all amine groups of PEI, including primary, secondary and tertiary amines of LGA-PEI polymers. From these data, we could understand the reliable reaction between B-PEI and PLGA through the amide bond formation in a dose-dependent fashion. In this study, LGA-PEI polymer (0.5:1 w/w or 1:1 mole/mole) was selected for further structure analyses, DNA loading and delivery experiments because at this condition, PLGA is completely broken down to the LGA single units, which covalently linked to B-PEI (25 kDa) molecule to form a truly new LGA-PEI polymer, while maintaining a relatively high primary amine content for nucleic acid loading and delivery.
Table 1. . Polyethylenimine content and primary amine percentage in different preparations of lactic-co-glycolic acid-modified polyethylenimine polymers.
| LGA/PEI polymer | PEI content (%) | LGA content (%) | Primary amine (%) |
|---|---|---|---|
| PEI |
100 |
0 |
100 |
| LGA-PEI (0.5:1 w/w) or (1:1 n/n)† |
64.7 ± 1.6 |
35.3 |
49 ± 3 |
| LGA-PEI (1:1 w/w) or (2:1 n/n) |
48.0 ± 0.8 |
52 |
9.5 ± 2.1 |
| LGA-PEI (2:1 w/w) or (4:1 n/n) |
32.3 ± 1.4 |
68 |
11.3 ± 0.1 |
| LGA-PEI (5:1 w/w) or (10/1 n/n) | 15.7 ± 0.6 | 84 | 8.2 ± 0.2 |
†In this study, LGA-PEI polymer (0.5:1 w/w or 1:1 n/n) was selected for DNA loading and delivery experiments.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; n/n: mole/mole; PEI: Polyethylenimine.
The structure of LGA-PEI (0.5:1 w/w) polymer was analyzed. The FT-IR spectroscopy of PLGA shows the ester C=O stretching absorption at 1746.8 cm-1. This peak disappeared in the spectroscopy of LGA-PEI polymer; instead, a new peak of amide C=O stretching at 1651.6 cm-1 appeared, indicating the formation of amide bonds (Figure 1B). Furthermore, the C-N stretching band at 1595 cm-1 in the pure B-PEI (25 kDa) sample disappeared, but a new peak appeared at 1530 cm-1 with greater intensity in the LGA-PEI sample. The FT-IR results indicate that the reaction was completed between PLGA and PEI polymers. NMR spectroscopy was also used to characterize the LGA-PEI (0.5:1 w/w) polymer. The chemical shifts of protons at 5.2, 4.8 and 1.56 ppm in the PLGA sample are assigned to CH, CH2 and CH3 protons, respectively [60] (Supplementary Figure 1A). In the PEI sample, the peak at chemical shift of 2.59 ppm was assigned to CH2 protons, and the broad peak at 2.8 ppm to NH and NH2 protons (Supplementary Figure 1B). In the LGA-PEI sample, many new peaks appeared which were different from those of PLGA or PEI individual samples (Figure 1C). The most important peak was the one at 7.75 ppm, assigned to amide RCONH proton [61], confirming the formation of an amide bond between LGA and PEI. Peaks at 1.8 ppm were assigned to –OH proton; at 1.22 ppm to –CH3; at 4 ppm to CH proton, and at 3.82 ppm to CH2 protons of the LGA unit, respectively. While peaks at 2.28 and 2.6 ppm were assigned to CH2 protons of -NCH2CH2NH2; peaks at 3.2 and 3.6 ppm to CH2 protons connected with amide and the broad peak at 3.45 ppm to amino protons of PEI unit [62]. The molecular mass of commercial B-PEI (25 kDa) and LGA-PEI (0.5:1 w/w) polymer was determined with Zetasizer Nano ZS90 instrument and Debye plot analysis. As expected, molecular weight of commercial B-PEI (25 kDa) was determined as 24.6 kDa (Supplementary Figure 2), and molecular weight of the LGA-PEI (0.5:1 w/w) polymer was determined as 42.6 kDa (Supplementary Figure 3).
Figure 1. . Synthesis of LGA-PEI polymer.
(A) Proposed mechanism for the formation of LGA-modified PEI polymers. (B) Fourier transform infrared spectroscopy (FT-IR) analysis of PLGA, PEI and LGA-PEI polymers. The formation of an amide bond between the PLGA and PEI is indicated by the -OC-NH- stretching absorption at 1651.6 cm-1. (C) NMR spectroscopy analysis.
DNA loading efficiency of LGA-PEI polymer
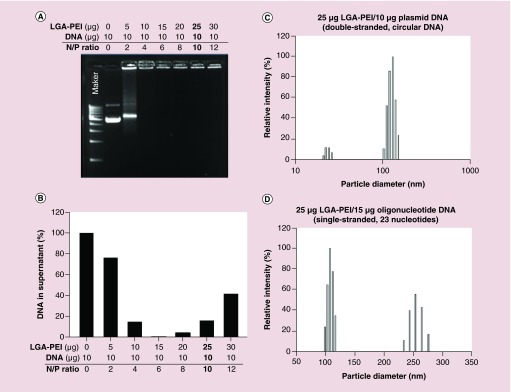
The positively charged LGA-PEI (0.5:1 w/w) polymer dissolved completely in water. LGA-PEI polymer and DNA were mixed at ratios that usually ranged from 1.5:1 to 25:1 by adding a plasmid DNA solution to a water solution of LGA-PEI polymer, and mixing the final solution with a vortex for 5 s. DNA loading was measured with both spectrophotometry and the gel retardation assay. Gel electrophoresis was performed on 0.8% agarose gels containing 25 nM ethidium bromide. At a 0.5:1 w/w ratio (2 N/P), LGA-PEI polymer retained most of the plasmid DNA, and at a low 1:1 w/w ratio (4 N/P), it was able to retain the DNA completely (Figure 2A). Therefore, at this ratio (1:1 w/w), the DNA loading is 100%. The more LGA-PEI polymer is available, the more cationically charged polymers bind to and condense DNA. Thus, LGA-PEI (15 μg or above) and plasmid DNA (10 μg) efficiently formed NPs. Optical measurements can determine free DNA or polymer/DNA NPs suspended in solution (Figure 2B). After LGA-PEI polymer (from 0 to 30 μg) and plasmid DNA (10 μg) was mixed and centrifuged, the DNA amount in the supernatants decreased from 100 to 0% as the polymer amount increased to 15 μg; however, when the polymer concentration increased further (20–30 μg), the DNA amount in the supernatants increased. This indicates that the negatively charged DNA molecules were neutralized by positively charged polymers, and that particles were formed and centrifuged down. However, when more polymers were present, the extra positive charge of the polymer made the particles soluble in the aqueous solution; the particle size became smaller and could not be centrifuged down under the conditions of the experiment. This was further confirmed by the zeta potentials of polymer/DNA NPs. The zeta potential of B-PEI (25 kDa) was up to only 90 mV, but decreased to 60 mV when PEI formed a complex with negatively charged DNA. The zeta potential of LGA-PEI/DNA was much smaller, around 20 mV; this indicated that the positive charge of PEI was effectively shielded by LGA single units (Table 2). The particles prepared at 1:1 w/w (4 N/P) ratio (i.e., 10 μg LGA-PEI with 10 μg DNA) aggregated into microparticles perhaps because of their low surface charge.
Figure 2. . Plasmid DNA loading efficiency of LGA-PEI polymer and particle size distribution.
LGA-PEI (0.5:1 w/w) polymer was used. (A) DNA gel retardation assay was performed on 0.8% agarose gels containing 25 nM ethidium bromide. Different amounts of LGA-PEI polymer (0, 5, 10, 15, 20, 25 or 30 μg) were mixed with 10 μg plasmid DNA. (B) Optical measurement. Free DNA or polymer/DNA NPs suspended in solution were detected with spectrophotometry. Plasmid DNA (10 μg) was mixed with 0 to 30 μg LGA-PEI polymer and centrifuged. (C) Size distribution of 25 μg LGA-PEI/10 μg plasmid DNA (double-stranded, circular DNA) NPs. The size of the particles in solution was determined with a Zeta Potential Analyzer. (D) Size distribution of 25 μg LGA-PEI/15 μg oligonucleotide DNA (single-stranded, 23 nucleotides).
