Abstract
Imaging mass spectrometry (IMS) has emerged as a powerful analytical tool enabling the direct molecular mapping of many types of tissue. Specifically, matrix-assisted laser desorption/ ionization (MALDI) represents one of the most broadly applicable IMS technologies. In recent years, advances in solid state laser technology, mass spectrometry instrumentation, computer technology, and experimental methodology have produced IMS systems capable of unprecedented data acquisition speeds (>50 pixels/second). In applications of this technology, throughput is an important consideration when designing an IMS experiment. As IMS becomes more widely adopted, continual improvements in experimental setups will be important to address biologically and clinically relevant time scales.
Keywords: Imaging mass spectrometry, MALDI, high-speed imaging, TOF
Introduction
Mass spectrometry (MS) has long been recognized for its high sensitivity, high throughput, and molecular specificity. Advances in MS instrumentation have enabled the use of MS technology as an imaging modality.1–2 Specifically, matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS), offers an untargeted approach to the regiospecific measurement of molecules in tissue specimens. The untargeted nature of this technology provides for the ability to simultaneously measure the complex array of molecular species present in biological tissue, producing molecular maps that can be correlated with anatomical tissue features without any required prior knowledge of the analytes present and without using any specialized reagents (e.g., antibodies). MALDI IMS analyses have been successfully applied to the study of eye physiology,3–7 neurology,8–9 skin cancer,10–11 pancreatic cancer,12 and breast cancer.13–14
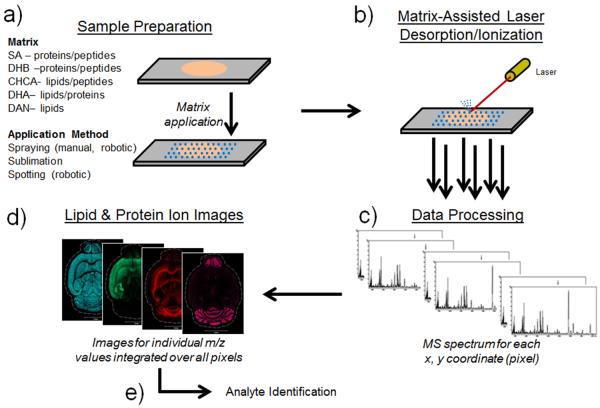
In a typical MALDI IMS experiment, a thinly sectioned tissue specimen is mounted onto a flat target, such as a microscope slide, and then coated with a MALDI matrix (Figure 1). The MALDI matrix is typically a small organic molecule with strong absorbance at the wavelength of the MALDI laser and is applied to the sample in a manner that preserves the spatial integrities of the analytes of interest (e.g., metabolites, lipids, peptides, proteins). A raster of the tissue surface is performed, generating a mass spectrum at each x, y coordinate (i.e., a single pixel in `microprobe' imaging mode).1, 15 Ion intensity maps can be constructed as a function of x, y position across the tissue surface for any ion of interest. Ions can then be identified through one or a combination of several techniques, including exact mass measurements,16–17 on-tissue tandem mass spectrometry (MS/MS),18–23 or off-tissue liquid chromatography-tandem mass spectrometry (LC-MS/MS).24
Figure 1. IMS workflow.
a) Specimens are prepared for analysis by mounting thinly cut tissue sections onto slides. Matrix application is then performed via any number of methods prior to b) MALDI analysis. c) Mass spectra generated at each x, y position are then used to d) construct intensity map images for any single ion of interest. e) Analyte identification can be performed by one or a combination of several techniques.
Early MALDI IMS experiments required 1–2 minutes/pixel for data acquisition, often requiring total analysis times of many hours or even days.1 However, recent advances in laser technology,25–27 data acquisition, and scanning methodology18, 23, 28–29 have provided for mass spectrometry systems which are capable of scanning at speeds as high as 50 pixels/second. The increasing use of IMS technology in many biological and clinical applications necessitates that continuing improvements in experimental times be achieved in order to perform IMS investigations on reasonable time scales.
Experimental
Transverse and coronal rat brain (Pel-Freez Biologicals, Rogers, AR) specimens were cryosectioned at 10 μm and thaw-mounted onto indium-tin oxide (ITO) slides. A custom built sublimation apparatus was used to apply a 1,5-diaminonapthalene (DAN, Sigma Aldrich, St. Louis, MO) matrix layer (110° C, 8 min, ~50 mtorr).30–31 IMS experiments were performed on either a SimulTOF 300 Tandem MALDI tandem time of flight (TOF/TOF) MS (SimulTOF Systems, Sudbury, MA) in positive ion reflectron MS mode or a RapifleX MALDI Tissuetyper TOF MS (Bruker Daltonics, Billerica, MA) in negative ion reflectron mode. The SimulTOF instrument is equipped with a 349 nm, diode-pumped, frequency-tripled Nd:YLF laser capable of laser repetition rates up to 5 kHz. This system employs continuous laser raster sampling and was scanned using a 1 mm/second stage speed, 1 kHz laser frequency, and 50 hardware averaged shots per spectrum.23 The RapifleX instrument is equipped with a Smartbeam 3D 10 kHz 355 nm Nd:YAG laser and was set to 200 shots per pixel using a single shot laser configuration and 85% focus setting.24
Results
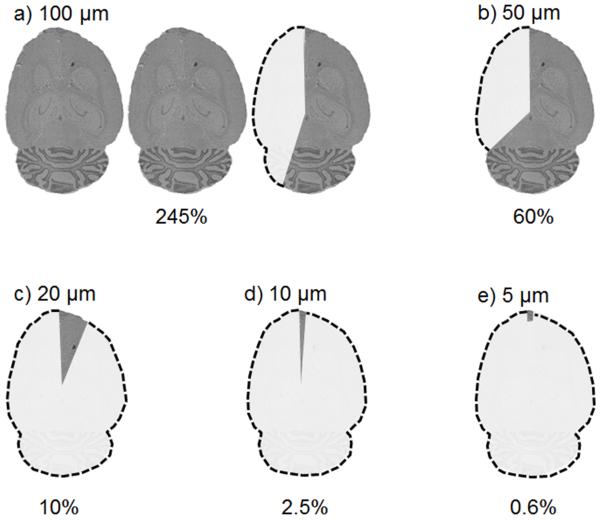
All analytical technologies experience trade-offs in various aspects of performance; as one particular characteristic is optimized, there are consequences that can affect the performance of other aspects of the technology. One such trade-off in IMS is the interplay between the amount of area sampled and the time required for data acquisition. The total number of pixels in an IMS experiment is a function of both the spatial resolution of the image (i.e., defined by the pitch or spacing between each pixel) and the total imaged area. As area is a square function, the number of pixels required to sample larger and larger amounts of tissue at a defined spatial resolution increases rapidly. For example, a 1 cm by 1 cm area of tissue sampled at 100 μm spatial resolution requires 10,000 pixels to analyze the entire region, whereas a 5 cm by 5 cm area of tissue analyzed at the same spatial resolution requires 250,000 pixels. Similarly, as the spatial resolution of an experiment is adjusted to a finer setting, the experimental time also increases. An example of the consequences of throughput is shown in Figure 2 and Table 1. Assuming a 1.5 cm diameter circular tissue section and a pixel acquisition speed of 1 pixel/second, a fairly common acquisition speed, the region of tissue that can be sampled in a reasonable 12 hour acquisition time decreases dramatically as the desired spatial resolution is altered (Figure 2). Similarly, the amount of time required to sample the entire tissue section using a 1 pixel/second acquisition speed at 5 μm spatial resolution can take over 81 days (Table 1)! Certainly, this acquisition time is not practical for most biological and clinical applications. However, at increased acquisition speeds, imaging the same area of tissue at finer spatial resolutions can again become practical. It is important to note that the acquisition times in Table 1 are reported for only a single tissue section; the effect on throughput is multiplicative when one considers the number of tissue sections in an experiment (e.g., normal versus disease state tissue comparisons), the number of technical replicates, and the number of biological replicates.
Figure 2.