Table 2. . Zeta potential of LGA-PEI/DNA nanoparticles.
| Nanoparticles | w/w ratio | N/P ratio | Zeta potential (mV)† |
|---|---|---|---|
| 5 μg LGA-PEI/10 μg DNA |
0.5:1 |
2 |
-38.9 ± 3.3 |
| 10 μg LGA-PEI/10 μg DNA |
1:1 |
4 |
N/A |
| 15 μg LGA-PEI/10 μg DNA |
1.5:1 |
6 |
30 ± 1.4 |
| 20 μg LGA-PEI/10 μg DNA |
2:1 |
8 |
22 ± 8 |
| 25 μg LGA-PEI/10 μg DNA |
2.5:1 |
10 |
21.4 ± 0.4 |
| 35 μg LGA-PEI/10 μg DNA |
3.5:1 |
14 |
27.5 ± 4.3 |
| 10 μg PEI/10 μg DNA |
1:1 |
6 |
57.5 ± 1.5 |
| 20 μg PEI/10 μg DNA | 2:1 | 12 | 63.8 ± 4.6 |
†LGA-PEI (0.5:1 w/w) polymer was used.
Zeta potential was measured in distilled water at ambient temperature.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine.
Self-assembly nanoparticles of the LGA-PEI polymers with nucleic acids
Mixing LGA-PEI (0.5:1 w/w) polymer and plasmid DNA in water resulted in the formation of LGA-PEI/DNA NPs. These NPs had sizes that depended on the ratios of LGA-PEI to DNA, and ranged from 100 to 130 nm, according to the dynamic light scattering results. A representative size distribution for 25 μg LGA-PEI/10 μg DNA is shown in Figure 2C. Polydispersity was between 0.1 and 0.3. In addition, the LGA-PEI delivery system was used to deliver oligonucleotide DNA or RNA for therapeutic purposes. LGA-PEI NPs were formulated with different amounts of single stranded oligo DNA primer (23 bases), and size distribution was determined with dynamic light scattering. We found that mixing 25 μg LGA-PEI with 5 μg oligo DNA did not form NPs; mixing it with 10 μg oligo DNA formed larger particles (250 and 900 nm), and mixing it with 15 oligo DNA formed smaller particles (100 and 250 nm) (Figure 2D).
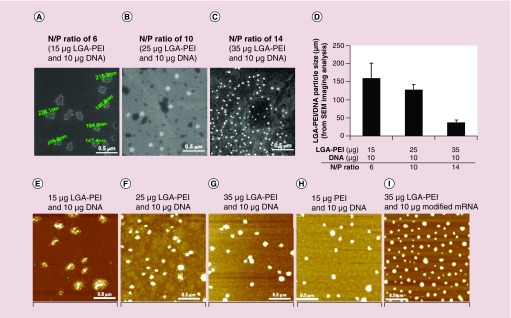
Particle images were taken with SEM and AFM. The samples were prepared by mixing LGA-PEI polymer and GFP plasmid DNA (5 kb) in nitrogen/phosphorus (N/P) ratios of 6 (1.5:1 w/w), 10 (2.5:1 w/w) and 14 (3.5:1 w/w). When DNA was condensed with LGA-PEI polymer at N/P ratio of 6 (1.5:1 w/w), SEM images clearly show that the DNA was wrapped by polymers and that the particles agglomerated and formed in sizes ranging from 150 to 230 nm (Figure 3A). The AFM images clearly show fused particles and free DNA chains (Figure 3E). With more LGA-PEI polymers, however, the particles look more solid and rigid, and the size decreases to 100–150 nm as N/P increased to 10 (2.5:1 w/w) (Figure 3B & F). When the N/P is increased to 14 (3.5:1 w/w), the particles become smaller to around 30–50 nm (Figure 3C, D & G). The AFM images show nonfused particles of about 100 nm in size for the higher N/P formulations (Figure 3F & G). Thus, LGA-PEI polymer condenses DNA effectively and controls the particle size. The more polymer is present, the smaller the size of the NPs is. As a control, we combined 15 μg PEI and 10 μg DNA to form NPs of less than 100 nm in size (Figure 3H). Furthermore, 35 μg LGA-PEI polymers and 10 μg modified mRNA (mmRNA) also formed NPs of less than 100 nm in size (Figure 3I). The NPs formed with LGA-PEI polymer and big plasmid DNA molecules (11.7 kb) were slightly bigger than those formed with smaller plasmid DNA (data not shown).
Figure 3. . Direct observation of LGA-PEI/DNA NPs with scanning electron microscopy and atomic force microscopy.
LGA-PEI polymer with 64% PEI corresponding to the LGA-PEI 0.5:1 w/w polymer was used. (A–C) are SEM images. (D) Average particle size of three different LGA-PEI/DNA N/P ratios (6, 10 and 14) in SEM images. LGA-PEI/DNA N/P ratios (6, 10 and 14) are correspondent to its w/w ratios (1.5:1, 2.5:1 and 3.5:1), respectively. (E–G) are AFM images of samples prepared with LGA-PEI polymers (64% PEI) and plasmid DNA (4.7 kb) at nitrogen/phosphorus (N/P) ratios of 6, 10 and 10. (H) Control: 15 μg PEI and 10 μg plasmid DNA. (I) Control: 35 μg LGA-PEI polymers and 10 μg modified mRNA of GFP.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; NP: Nanoparticle; PEI: Polyethylenimine.
Cytotoxicity of LGA-PEI/DNA NPs
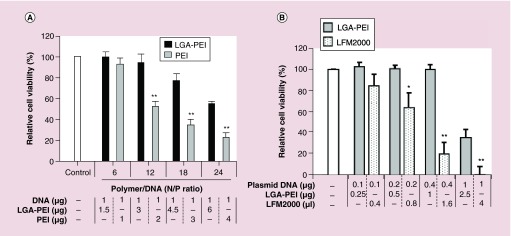
The cytotoxicity of LGA-PEI/DNA NPs and PEI/DNA complexes was investigated in a human pancreatic cancer cell line (PANC-1). The cells were treated overnight with NPs in 10% FBS DMEM medium, then cell viability was determined with the MTT assay. The amount of plasmid DNA was fixed at 1 μg/well in 96-well plates, while different amounts of LGA-PEI polymers or PEI were used (Figure 4A). PEI content in LGA-PEI polymers was the same as the amount (N/P) of unmodified PEI used in the experiments. We studied PEI/DNA and LGA-PEI/DNA N/P ratios at 6, 12, 18 and 24. The MTT test results indicated that the treatment with LGA-PEI/DNA NPs at 12 N/P produced either no or minor toxicity when compared with no treatment. When we used PEI/DNA complexes as controls, however, approximately half of the cells died when the PEI/DNA N/P ratio was 12. Thus, the LGA component of LGA-PEI polymers has a protective effect against PEI-induced cytotoxicity. The LGA-PEI/DNA delivery system is much less toxic than the PEI/DNA delivery system at the same PEI and DNA contents and experimental conditions. PEI to DNA ratio (1.6:1 w/w; 10 N/P) was equivalent to a NP of 25 μg LGA-PEI/10 μg DNA (2.5:1 w/w; 10 N/P), which was the most commonly used condition in the current study. In addition, we compared the cytotoxicity of LGA-PEI and lipofectamine 2000 (4 μl for 1 μg DNA as industry recommended) by loading serial amounts of plasmid DNA for delivery in PANC-1 cells. The results showed that LGA-PEI polymer delivering same amounts of DNA with lipofectamine 2000 showed a less toxicity than lipofectamine 2000 in PANC-1 cells (Figure 4B).
Figure 4. . Cytotoxicity of LGA-PEI/plasmid DNA NPs in vitro.
LGA-PEI (0.5:1 w/w) polymer was used. (A) Comparison between LGA-PEI polymer and unmodified PEI. Different amounts of LGA-PEI polymer were mixed with plasmid DNA (1 μg/well in 96-well plates). The amount of PEI in LGA-PEI and unmodified PEI was the same. Pancreatic cancer cell line (PANC-1) cells were incubated overnight with nanoparticles in 10% FBS DMEM medium; cell viability was determined with the MTT assay. n = 3/group. (B) Comparison between LGA-PEI polymer and commercial lipofectamine 2000. NPs of LGA-PEI/DNA (2.5/1) or lipofectamine 2000 delivered different amounts of plasmid DNA into PANC-1 cells.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
LGA-PEI polymer efficiently delivers nucleic acids to different cell types in vitro
PANC-1 cells cultured in 96-well plates were treated overnight with NPs in 10% FBS DMEM medium. Then, the medium was replaced with fresh medium, and the cells incubated for another day. Green fluorescence was evaluated for the transfection efficacy of the LGA-PEI/GFP DNA NPs and compared with that of lipofectamine 2000/DNA and PEI/DNA delivery systems (Figure 5A). Under the same experimental conditions, LGA-PEI/DNA NPs had a transfection activity that was better than that of PEI/DNA polyplexes and similar to that of lipofectamine 2000/DNA in PANC-1 cells, while the cell density in LGA-PEI/DNA NP-treated cells was greater than that in both PEI/DNA and lipofectamine 2000/DNA groups, indicating less cytotoxicity. These results were confirmed in PANC-1 cultures in 12-well plates (Supplementary Figure 4A). PANC-1 cells treated with LGA-PEI/DNA expressed GFP for a longer time than the cells treated with lipofectamine/DNA. GFP fluorescence was still observed in days 16 and 28 in cells transfected with LGA-PEI/DNA, while cells transfected with lipofectamine/DNA showed very weak GFP fluorescence (Supplementary Figure 4B). LGA-PEI polymer kept at room temperature from 2 weeks to 3 months did not lose its plasmid DNA delivery capability, demonstrating the stability of the polymer (Supplementary Figure 4C). Transfection rate of LGA-PEI/GFP DNA NPs was also high in another pancreatic cancer cell line (AsPC-1) (Supplementary Figure 4D). Interestingly, LGA-PEI polymer was able to successfully deliver a modified mouse artificial chromosome DNA (∼200 kb) containing a GFP gene to PANC-1 cells (Supplementary Figure 5).
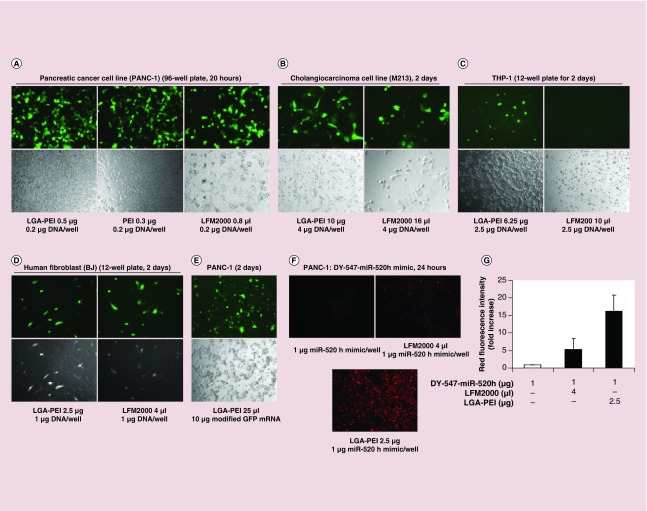
Figure 5. . Gene delivery efficiency of LGA-PEI/DNA or RNA NPs in vitro determined by immunofluorescence.
LGA-PEI (0.5:1 w/w) polymer was used. (A) Delivery of plasmid DNA to pancreatic cancer cell line (PANC-1). LGA-PEI/plasmid DNA (0.2 μg/well in 96-well plates), and lipofectamine 2000/DNA or PEI/DNA controls containing GFP gene, were incubated with PANC-1 cells for 20 h. (B) Delivery of GFP gene-containing plasmid DNA to a cholangiocarcinoma cell line (M213). LGA-PEI polymers or lipofectamine 2000 control with GFP gene-containing plasmid (4 μg/well in the 6-well plate) were incubated with M213 cells for 48 h. (C) Delivery of DNA into THP-1 (suspension cell culture). LGA-PEI or lipofectamine 2000 delivered 2.5 μg DNA/well in 12-well plates for 48 h. (D) Human fibroblasts (BJ). LGA-PEI and lipofectamine 2000 delivered 1 μg DNA/well in 12-well plates for 48 h. (E) Delivery of modified GFP mRNA (mmRNA) into PANC-1 cells (10 μg RNA/well in 12-well plates) in 48 h. (F) Delivery of synthetic miR-520h mimic labeled with DY-547 (1 μg double stranded RNA/well in 8-well chamber slides) by LGA-PEI or lipofectamine 2000 into PANC-1 cells in 24 h. Control: miR-520h mimic alone. (G) Fluorescence density analysis of delivery of synthetic miR-520h mimic labeled with DY-547 (1 μg double-stranded RNA/well in 8-well chamber slides) into PANC-1 cells in 24 h by LGA-PEI copolymer and lipofectamine 2000.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
Gene transfection efficiency of LGA-PEI polymer was tested in cholangiocarcinoma cell lines (M213; Figure 5B; M055, M214 and M139; Supplementary Figure 6) with a GFP gene-containing plasmid and compared with that of the lipofectamine 2000 delivery system. Under the same culture conditions, the transfection rate of the LGA-PEI polymer was slightly better than that of lipofectamine 2000. Importantly, the LGA-PEI polymer was less toxic than lipofectamine 2000 as demonstrated by a higher cell density (Figure 5B). LGA-PEI polymer was able to deliver GFP plasmid DNA into THP-1 cells in suspension with no or limited toxicity, while lipofectamine 2000 was unable to do so and caused significant cell death (Figure 5C). Although both the LGA-PEI polymer and lipofectamine 2000 transfected GFP plasmids into human fibroblasts (BJ) with similar transfection efficiency, the LGA-PEI polymer was much less toxic (Figure 5D). In addition, the gene transfection efficiency of LGA-PEI/DNA NPs was also tested in culture media with or without serum. We found that serum did not affect gene delivery of LGA-PEI polymers (Supplementary Figure 7).
The LGA-PEI polymer efficiently delivered modified GFP mRNA into PANC-1 cells (Figure 5E). LGA-PEI's delivery of synthetic miR-520h mimic (double-stranded RNA) labeled with fluorescent dye DY-547 was also effective. After incubation with miR-520h mimic (control), lipofectamine 2000/miR-520h, or LGA-PEI/miR-520h mimic for 24 h, the cells with the LGA-PEI/miR-520h mimic NPs showed red fluorescence that was much stronger than that of control cells with miR-520h mimic alone and with lipofectamine 2000/miR-520h (Figure 5F, G).