A depiction of an IMS experiment showing that, given a defined circular tissue area (d=1.5 cm, represented by one cartoon brain image, images not shown to scale) and a 1 pixel/second acquisition speed, the amount of tissue that can be sampled in 12 hours is dependent upon the spatial resolution of the experiment. Percentages indicate the approximate proportion of one brain section that can be measured at the indicated spatial resolution.
Table 1.
The number of pixels required to sample a defined circular tissue area is determined by the spatial resolution of the experiment. Depending on the type of IMS instrumentation utilized, acquisition speeds can vary and will have a dramatic influence on the total analysis time.
| Spatial Resolution (μm) | Number of Pixels Required to Sample Entire Tissue (d=1.5 cm) | Analysis Time (1 pixel/sec acquisition) | Analysis Time (40 pixels/sec acquisition) |
|---|---|---|---|
| 100 | ~17,670 | 4.9 hours | 7.4 minutes |
| 50 | ~70,690 | 19.6 hours | 29.5 minutes |
| 20 | ~441,790 | 122.7 hours | 3.1 hours |
| 10 | ~1,767,150 | 20.5 days | 12.3 hours |
| 5 | ~7,068,580 | 81.8 days | 49.1 hours |
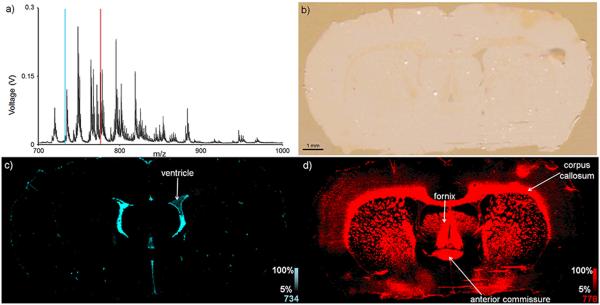
New advances in MS instrumentation have provided for systems capable of imaging at well above single pixel/second acquisition speeds. One such system utilizes continuous laser raster sampling to acquire data while continually firing the laser and scanning the stage (SimulTOF 300 Tandem).23 This is in contrast to more traditional scanning methods that move the stage in discrete steps under a stationary laser, pausing the stage to fire the laser before turning the laser off and moving to the next raster step. By using continuous raster sampling, a 39,073 pixel image of a coronal rat brain section sampled at 50 μm spatial resolution was acquired in approximately 45 minutes (Figure 3). This corresponds to an acquisition rate of roughly 15 pixels/second. In this image, many brain substructures are clearly visible, including the corpus callosum, fornix, anterior commissure, and ventricles.32 It is also evident that various ions in the mass spectrum display quite different spatial localizations. For example, the ion at m/z 734 is localized primarily to the ventricles (Figure 3c) while the ion at m/z 778 is primarily localized to the corpus callosum, fornix, and anterior commissure (Figure 3d). Given that this image was acquired in positive ion mode under sample conditions conducive to lipid ionization, these ions are likely forms of phosphatidylcholine ions, a class of glycerophospholipids that constitutes the major phospholipid present in most cellular membranes.33 While the identity of these two ions can be hypothesized based on the experimental polarity and the nominal mass of the peak, further rigor should be employed when assigning identities to ions from these types of experiments (vide infra).
Figure 3.
A 39,073 pixel MALDI image of a coronal rat brain section was sampled at 50 μm spatial resolution and was acquired in approximately 45 minutes. a) An average mass spectrum of 100 pixels shows the various lipid ions that were detected. b) A scanned optical image shows the tissue section following MALDI matrix application. Ion images of nominal masses c) m/z 734 and d) m/z 778 are plotted as m/z 734.6±0.6 and m/z 778.6±0.6, respectively, and show differential localization in brain substructures.