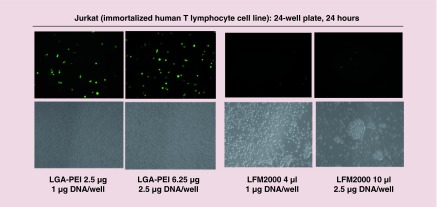
It is known that cell density affects gene transfection of the lipofectamine 2000/DNA delivery system. We confirmed that both LGA-PEI polymer and lipofectamine 2000 effectively delivered the GFP gene-containing plasmid into human embryonic kidney 293 (HEK293) cells (Supplementary Figure 8A). In order to determine whether cell density influences GFP expression in the LGA-PEI/DNA system, HEK293 cells that were 80–90%, 40% and 20% confluent were treated with LGA-PEI/DNA NPs. We found that the rate of GFP expression in these cells was not dependent on cell confluency (Supplementary Figure 8B); this indicates that cell density had no effect on the gene transfection of the LGA-PEI/DNA delivery system. LGA-PEI/DNA NPs had a transfection activity in a breast cancer cell line (MCF-7) that was similar to that of lipofectamine 2000/DNA in these cells (Supplementary Figure 9). Furthermore, we showed that LGA-PEI polymer was able to deliver GFP gene into difficult-to-transfect cell lines such as THP-1 (Figure 5C), HUVECs (Supplementary Figure 10), primary mouse embryo fibroblasts (Supplementary Figure 11A) and mouse embryo stem cells (Supplementary Figure 11B) although the transfection rate was limited in a low to moderate level at the current experimental condition. Jurkat cell is another difficult-to-transfect cell line. Compared to lipofectamine 2000 delivery system, LGA-PEI polymer was much more efficient to deliver GFP gene into Jurkat cells, and it also showed less cytotoxicity in Jurkat cells (Figure 6).
Figure 6. . Comparison of gene delivery by LGA-PEI polymer and lipofectamine 2000 (LFM2000) in Jurkat cells.
LGA-PEI delivery of a GFP gene-containing plasmid (1 and 2.5 μg DNA/well) into Jurkat cells in 24-well plates for 24 h. The same amount of plasmid DNA with LFM2000 delivery was used for the comparison. Fluorescent images and cell density images were observed at 24 h.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
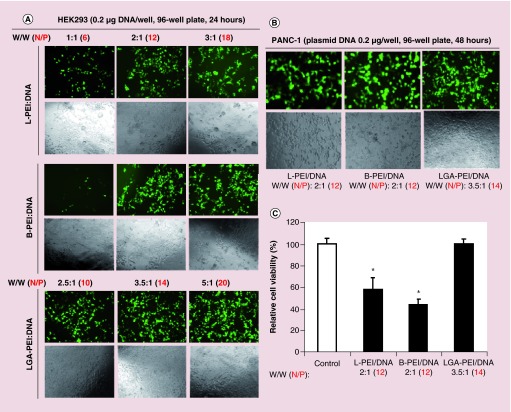
Since L-PEI is often used as an effective transfection agent, we performed separate experiments to compare the transfection efficiency and cytotoxicity of L-PEI (25 kDa), B-PEI (25 kDa) and our new LGA-PEI (0.5:1 w/w) polymer at comparable transfection conditions. Commercial L-PEI (25 kDa) is recommended to use L-PEI/DNA weight ratios of 1:1, 2:1 and 3:1 for its optimal condition of transfection experiments in many types of cells such as HEK293 cells. Thus, we chose HEK293 cells for our initial experiments. HEK293 cells (5000/well) were seeded onto the 96-well plate overnight with DEME medium and 10% FBS; three concentrations of delivery agents (1:1, 2:1 and 3:1 w/w for L-PEI and B-PEI; 2.5:1, 3.5:1 and 5:1 w/w for LGA-PEI polymer) and 0.2 μg GFP plasmid DNA and opti-MEM medium were added to each well and cultured for 24 h. Green florescence signal as transfection efficiency was examined under a fluorescence microscope, and cellular density as potential cytotoxicity was evaluated with phase contrast microscopy analysis. LGA-PEI polymers showed higher transfection efficiency and cell density than both L-PEI and B-PEI delivery agents at all three concentrations. B-PEI had slightly higher transfection efficiency than L-PEI at two concentrations (2:1 and 3:1 w/w). Both L-PEI and B-PEI showed cytotoxicity at 2:1 and 3:1 (w/w) concentrations as the cell density was reduced (Figure 7A). In addition, we performed a similar transfection experiment in pancreatic cancer cell line PANC-1 cells (48 h). Although three delivery agents had similar transfection efficiency, LGA-PEI showed a higher cell density, indicating much less cytotoxicity than L-PEI and B-PEI at the comparable condition (Figure 7B). Furthermore, we performed the MTT assay for PANC-1 cells to further confirm the cytotoxicity of three delivery agents with identical transfection conditions. LGA-PEI polymer did not show any cytotoxicity at the (3.5:1 w/w) concentration as compared with the negative control. However, L-PEI and B-PEI at the (2:1 w/w) concentration significantly reduced cell viability by 42 and 56%, respectively, as compared with the control (p < 0.05, n = 6) (Figure 7C). Thus, LGA-PEI polymer is superior to both L-PEI and B-PEI delivery agents in these cell transfection studies.
Figure 7. . Comparison of gene delivery efficiency and cytotoxicity of L-PEI (25 kDa), B-PEI (25 kDa) and LGA-PEI polymer in HEK293 and PANC-1 cells.
(A) Delivering GFP containing plasmid to HEK293 cells. Cells were seeded onto the 96-well plate overnight. Three concentrations of delivery agents (1:1, 2:1 and 3:1 w/w for L-PEI and B-PEI; 2.5:1, 3.5:1 and 5:1 w/w for LGA-PEI polymer) and 0.2 μg GFP plasmid DNA/well were used for transfection for 24 h. Green florescence signal as transfection efficiency was examined under a fluorescence microscope, and cellular density as potential cytotoxicity was evaluated with phase contrast microscopy analysis. (B) Delivering GFP containing plasmid to PANC-1 cells. L-PEI (2:1 w/w), B-PEI (2:1 w/w) or LGA-PEI (3.5:1 w/w) and 0.2 μg GFP plasmid DNA/well were used for transfection in the 96-well plate for 48 h. Green florescence signal and cellular density were recorded. (C) Cell viability after gene delivering in PANC-1 cells. L-PEI (2:1 w/w), B-PEI (2:1 w/w) or LGA-PEI (3.5:1 w/w) and 0.2 μg GFP plasmid DNA/well were used for transfection in the 96-well plate for 48 h. Cell viability was determined by MTT assay.
*p < 0.05, n = 6.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
In vivo toxicity & DNA delivery
Lipofectamine 2000 is not recommended for in vivo use because it is toxic to red blood cells and other cells. Only some liposome formulations and PEI can be used in vivo, but their toxicity is well recognized [3,25]. In this study, Balb/c mice received iv. injection of B-PEI (25 kDa) at doses of 8, 6.5 and 6 mg/kg body weight died, while the mice received B-PEI at 5.5 and 5 mg/kg survived. Balb/c mice injected with LGA-PEI (0.5:1 w/w) polymers at doses of 29, 26, 23, 22 and 21 mg/kg body weight died, while the mouse received LGA-PEI at 20.5 mg/kg survived (Supplementary Table 1). Balb/c mice showed a maximum tolerate dose of B-PEI (25 kDa) of about 5 mg/kg. By contrast, the LGA-PEI (0.5:1 w/w) polymer showed a maximum tolerate dose of about 20 mg/kg, which corresponds to a PEI dose of 12 mg/kg. For plasmid DNA delivery, all mice (n = 3) died after they were injected via tail vein with PEI (75 μg)/DNA (50 μg) complexes (1.5:1 w/w; 9.4 N/P). On the other hand, all mice (n = 3) injected with LGA-PEI (125 μg)/DNA (50 μg) (2.5:1 w/w; 10 N/P) NPs survived the treatment. Furthermore, all mice (n = 4) that received LGA-PEI (250 μg)/DNA (100 μg) NPs survived, and half the number of mice (n = 4) that received LGA-PEI (500 μg)/DNA (200 μg) NPs survived (Table 3). Thus, LGA-PEI and LGA-PEI/DNA NPs are much less toxic in vivo than PEI and PEI/DNA complexes.
Table 3. . Survival rate of mice after tail vein injection of PEI/DNA or LGA-PEI/DNA nanoparticles.
| PEI/DNA delivery† (n = 3) (μg DNA) | Survival rate (%) | LGA-PEI/DNA (2.5:1 w/w) delivery (n = 4) (μg DNA) | Survival rate (%) |
|---|---|---|---|
| 30 |
100 |
30 |
100 |
| 50 |
0 |
50 |
100 |
| 100 |
- |
100 |
100 |
| 200 | - | 200 | 50 |
†LGA-PEI (0.5:1 w/w) polymer was used.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
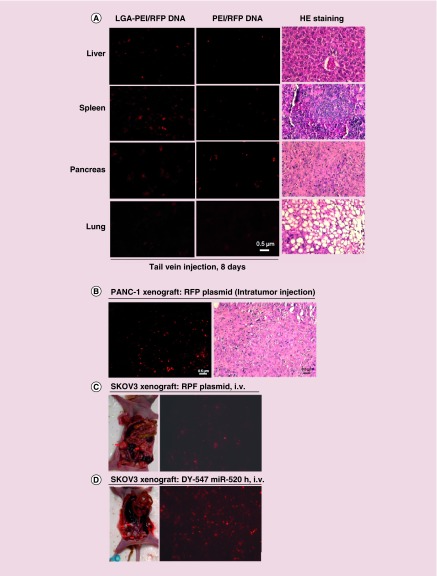
The LGA-PEI polymer was used to deliver RFP plasmid DNA to Balb/c mice via tail vein injection to study the DNA transfection efficiency in mouse organs and to compare it with that of PEI/DNA delivery. Plasmid DNA (30 μg) with either LGA-PEI polymer or B-PEI (25 kDa) was injected into mice three-times in 5 days, then the mice were sacrificed on day 8 (n = 3) (Figure 8A) and day 19 (n = 3) (Supplementary Figure 12), respectively. In LGA-PEI/DNA NPs-treated mice, the liver, spleen and pancreas showed strong red fluorescence, while the heart, lungs and kidneys showed no red fluorescence. PEI/DNA-treated mice showed only weak red fluorescence in liver, spleen and pancreas (Figure 8A). Thus, the LGA-PEI delivery system is more efficient than the PEI-based delivery system in vivo. In addition, nude mice (n = 3) were injected subcutaneously with a pancreatic cancer cell line (AsPC-1). When the tumors were established after 2 weeks, LGA-PEI (25 μg)/RFP DNA (10 μg) was directly injected into the tumor once and the tissue harvested 3 days later. We observed strong red fluorescence in the tumors indicating high DNA transfection inside the tumor tissue (Figure 8B). We also studied the systemic delivery of RFP plasmid DNA and miRNA mimic into ovarian cancer in a xenograft mouse model. Ovarian cancer cell line (SKOV3) was orthotopically injected into bilateral mouse ovaries and the tumor was established after 5 weeks. LGA-PEI (75 μg)/RFP DNA (30 μg) NPs were injected into each mouse (n = 5) via tail vein three-times in 5 days. Five days after the last injection, the mice were sacrificed for analysis; red florescence signal was readily detected in the xenograft tissues (Figure 8C). In a separate experiment, LGA-PEI (37.5 μg)/miR-520h mimic with DY-547 tag (15 μg) NPs were injected into each of the SKOV3 xenografted mice (n = 3) via tail vein once. Five days later, the mice were sacrificed for analysis; red fluorescence was also observed in ovarian cancer tissues (Figure 8D). Thus, the LGA-PEI polymer is an effective system to deliver plasmid DNA or RNA into tumors in the mouse xenograft model.
Figure 8. . Delivery efficiency of LGA-PEI polymer/nucleic acids into mouse models.
LGA-PEI (0.5:1 w/w) polymer was used. (A) Organ distribution. Plasmid DNA (30 μg), with either LGA-PEI polymer or PEI, was injected via tail vein into mice three-times during 5 days; mice were sacrificed on day 8 (n = 3). Fluorescence microscope imaging was performed for liver, spleen, pancreas and lungs. H&E staining for these organs was performed for observation of the tissue structure, which is normal. (B) Pancreatic xenograft tumor in a nude mouse model. Nude mice (n = 3) were injected subcutaneously with pancreatic cancer cell line AsPC-1 for 2 weeks. LGA-PEI/RFP DNA (10 μg) NPs were directly injected into the tumor. Three days later, the tumors were harvested and the red fluorescence analyzed. H&E staining showed xenograft cellular morphology. (C) Human ovarian cancer cell line (SKOV3) was orthotopically injected into the bilateral ovaries of female nude mice (n = 5). When xenograft tumors developed in about 5 weeks, LGA-PEI/RFP DNA (30 μg) NPs were injected via tail vein into the nude mice three-times in 5 days. Five days later, the tumors were harvested and the red fluorescence analyzed. (D) Orthotopic SKOV3 tumors in nude mice (n = 3) were established in 5 weeks, then LGA-PEI/miR-520h mimic (DY-547 tag, 15 μg) NPs were injected into the mouse via tail vein once. Five days later, the tumors were harvested and the red fluorescence was analyzed.
LGA-PEI: Lactic-co-glycolic acid-modified polyethylenimine; PEI: Polyethylenimine.
Discussion
In the current study, we have proved our hypothesis that the properties of B-PEI (25 kDa) could be improved by covalently linking it to LGA single unites for nucleic acid delivery. The mechanism of the reaction between PLGA and PEI is via nucleophilic attack by the primary amine of PEI on the carbonyl carbon of PLGA to form a positively charged tetrahedral intermediate, followed by the removal of an alcohol group (RC-O-) [63]. This reaction is favorable in organic solvent because the overall activation energy of the aminolysis reaction is often negative [63,64]. Under current experimental conditions, PLGA is completely broken down into LGA single units by PEI and forms a new LGA-PEI (0.5:1 w/w) polymer. Based on the number of ester bonds, the PEI content and the primary amine percentage, we estimate that each LGA-PEI polymer contains about 109 LGA single units and 1 PEI molecule (Supplementary Figure 13A). To the best of our knowledge, LGA monomer or LGA ester is not commercially available. Theoretically, monomer LGA ester can be coupled to B-PEI (25 kDa), producing a similar LGA-PEI polymer that is obtained by the reaction between PLGA and PEI as shown in the current study. Simple lactate ester or glycolide ester can also react with primary amines of PEI to form lactate-PEI or glycolide-PEI polymers, but not LGA-PEI polymers.
This LGA-PEI polymer is fundamentally different from other PLGA and PEI preparations previously reported in the literature. Several studies reported preparing PLGA NPs bearing PEI on their surface by mixing first the PLGA NP with the water-in-oil-in-water technique in PVI solution, and then adding PEI into the solution to form a PEI-coated PLGA NPs [44,54,65–71]. In these studies, PLGA NPs are the core, and PEI attached to the surface makes the NPs cationic. All the methods reported in the literature used less than 15% PEI to prepare PLGA-PEI and less than 2 h reaction time. In our study, however, PLGA molecules are fragmented to LGA single unites by PEI and these LGA units are covalently conjugated with PEI through the amide linkage forming completely a new LGA-PEI polymer (0.5:1 w/w and 48 h reaction time), which contains 67% PEI. In fact, intact PLGA no longer exists in our current LGA-PEI polymer. Two important reaction factors that determine the structure difference of our LGA-PEI polymer with other polymers that include PLGA and PEI are the reaction time of PLGA with PEI and the PLGA to PEI ratio. Thus, our LGA-PEI polymer is a new material, which has differences in chemical structure, properties, functions and potential applications from those reported in literature [44,54,65–71].