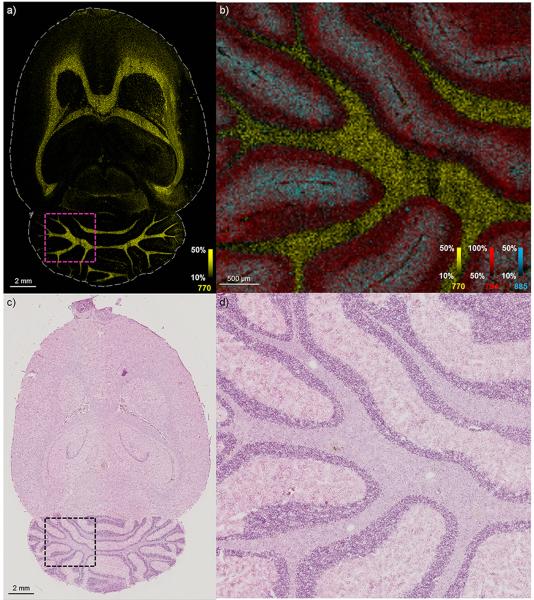
Another high-speed system employs a 10 kHz laser and moves the laser and the stage independently from one another, allowing the stage to be moved continuously while still acquiring discrete pixel spots (RapifleX Tissuetyper).24 Using this setup, a 644,134 pixel image of a transverse rat brain section sampled at 20 μm spatial resolution was acquired in approximately 345 minutes (Figure 4a). This corresponds to an acquisition rate of roughly 30 pixels/second. At this fine spatial resolution, the gray and white matter regions of the cerebellum are clearly well resolved (Figure 4b). However, when conducting an IMS experiment, the user should not simply ask: “how high of a spatial resolution can I achieve?” Rather, the user should consider the experimental question for the biological or clinical task at hand and ask: “what spatial resolution is sufficient for this experiment?” By considering the level of image detail required and the area required to be imaged, the user can define a reasonable spatial resolution and analysis time.
Figure 4.
A 644,134 pixel MALDI image of a transverse rat brain section was sampled at 20 μm spatial resolution and was acquired in approximately 345 minutes. a) An ion image of nominal mass m/z 770 depicts the entire brain. b) Ion images of nominal masses m/z 770, m/z 794, and m/z 885 are shown in an enlarged region of the cerebellum highlighted in (a) using the pink box and are plotted as m/z 770.6±0.5, 794.7±0.5 and m/z 885.6±0.5, respectively, using root mean square (RMS) normalization. c) A hematoxylin and eosin stain of the tissue section is performed following MALDI IMS analysis. d) The enlarged region of the cerebellum is highlighted in (c) using the black box.
Such considerations not only have implications on throughput, but also on other aspects of IMS performance. For example, as the number of pixels in an image increases, the data file size also increases.34–36 In some cases, file sizes can be well over a terabyte in size! Additionally, at smaller laser diameters required for high-spatial resolution analyses, sensitivity can become a concern.37–39 While the images shown here were acquired on high-speed TOF systems, other systems with slower acquisition speeds can be extremely important for high-quality IMS studies. As mentioned above, the ability to identify an ion of interest should not rely solely on the measured nominal mass of the ion. Exact mass measurements using high-mass accuracy, high-resolving power mass spectrometers such as Fourier transform ion cyclotron resonance (FTICR) instruments, Orbitrap instruments, and quadrupole-TOF (Q-TOF) instruments can be used to identify the empirical formula of an ion of interest, facilitating analyte identification.16–17,40 For example, the ion of nominal m/z 885 in Figure 4b was determined to have an exact m/z of 885.550 by tissue profiling on a 15T FTICR MS (Bruker Daltonics, Billerica, MA) (data not shown). Searching this experimental exact mass through an online lipid database (LIPID MAPS, Lipidomics Gateway, www.lipidmaps.org) allows for the identification of this lipid with an accuracy of <1 ppm as a phosphatidylinositol, PI(38:4), a glycerophospholipid commonly found in brain tissue.41 While extremely powerful, high-resolving power instrument platforms are often the most expensive and have larger data file sizes. MS/MS enabled ion trap, TOF, and hybrid instruments18, 20–22, 40, 42–43 as well as ion mobility-mass spectrometry (IM-MS) instruments40, 44 can also be used to provide structural information to aid in identification. While not always the case, many of these instruments have IMS acquisition speeds closer to 1 pixel/second, making them much slower than the high-speed TOF systems detailed above. However, diminished throughput is countered by an increase in molecular specificity with these instruments, an extremely important aspect of any MS experiment.