With one simple step reaction, our LGA-PEI polymer can load DNA completely at a low polymer/DNA ratio of 1:1 w/w (4 N/P), which corresponds to a loading capacity of 1 μg polymer loading 1 μg DNA. This indicates that LGA modification does not change the loading efficiency and capacity of PEI. LGA-PEI polymers condense DNA molecules to form a particle. According to our estimation, the LGA-PEI/DNA weight ratio of 1.5:1 (6 N/P) corresponds to a LGA-PEI to DNA molecular ratio of about 180:1. At this ratio LGA-PEI polymers may form particles that have several plasmid DNA molecules between 150 and 230 nm in size. In the case of the higher LGA-PEI/DNA weight ratio, such as 3.5:1 (14 N/P), more LGA-PEI polymer molecules may separate DNA molecules and form particles with fewer DNA molecules than in the case of the polymer/DNA weight ratio of 1.5:1 (6 N/P). Thus, LGA-PEI polymers can further condense DNA molecule(s) to particles as small as 50 nm. We believe this explains our observation that the higher the polymer/DNA ratio is, the smaller the particle sizes are, as determined by SEM. Based on the calculation of the molecular weight of LGA-PEI polymer (42 kDa), plasmid DNA (5 kb, 3300 kDa), and size as well as volume of the NPs, we can estimate the contents of different NPs. Smaller particles (36 nm) formed with 35 μg LGA-PEI/10 μg DNA may contain 1 plasmid DNA molecule and 275 LGA-PEI polymers (Supplementary Figure 13B). The larger particles (100 nm) formed with 25 μg LGA-PEI and 10 μg DNA may contain about 15 plasmid DNA molecules and 2955 LGA-PEI polymers (Supplementary Figure 13C).
The LGA-PEI polymer allows for sufficient DNA binding through electrostatic forces between the amines of PEI and the phosphate DNA backbones, but the LGA units might be spared and form a barrier that may shield the PEI/DNA complex from nonspecific interactions. Indeed, we found that the LGA-modified PEI significantly changed some properties of PEI, for example, it lowered the zeta potential from 60 mV (of PEI/DNA) to 20–30 mV (of LGA-PEI/DNA). LGA-PEI polymer may shield nucleic acids better than PEI. Interestingly, the LGA-PEI polymer can condense and deliver a large artificial chromosome DNA (∼200 kb) into cells. It is also a better DNA delivery system for difficult-to-transfect cells, such as THP-1 cells and Jurkat cells in suspension and HUVECs, than commercial delivery reagents such as lipofectamine 2000. In addition, we showed that LGA-PEI polymer could be used for primary fibroblasts and stem cells. Thus, the LGA-PEI polymer condenses the DNA effectively, controls particle size and may have broad applications for gene delivery in different types of cells.
The LGA-modified PEI significantly decreased the cytotoxicity of PEI/DNA in vitro. In PANC-1 cells, LGA-PEI/DNA showed either no or minor toxicity at LGA-PEI to DNA weight ratios up to 3:1 (N/P 12) when compared with untreated cells, while PEI/DNA complexes at N/P 12 killed half of the cells. Most importantly, LGA modification remarkably reduced PEI-associated toxicity in the mouse model. The maximum tolerated dose of LGA-PEI (20 mg/kg) was much higher than that of PEI (5 mg/kg). Furthermore, the LGA-PEI polymer could deliver up to 200 μg DNA to mice without killing them; in contrast, all mice died when PEI delivered 50 μg DNA. Having lower toxicity is one of the major advantages of LGA-PEI polymers for in vitro and in vivo applications of gene delivery. It is known that the transfection efficiency of PEI/DNA increases with increased molecular weight of PEI, while high molecular weight PEI alone results in higher cytotoxicity [72]. The high toxicity of PEI also depends on the polydispersity, and the surface charge density of the polymers. Fischer et al. observed that the higher cytotoxicity of high molecular weight PEI was caused by high affinity binding to the cell surface which resulted in massive necrosis [15]. In vivo, high molecular weight PEI may accumulate due to either a lack of a degradation pathway or a mechanism of excretion for such molecules [40]. Our LGA-PEI polymer is less toxic than the unmodified PEI. Further study is necessary to investigate the underline mechanism of less toxic LGA-PEI polymers.
In the current study, we did not study the properties of lipofectamine 2000 with static light-scattering technology and other technologies. Lipofectamine 2000 was used as a positive control of gene transfection experiments in vitro. Since LGA-PEI polymer has completely different chemical structures and mechanisms of nucleic acid delivery with lipofectamine 2000, we only compare its delivery efficiency and cellular toxicity with the LGA-PEI polymer. We did not determine the optimal transfection conditions of lipofectamine 2000 for all cell lines used in this study. It is well known that different cell types may have different optimal vector/DNA ratios for effective transfection. Based on the instructions provided from the company, we only used 4 μl lipofectamine 2000 per 1 μg DNA or RNA for standard transfection experiments. Thus, comparison of the transfection efficiency between LGA-PEI and lipofectamine 2000 in the current study may have limitations to draw solid conclusions. High efficiency of the delivery of double stranded RNAs into cells or tissues shown in the current study does not necessarily mean a high functional efficiency of these RNAs; functions of these therapeutic RNAs are also dependent on cellular process such as RNA release from polymers and their stability in cytoplasm. In the study, we did not examine the functions of these delivered RNAs. More experiments are warranted to address these important issues.
Conclusion
We have designed, synthesized and characterized a new LGA-modified PEI polymer (42.6 kDa), which can self-assemble into NPs and deliver nucleic acids into cells in both cell culture and animal models. The LGA-PEI polymer has higher nucleic acid loading capacity and lower toxicity than PEI, and it condenses the nucleic acids into small-sized NPs. The LGA-PEI/DNA or RNA NPs have high transfection efficiency in PANC-1 cells and effectively deliver DNA into difficult-to-transfect cells and organs or tumors in animal models. Thus, new LGA-PEI polymer as an improved delivery system for nucleic acids could have broad clinical applications of gene or nucleotide therapies for cancer and many other diseases.
Executive summary.
Current gene delivery systems are less successful for clinical applications because they often have low efficacy and high toxicity.
We report the development of an improved delivery system for nucleic acids. We have designed, synthesized and characterized a new polymer of lactic-co-glycolic acid-modified polyethylenimine (LGA-PEI), which spontaneously forms nanoparticles (NPs) with nucleic acids.
These LGA-PEI/plasmid DNA NPs showed higher DNA loading efficiency, higher DNA transfection efficacy and lower cytotoxicity in several cell types including PANC-1 and Jurkat cells, when compared with those NPs formed of lipofectamine 2000 and PEI.
This LGA-PEI polymer effectively delivered plasmid DNA, modified mRNA, miRNA and large artificial chromosome DNA (∼200 kb) into cells.
In nude mouse models, LGA-PEI polymer effectively delivered plasmid DNA or miRNA mimic into pancreatic and ovarian xenograft tumors.
In mouse models, LGA-PEI/DNA NPs showed higher delivery efficiency into major organs, such as liver, spleen and pancreas, and much lower toxicity than control PEI NPs.
Supplementary Material
Acknowledgements
Both miR-520h mimic and miR-520h plasmid DNA containing RFP were provided by M Anderson (Baylor College of Medicine). Artificially modified GFP mRNA (mmRNA) was provided by E Yakubov (Houston Methodist Research Institute). A modified mouse artificial chromosome DNA containing a GFP gene (∼200 kb) was provided by J Yang (Baylor College of Medicine). The authors would like to thank V Zaitsev, Assistant Professor at Department of Chemistry, University of Houston, for his assistance in measuring the FT-IR and NMR spectroscopy of polymers and H Wang and X Xu (Baylor College of Medicine) for their assistance in the experiments of primary mouse embryo fibroblasts and mouse embryo stem cells. The authors would also like to thank AM Rodríguez, PhD, a member of the Michael E. DeBakey Department of Surgery Research Core Team, for her editorial assistance during the preparation of this manuscript.