Conclusions
As IMS workflows are extended to more and more applications, it will be imperative that experimental setups provide biologically or clinically relevant time scales. In addition to continuing advances in instrumentation, advances in data acquisition also provide promising avenues for increasing experimental throughput. Histology-directed MS profiling experiments, where MS data is only acquired from several small predefined regions of the tissue, allow for dramatic increases in throughput.45 Additionally, predictive imaging modalities that employ multivariate regression46 or pan-sharpening algorithms47 can allow for the mathematical combination of multiple imaging datasets, allowing for poorer spatial resolution IMS experiments to be sharpened to finer resolution IMS experiments. This `image fusion' approach can also be used to provide out-of-sample ion distribution predictions for regions of unanalyzed tissue.46 Combined with high-speed MS instrumentation, these methods and bioinformatics tools will be vital to the widespread adoption of IMS technology.
Acknowledgements
This work was sponsored by the National Institutes of Health/National Institute of General Medical Sciences under Award 5P41 GM103391-05. B.M.P. was supported by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases under Award F32 FDK105841A.
References
- 1.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Analytical Chemistry. 1997;69:4751–4760. doi: 10.1021/ac970888i. http://dx.doi.org/10.1021/ac970888i PMid:9406525. [DOI] [PubMed] [Google Scholar]
- 2.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chemical Reviews. 2013;113(4):2309–2342. doi: 10.1021/cr3004295. http://dx.doi.org/10.1021/cr3004295 PMid:23394164 PMCid:PMC3624074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibault D, Gillam C, Grey A, Han J, Schey K. MALDI tissue profiling of integral membrane proteins from ocular tissues. Journal of the American Society for Mass Spectrometry. 2008;19(6):814–822. doi: 10.1016/j.jasms.2008.03.002. http://dx.doi.org/10.1016/j.jasms.2008.03.002 PMid:18396059 PMCid:PMC2430993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grey A, Schey K. Distribution of bovine and rabbit lens alpha-crystallin products by MALDI imaging mass spectrometry. Molecular Vision. 2008;14:171–179. PMid:18334935 PMCid:PMC2254960. [PMC free article] [PubMed] [Google Scholar]
- 5.Pól J, Vidová V, Hyötyläinen T, Volný M, Novák P, Strohalm M, et al. Spatial distribution of glycerophospholipids in the ocular lens. PloS one. 2011;6(4) doi: 10.1371/journal.pone.0019441. http://dx.doi.org/10.1371/journal.pone.0019441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada Y, Hidefumi K, Shion H, Oshikata M, Haramaki Y. Distribution of chloroquine in ocular tissue of pigmented rat using matrix-assisted laser desorption/ionization imaging quadrupole time-of-flight tandem mass spectrometry. Rapid Communications in Mass Spectrometry. 2011;25(11):1600–1608. doi: 10.1002/rcm.5021. http://dx.doi.org/10.1002/rcm.5021 PMid:21594935. [DOI] [PubMed] [Google Scholar]
- 7.Ronci M, Sharma S, Chataway T, Burdon K, Martin S, Craig J, Voelcker N. MALDI-MS-imaging of whole human lens capsule. Journal of Proteome Research. 2011;10(8):3522–3529. doi: 10.1021/pr200148k. http://dx.doi.org/10.1021/pr200148k PMid:21663315. [DOI] [PubMed] [Google Scholar]
- 8.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. Journal of the American Society for Mass Spectrometry. 2005;16(7):1093–1099. doi: 10.1016/j.jasms.2005.02.026. http://dx.doi.org/10.1016/j.jasms.2005.02.026 PMid:15923124. [DOI] [PubMed] [Google Scholar]
- 9.Ljungdahl A, Hanrieder J, Fälth M, Bergquist J, Andersson M. Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L-DOPA-induced dyskinesia in rat model of Parkinson's disease. PloS one. 2011;6(9) doi: 10.1371/journal.pone.0025653. http://dx.doi.org/10.1371/journal.pone.0025653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taverna D, Nanney LB, Pollins AC, Sindona G, Caprioli RM. Spatial mapping by imaging mass spectrometry offers advancements for rapid definition of human skin proteomic signatures. Experimental Dermatology. 2011;20(8):642–647. doi: 10.1111/j.1600-0625.2011.01289.x. http://dx.doi.org/10.1111/j.1600-0625.2011.01289.x PMid:21545539 PMCid:PMC3135742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazova R, Seeley EH, Keenan M, Gueorguieva R, Caprioli RM. Imaging mass spectrometry--a new and promising method to differentiate Spitz nevi from Spitzoid malignant melanomas. The American Journal of Dermatopathology. 2012;34(1):82–90. doi: 10.1097/DAD.0b013e31823df1e2. http://dx.doi.org/10.1097/DAD.0b013e31823df1e2 PMid:22197864 PMCid:PMC3263382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grüner BM, Hahne H, Mazur PK, Trajkovic-Arsic M, Maier S, Esposito I, et al. MALDI imaging mass spectrometry for in situ proteomic analysis of preneoplastic lesions in pancreatic cancer. PloS one. 2012;7(6) doi: 10.1371/journal.pone.0039424. http://dx.doi.org/10.1371/journal.pone.0039424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balluff B, Elsner M, Kowarsch A, Rauser S, Meding S, Schuhmacher C, et al. Classification of HER2/neu status in gastric cancer using a breast-cancer derived proteome classifier. Journal of Proteome Research. 2010;9(12):6317–6322. doi: 10.1021/pr100573s. http://dx.doi.org/10.1021/pr100573s PMid:21058730. [DOI] [PubMed] [Google Scholar]
- 14.Rauser S, Marquardt C, Balluff B, Deininger S-O, Albers C, Belau E, et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. Journal of Proteome Research. 2010;9(4):1854–1863. doi: 10.1021/pr901008d. http://dx.doi.org/10.1021/pr901008d PMid:20170166. [DOI] [PubMed] [Google Scholar]
- 15.Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. Journal of the American Society for Mass Spectrometry. 2002;13(6):735–748. doi: 10.1016/S1044-0305(02)00376-8. http://dx.doi.org/10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 16.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Analytical Chemistry. 2008;80(14):5648–5653. doi: 10.1021/ac800617s. http://dx.doi.org/10.1021/ac800617s PMid:18564854 PMCid:PMC2924168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3(21):2427–2441. doi: 10.4155/bio.11.232. http://dx.doi.org/10.4155/bio.11.232 PMid:22074284. [DOI] [PubMed] [Google Scholar]
- 18.Hopfgartner G, Varesio E, Stoeckli M. Matrix-assisted laser desorption/ionization mass spectrometric imaging of complete rat sections using a triple quadrupole linear ion trap. Rapid communications in mass spectrometry : RCM. 2009;23(6):733–736. doi: 10.1002/rcm.3934. PMid:19206086. [DOI] [PubMed] [Google Scholar]
- 19.Landgraf RR, Prieto Conaway MC, Garrett TJ, Stacpoole PW, Yost RA. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Analytical Chemistry. 2009;81(20):8488–8495. doi: 10.1021/ac901387u. http://dx.doi.org/10.1021/ac901387u PMid:19751051 PMCid:PMC2767259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perdian DC, Lee YJ. Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-orbitrap mass spectrometer. Analytical Chemistry. 2010;82(22):9393–9400. doi: 10.1021/ac102017q. http://dx.doi.org/10.1021/ac102017q PMid:20977220. [DOI] [PubMed] [Google Scholar]