Footnotes
Financial & competing interests disclosure
This study was partially supported by the Michael E DeBakey Department of Surgery, by an Alkek Award from Baylor College of Medicine, and by a grant (R01 CA183984, Q Yao) from the NIH. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Wei Y, Gong C. Polymeric nanocarriers for non-viral gene delivery. J. Biomed. Nanotechnol. 2015;11:739–770. doi: 10.1166/jbn.2015.2069. [DOI] [PubMed] [Google Scholar]
- 4.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15(8):541–55. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 5.Ohana P, Gofrit O, Ayesh S, et al. Regulatory sequences of the H19 gene in DNA based therapy of bladder cancer. Gene Ther. Mol. Biol. 2004;8:181–192. [Google Scholar]
- 6.Lisziewicz J, Trocio J, Whitman L, et al. DermaVir: a novel topical vaccine for HIV/AIDS. J. Invest. Dermat. 2005;124:160–169. doi: 10.1111/j.0022-202X.2004.23535.x. [DOI] [PubMed] [Google Scholar]
- 7.Lisziewicz J, Trocio J, Xu J, et al. Control of viral rebound through therapeutic immunization with DermaVir. AIDS. 2005;19:35–43. doi: 10.1097/00002030-200501030-00004. [DOI] [PubMed] [Google Scholar]
- 8.Basarkar A, Devineni D, Palaniappan R, Singh J. Preparation, characterization, cytotoxicity and transfection efficiency of poly(dl-lactide-co-glycolide) and poly(dl-lactic acid) cationic nanoparticles for controlled delivery of plasmid DNA. Int. J. Pharm. 2007;343:247–254. doi: 10.1016/j.ijpharm.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumakova OV, Liopo AV, Andreev VG, et al. Composition of PLGA and PEI/DNA nanoparticles improves ultrasound-mediated gene delivery in solid tumors in vivo . Cancer Lett. 2008;261:215–225. doi: 10.1016/j.canlet.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Boussif O, Lezoualc'h F, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J. Control Release. 1990;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami S, Ito Y, Charoensit P, Yamashita F, Hashida M. Evaluation of proinflammatory cytokine production induced by linear and branched polyethylenimine/plasmid DNA complexes in mice. J. Pharmacol. Exp. Ther. 2006;3(317):1382–1390. doi: 10.1124/jpet.105.100669. [DOI] [PubMed] [Google Scholar]
- 13.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol. Ther. 2005;6(11):990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Merkel OM, Urbanics R, Bedocs P, et al. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;21(32):4936–4942. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Fischer D, Bieber T, Li YX, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999;8(16):1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 16.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv. Drug Deliv. Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A, Moghimi SM. Low and high molecular weight poly(L-lysine)s/poly(L-lysine)-DNA complexes initiate mitochondrial-mediated apoptosis differently. FEBS Lett. 2005;579:6191–6198. doi: 10.1016/j.febslet.2005.09.092. [DOI] [PubMed] [Google Scholar]
- 18.Lin CW, Jan MS, Kuo JHS, Hsu LJ, Lin YS. Protective role of autophagy in branched polyethylenimine (25K)- and poly(L-lysine) (30–70K)-induced cell death. Eur. J. Pharm. Sci. 2012;47:865–874. doi: 10.1016/j.ejps.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Grandinetti G, Ingle NP, Reineke TM. Interaction of poly(ethylenimine)–DNA polyplexes with mitochondria: implications for a mechanism of cytotoxicity. Mol. Pharm. 2011;8:1709–1719. doi: 10.1021/mp200078n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao XL, Yao L, Song QX, et al. The association of autophagy with polyethylenimine-induced cytotoxity in nephritic and hepatic cell lines. Biomaterials. 2011;32:8613–8625. doi: 10.1016/j.biomaterials.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 21.Hall A, Larsen AK, Parhamifar L, Meyle KD, Wu LP, Moghimi SM. High resolution respirometry analysis of polyethylenimine-mediated mitochondrial energy crisis and cellular stress: mitochondrial proton leak and inhibition of the electron transport system. Biochim. Biophys. Acta. 2013;1827(10):1213–1225. doi: 10.1016/j.bbabio.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Wightman L, Kircheis R, Rössler V, et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo . J. Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 23.Zou AM, Erbacher P, Remy JS, Behr JP. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. J. Gene Med. 2000;2:128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Huha SH, Dob HJ, Lima HY, et al. Optimization of 25 kDa linear polyethylenimine for efficient gene delivery. Biologicals. 2007;35(3):165–171. doi: 10.1016/j.biologicals.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo . J. Control Release. 2008;130(2):129–138. doi: 10.1016/j.jconrel.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Yadava P, Hughes J. Polyethylenimine strategies for plasmid delivery to brain-derived cells. Methods. 2004;33(2):144–150. doi: 10.1016/j.ymeth.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs, and cationic PAMAM dendrimers in B16f10 melanoma cells. J. Control Release. 2007;117(3):291–300. doi: 10.1016/j.jconrel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Biophysical characterization of PEI/DNA complexes. J. Pharm. Sci. 2003;92(8):1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 29.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo . Gene Ther. 2003;10(19):1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 30.Rettig GR, McAnuff M, Liu DJ, Kim JS, Rice KG. Quantitative bioluminescence imaging of transgene expression in vivo . Anal. Biochem. 2003;355(1):90–94. doi: 10.1016/j.ab.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethyleniminemediated siRNA delivery in vitro . Pharm. Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 32.Goula D, Remy JS, Erbacher P, et al. Size, diffusibility and transfection performance of linear PEI/DNA complexes in the mouse central nervous system. Gene Ther. 1998;5(5):712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 33.Goula D, Becker N, Lemkine GF, et al. Rapid crossing of the pulmonary endothelial barrier by polyethylenimine/DNA complexes. Gene Ther. 2000;7(6):499–504. doi: 10.1038/sj.gt.3301113. [DOI] [PubMed] [Google Scholar]
- 34.Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 1998;5(9):1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 35.Bragonzi A, Boletta A, Biffi A, et al. Comparison between cationic polymers and lipids in mediating systemic gene delivery to the lungs. Gene Ther. 1999;6(12):1995–2004. doi: 10.1038/sj.gt.3301039. [DOI] [PubMed] [Google Scholar]
- 36.Lampela P, Räisänen J, Männistö PT, Ylä-Herttuala S, Raasmaja A. The use of low-molecular-weight PEIs as gene carriers in the monkey fibroblastoma and rabbit smooth muscle cell cultures. J. Gene Med. 2002;4(2):205–214. doi: 10.1002/jgm.245. [DOI] [PubMed] [Google Scholar]
- 37.Grosse S, Thévenot G, Aron Y, et al. In vivo gene delivery in the mouse lung with lactosylated polyethylenimine, questioning the relevance of in vitro experiments. J. Control Release. 2008;132(2):105–112. doi: 10.1016/j.jconrel.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Choi S, Lee KD. Enhanced gene delivery using disulfide-crosslinked low molecular weight polyethylenimine with listeriolysin o-polyethylenimine disulfide conjugate. J. Control Release. 2008;131(1):70–76. doi: 10.1016/j.jconrel.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Jere D, Jiang HL, Arote R, et al. Degradable polyethylenimines as DNA and small interfering RNA carriers. Expert Opin. Drug Deliv. 2009;6(8):827–34. doi: 10.1517/17425240903029183. [DOI] [PubMed] [Google Scholar]