- 21.Prideaux B, Dartois V, Staab D, Weiner D, Goh A, Via L, Barry C, Stoeckli M. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Analytical Chemistry. 2011;83(6):2112–2118. doi: 10.1021/ac1029049. http://dx.doi.org/10.1021/ac1029049 PMid:21332183 PMCid:PMC3158846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, et al. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Analytical Chemistry. 2013;85(20):9596–9603. doi: 10.1021/ac401760s. http://dx.doi.org/10.1021/ac401760s PMid:24040919 PMCid:PMC3867692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prentice BM, Chumbly CW, Caprioli RM. High-speed MALDI TOF/TOF imaging mass spectrometry using continuous raster sampling. Journal of Mass Spectrometry. 2015;50(4):703–710. doi: 10.1002/jms.3579. http://dx.doi.org/10.1002/jms.3579 PMid:26149115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spraggins JMR, David G, Moore Jessica L., Rose Kristie L., Hammer Neal D., Skaar Eric P., Caprioli Richard M. MALDI FTICR IMS of intact proteins: using mass accuracy to link protein images with proteomics data. Journal of the American Society for Mass Spectrometry. 2015;26:974–985. doi: 10.1007/s13361-015-1147-5. http://dx.doi.org/10.1007/s13361-015-1147-5 PMid:25904064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther S, Koestler M, Schulz O, Spengler B. Laser spot size and laser power dependence of ion formation in high resolution MALDI imaging. International Journal of Mass Spectrometry. 2010;294:7–15. [Google Scholar]
- 26.Holle A, Haase A, Kayser M, Höhndorf J. Optimizing UV laser focus profiles for improved MALDI performance. Journal of Mass Spectrometry. 2006;41(6):705–716. doi: 10.1002/jms.1041. http://dx.doi.org/10.1002/jms.1041 PMid:16718638. [DOI] [PubMed] [Google Scholar]
- 27.Trim PJ, Djidja M-C, Atkinson SJ, Oakes K, Cole LM, Anderson DMG, Hart PJ, Francese S, Clench MR. Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MALDI-MS imaging. Analytical and Bioanalytical Chemistry. 2010;397(8):3409–3419. doi: 10.1007/s00216-010-3874-6. http://dx.doi.org/10.1007/s00216-010-3874-6 PMid:20635080. [DOI] [PubMed] [Google Scholar]
- 28.Simmons DA. Applied Biosystems Technical Note. 2008. Improved MALDI-MS imaging performance using continuous laser rastering; pp. 1–5. [Google Scholar]
- 29.Spraggins JM, Caprioli RM. High-speed MALDI-TOF imaging mass spectrometry: rapid ion image acquisition and considerations for next generation instrumentation. Journal of the American Society for Mass Spectrometry. 2011;22(6):1022–1031. doi: 10.1007/s13361-011-0121-0. http://dx.doi.org/10.1007/s13361-011-0121-0 PMid:21953043 PMCid:PMC3514015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. Journal of the American Society for Mass Spectrometry. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. http://dx.doi.org/10.1016/j.jasms.2007.06.010 PMid:17659880 PMCid:PMC2042488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Analytical Chemistry. 2011;83(14):5728–5734. doi: 10.1021/ac200998a. http://dx.doi.org/10.1021/ac200998a PMid:21639088 PMCid:PMC3136623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lein ESH, Michael J, Ao Nancy, Ayres Mikael, Bensinger Amy, Bernard Amy, Boe Andrew F., Boguski Mark S., Brockway Kevin S., Byrnes Emi J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. http://dx.doi.org/10.1038/nature05453 PMid:17151600. [DOI] [PubMed] [Google Scholar]
- 33.Billah MMA, John C. The regulation and cellular functions of phosphatidylcholine hydrolysis. Biochemical Journal. 1990;269:281–291. doi: 10.1042/bj2690281. http://dx.doi.org/10.1042/bj2690281 PMid:2201284 PMCid:PMC1131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van de Plas RDM, Waelkens Bart. Etienne In Discrete wavelet transform-based multivariate exploration of tissue via imaging mass spectrometry. Proceedings of the 2008 ACM Symposium on Applied Computing, Fortaleza, Brazil, Fortaleza, Brazil. 2008:1307–1308. http://dx.doi.org/10.1145/1363686.1363989.
- 35.McDonnell L. A. v. R., de Velde Alexandra, van Zeijl Nico, Rene JM, Deelder Andre M. Imaging mass spectrometry data reduction: automated feature identification and extraction. Journal of the American Society for Mass Spectrometry. 2010;21:1969–1978. doi: 10.1016/j.jasms.2010.08.008. http://dx.doi.org/10.1016/j.jasms.2010.08.008 PMid:20850341. [DOI] [PubMed] [Google Scholar]
- 36.Rompp AS, Hester Thorsten, Klinkert Alfons, Both Ivo, Heeren Jean-Pierre, Ron MA, Stockli Marcus, Spengler Bernhard. imzML: imaging mass spectrometry markup language: a common data format for mass spectrometry imaging. In: Hamacher ME, Stephan Martin, Christian, editors. Methods in Molecular Biology. Vol. 696. Springer Science+Business Media, LLC; New York Dordrecht Heidelberg London: 2010. [DOI] [PubMed] [Google Scholar]
- 37.Dreisewerd KS, Karas Michael Martin, Hillenkamp Franz. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix-assisted laser desorption/ionization with a uniform beam profile. International Journal of Mass Spectrometry and Ion Processes. 1995;141(2):127–148. http://dx.doi.org/10.1016/0168-1176(94)04108-J. [Google Scholar]
- 38.Guenther SK, Schulz Martin, Spengler Oliver, Bernhard Laser spot size and laser power dependence of ion formation in high resolution MALDI imaging. International Journal of Mass Spectrometry. 2010;294(1):7–15. http://dx.doi.org/10.1016/j.ijms.2010.03.014. [Google Scholar]
- 39.Qiao HS, Ens Victor, Werner The effect of laser profile, fluence, and spot size on sensitivity in orthogonal-injection matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22(18):2779–2790. doi: 10.1002/rcm.3675. http://dx.doi.org/10.1002/rcm.3675 PMid:18697229. [DOI] [PubMed] [Google Scholar]
- 40.Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, et al. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Analytical Chemistry. 2008;80(22):8628–8634. doi: 10.1021/ac8015467. http://dx.doi.org/10.1021/ac8015467 PMid:18847214. [DOI] [PubMed] [Google Scholar]
- 41.Zemski Berry KAH, Barkley Joseph A., Spraggins Robert A., Caprioli Jeffery M., Murphy Richard M., Robert C. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chemical Reviews. 2011;111(10):6491–6512. doi: 10.1021/cr200280p. http://dx.doi.org/10.1021/cr200280p PMid:21942646 PMCid:PMC3199966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debois D, Bertrand V, Quinton L, De Pauw-Gillet M-C, De Pauw E. MALDI-in source decay applied to mass spectrometry imaging: a new tool for protein identification. Analytical Chemistry. 2010;82(10):4036–4045. doi: 10.1021/ac902875q. http://dx.doi.org/10.1021/ac902875q PMid:20397712. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andre PE. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One. 2010;5(7):e11411. doi: 10.1371/journal.pone.0011411. http://dx.doi.org/10.1371/journal.pone.0011411 PMid:20644728 PMCid:PMC2904372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. Journal of Mass Spectrometry. 2007;42(8):1099–1105. doi: 10.1002/jms.1254. http://dx.doi.org/10.1002/jms.1254 PMid:17621390. [DOI] [PubMed] [Google Scholar]
- 45.Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, Caprioli RM. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Molecular & Cellular Proteomics. 2006;5(10):1975–1983. doi: 10.1074/mcp.M600119-MCP200. http://dx.doi.org/10.1074/mcp.M600119-MCP200 PMid:1689436. [DOI] [PubMed] [Google Scholar]
- 46.Van de Plas RY, Spraggins Junhai, Caprioli Jeffery, Richard M. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nature Methods. 2015;12(4):366–372. doi: 10.1038/nmeth.3296. http://dx.doi.org/10.1038/nmeth.3296 PMid:25707028 PMCid:PMC4382398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarolli JGJ, Lauren M. Winograd, Nicholas, Improving secondary ion mass spectrometry image quality with image fusion. Journal of the American Society for Mass Spectrometry. 2014;25(12):2154–2162. doi: 10.1007/s13361-014-0927-7. http://dx.doi.org/10.1007/s13361-014-0927-7 PMid:24912432 MCid:PMC4224624. [DOI] [PMC free article] [PubMed] [Google Scholar]