- 41.Lungu CN, Diudea MV, Putz MV, Grudziński IP. Linear and branched PEIs (polyethylenimines) and their property apace. Int. J. Mol. Sci. 2016;17(4):555. doi: 10.3390/ijms17040555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas M, Klibanov AM. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortune JA, Novobrantseva TI, Klibanov AM. Highly effective gene transfection in vivo by alkylated polyethylenimine. J. Drug Deliv. 2011;2011:204058. doi: 10.1155/2011/204058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shau MD, Shih MF, Lin CC, et al. A one-step process in preparation of cationic nanoparticles with poly(lactide-co-glycolide)-containing polyethylenimine gives efficient gene delivery. Eur. J. Pharm. Sci. 2013;5(46):522–529. doi: 10.1016/j.ejps.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Zeng P, Xu Y, Zeng CH, Ren H, Peng ML. Chitosan-modified poly(d, l-lactide-co-glycolide) nanospheres for plasmid DNA delivery and HBV gene-silencing. Int. J. Pharm. 2011;415(1–2):259–266. doi: 10.1016/j.ijpharm.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 46.Bergen JM, Pun SH. Analysis of the intracellular barriers encountered by nonviral gene carriers in a model of spatially controlled delivery to neurons. J. Gene Med. 2008;10(2):187–197. doi: 10.1002/jgm.1137. [DOI] [PubMed] [Google Scholar]
- 47.Moreno MR, Giudici M, Villalain J. The membranotropic regions of the endo and ecto domains of HIV gp41 envelope glycoprotein. Biochim. Biophys. Acta. 2006;1758(1):111–123. doi: 10.1016/j.bbamem.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Kwon EJ, Bergen JM, Park IK, Pun SH. Peptide-modified vectors for nucleic acid delivery to neurons. J. Control Release. 2008;132(3):230–235. doi: 10.1016/j.jconrel.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Creusat G, Zuber G. Self-assembling polyethylenimine derivatives mediate efficient siRNA delivery in mammalian cells. Chembiochem. 2008;9:2787–2789. doi: 10.1002/cbic.200800540. [DOI] [PubMed] [Google Scholar]
- 50.De Laporte L, Rea JC, Shea LD. Design of modular non-viral gene therapy vectors. Biomaterials. 2006;7(27):947–954. doi: 10.1016/j.biomaterials.2005.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown MD, Schatzlein AG, Uchegbu IF. Gene delivery with synthetic (non-viral) carriers. Int. J. Pharm. 2001;229(1–2):1–21. doi: 10.1016/s0378-5173(01)00861-4. [DOI] [PubMed] [Google Scholar]
- 52.Lu JM, Wang X, Marin-Muller C, et al. Current advances in research and clinical applications of PLGA-based nanotechnology. Expert Rev. Mol. Diagn. 2009;9:325–341. doi: 10.1586/erm.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J. Control Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 54.Lim HJ, Kim JK, Park JS. Complexation of poptotic genes with polyethyleneimine (PEI)-coated poly-(DL)-lactic-co-glycolic acid nanoparticles for cancer cell apoptosis. J. Biomed. Nanotechnol. 2015;11:211–225. doi: 10.1166/jbn.2015.1880. [DOI] [PubMed] [Google Scholar]
- 55.von Harpe A, Petersen H, Li Y, Kissel T. Characterization of commercially available and synthesized polyethylenimines for gene delivery. J. Control Release. 2000;69:309–322. doi: 10.1016/s0168-3659(00)00317-5. [DOI] [PubMed] [Google Scholar]; •• Describes the method to determine the polyethylenimine (PEI) amount in the lactic-co-glycolic acid-modified polyethylenimine (LGA-PEI) polymer.
- 56.Bullock J, Chowdhury S, Severdia A, Sweeney J, Johnston D, Pachla L. Comparison of results of various methods used to determine the extent of modification of methoxy polyethylene glycol 5000-modified bovine cupri-zinc superoxide dismutase. Anal. Biochem. 1997;254:254–262. doi: 10.1006/abio.1997.2405. [DOI] [PubMed] [Google Scholar]; •• Describes the method to determine the primary amine content of the LGA-PEI polymer.
- 57.Ekhorutomwen SA, Sawan SP, Smith BF, Robison TW, Wilson KV. Solution behavior of modified polyethylenimine (PEI) polymers by light scattering investigations. 2004. www.osti.gov/scitech/servlets/purl/822405/; •• Describes the method to determine the molecular mass of PEI LGA-PEI (0.5:1 w/w) polymer.
- 58.Sribenja S, Sawanyawisuth K, Kraiklang R, et al. Suppression of thymosin beta10 increases cell migration and metastasis of cholangiocarcinoma. BMC Cancer. 2013;13:430. doi: 10.1186/1471-2407-13-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Hendricks W, Liu G, et al. A nanoparticle formulation that selectively transfects metastatic tumors in mice. Proc. Natl Acad. Sci. USA. 2013;110:14717–14722. doi: 10.1073/pnas.1313330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erbetta CDC, Alves RJ, Resende JM, de Souza Freitas Roberto F, de Sousa RG. Synthesis and characterization of poly(D,L-lactide-co-glycolide) copolymer. J. Biomat. Nanobiotech. 2012;3:208–225. [Google Scholar]; • Provides information to understand chemical reaction and structure of LGA-PEI polymer by NMR spectroscopy analysis.
- 61.Smith JG. Organic Chemistry (3rd Edition) McGraw-Hill; 2011. [Google Scholar]; • Provides information to measure amide RCONH proton of the LGA-PEI polymer by NMR spectroscopy analysis.
- 62.Wang YQ, Su J, Wu F, et al. Biscarbamate cross-linked polyethylenimine derivative with low molecular weight, low cytotoxicity, and high efficiency for gene delivery. Int. J. Nanomedicine. 2012;7:693–704. doi: 10.2147/IJN.S27849. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides information to understand chemical structure of the LGA-PEI polymer by NMR spectroscopy analysis.
- 63.Croll TI, O'Connor AJ, Stevens GW, Cooper-White JJ. Controllable surface modification of poly(lactic-co-glycolic acid) (PLGA) by hydrolysis or aminolysis I: physical, chemical, and theoretical aspects. Biomacromolecules. 2004;5:463–473. doi: 10.1021/bm0343040. [DOI] [PubMed] [Google Scholar]; • Provides information to understand chemical reaction between PLGA and branched PEI.
- 64.Tuulmets A, Talvik AT. Activation energies of aminolysis of aliphatic esters in aprotic media. Ach. Models Chem. 2000;137:111–119. [Google Scholar]; • Provides information to understand chemical reaction between PLGA and branched PEI.
- 65.Bivas-Benita M, Romeijn S, Junginger HE, Borchard G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. Eur. J. Pharm. Biopharm. 2004;58:1–6. doi: 10.1016/j.ejpb.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gargouri M, Sapin A, Arica-Yegin B, Merlin JL, Becuwe P, Maincent P. Photochemical internalization for pDNA transfection: evaluation of poly(d, l-lactide-co-glycolide) and poly(ethylenimine) nanoparticles. Int. J. Pharm. 2011;403:276–284. doi: 10.1016/j.ijpharm.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 67.Nam YS, Kang HS, Park JY, Park TG, Han SH, Chang IS. New micelle-like polymer aggregates made from PEI-PLGA diblock copolymers: micellar characteristics and cellular uptake. Biomaterials. 2003;24:2053–2059. doi: 10.1016/s0142-9612(02)00641-5. [DOI] [PubMed] [Google Scholar]
- 68.Katas H, Cevher E, Alpar HO. Preparation of polyethyleneimine incorporated poly(D,L-lactide-co-glycolide) nanoparticles by spontaneous emulsion diffusion method for small interfering RNA delivery. Int. J. Pharm. 2009;369:144–154. doi: 10.1016/j.ijpharm.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 69.Son S, Kim WJ. Biodegradable nanoparticles modified by branched polyethylenimine for plasmid DNA delivery. Biomaterials. 2010;31:133–43. doi: 10.1016/j.biomaterials.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 70.Bivas-Benita M, Lin MY, Bal SM, et al. Pulmonary delivery of DNA encoding Mycobacterium tuberculosis latency antigen Rv1733c associated to PLGA-PEI nanoparticles enhances T cell responses in a DNA prime/protein boost vaccination regimen in mice. Vaccine. 2009;27:4010–4017. doi: 10.1016/j.vaccine.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 71.Wolfert MA, Dash PR, Nazarova O, et al. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug. Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- 72.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.