Abstract
Antidepressants are often only moderately successful in decreasing the severity of depressive symptoms. In part, antidepressant treatment response in patients with depression is genetically determined. However, although a large number of studies have been conducted aiming to identify genetic variants associated with antidepressant drug response in depression, only a few variants have been repeatedly identified. Within the present review, we will discuss the methodological challenges and limitations of the studies that have been conducted on this topic to date (e.g., ‘treated-only design’, statistical power) and we will discuss how specifically drug–gene interaction models can be used to be better able to identify genetic variants associated with antidepressant drug response in depression.
Keywords: : antidepressive agents, drug–gene interaction models, pharmacogenomics
Background
The lifetime prevalence of major depressive disorder (MDD) is approximately 12–18% in American and European populations [1–4], with a higher disease burden in female (notably two-times as high as in men) and older populations [1,5]. MDD is usually treated with psychotherapy or with antidepressant drug treatment, the latter being frequently divided in three main drug classes that affect different neurotransmitter pathways by inhibiting or enhancing signal transduction across neurons: classical tricyclic antidepressants (TCAs); selective serotonin reuptake inhibitors (SSRIs); and other antidepressants (e.g., venlafaxine and mirtazapine). For example, SSRIs affect the serotonergic signaling pathway by inhibiting serotonin reuptake from the synaptic cleft of serotonergic presynaptic neurons. This inhibition of serotonin reuptake results in a higher bioavailability of serotonin to postsynaptic neurotransmitter signaling pathways. Alternatively, TCAs affect several neurotransmitter signaling pathways, including dopaminergic, noradrenergic and serotonergic signaling pathways. As TCAs work through multiple neurotransmitter pathways, adverse drug reactions are more common with TCAs than with SSRIs. Because of the lower risk of adverse drug reactions during the use of SSRIs than during the use of TCAs, SSRIs are also more frequently used for the treatment of depressive indications [6]. Out of these two main antidepressant drug classes, most recent studies have been focused on the SSRIs.
Despite the fact that multiple antidepressant drugs have been launched on the market during the past decades, failure of antidepressant treatment, defined as insufficient decrease in depressive symptoms within a certain period of time, is common [7], and may be partly genetically determined [8–10]. Genes affecting antidepressant drug response have all been identified in candidate gene studies, in which a gene was tested in relation with antidepressant drug response in depression on the basis of a predefined hypothesis [11,12]. Within these efforts, multiple genes have been identified that potentially affect antidepressant treatment response in depression [11,12]. However, heterogeneity of the findings between studies was large [12], and only few genes were well replicated in more recent studies or were only associated with antidepressant drug response in depression in subsequent studies comprising patients with specific subtypes of depression (e.g., melancholic or psychotic depression [13,14]). For this reason, only few genes have been repeatedly associated with antidepressant drug response in depression in meta-analyses [12].
There are two biological processes governing the success of antidepressant drug treatment: pharmacokinetics and pharmacodynamics. For this reason, genetic studies on antidepressant drug response in depression have primarily evaluated genes encoding proteins that affect the pharmacokinetics of an antidepressant drug (e.g., drug metabolism) or genes encoding proteins that affect pharmacodynamics (e.g., treatment target). With respect to genes that affect antidepressant pharmacokinetics, two independent genes have to date been repeatedly identified in relation to antidepressant drug response in depression, CYP2D6 and ABCB1 [11,12]. In addition to antidepressant drug response in depression, these two genes have also been associated with the risk of adverse drug reactions during antidepressant drug treatment (e.g., [15–19]). However, even though genetic variation in CYP2D6 was one of the robust genetic variants associated with antidepressant drug response in the meta-analyses [11,12], studies conducted in both the STAR*D and GENDEP trials did not confirm these results [20,21], although GENDEP showed that genetic variation in CYP2D6 was associated with serum concentrations of both escitalopram and nortriptyline [20].
In addition, also two independent genes that are related to antidepressant pharmacodynamics have been identified in relation to antidepressant drug response, SLC6A4 and HTR2A [11,12,22]. Both genes encode for proteins that affect the synaptic cleft of serotonergic neurons, and SLC6A4 encodes the serotonin reuptake transporter, the main pharmacological target of SSRIs [11,12,22]. Nevertheless, more genes have been identified in studies in relation to antidepressant drug response in depression, including for example FKBP5 and BDNF, but evidence is weaker (e.g., heterogeneity between studies is larger or studies were conducted with limited sample size) and requires additional studies to fully elucidate their role in antidepressant drug response in depression. Although a small number of genes have been repeatedly associated with antidepressant drug response in depression, the clinical benefit of knowing the status of these genes is considered to be limited at this time [12]. Furthermore, genome-wide association studies (GWAS), that aimed to identify genetic variants in relation to antidepressant drug response across the genome without having a predefined hypothesis [23–27], have not detected genetic loci associated with antidepressant drug response that reached the level of statistical significance, not even the above-mentioned genes that have been previously reported in relation to antidepressant drug response in depression.
Within the present review, our main aim is to provide an overview of the methodological limitations and challenges associated with genetic studies of antidepressant response in depression. Next, we will discuss how specifically drug–gene interaction models in studies can contribute to identify genetic variants affecting antidepressant drug response in depression.
Challenges to identify genetic loci associated with antidepressant response in depression
One of the major challenges to the identification of genetic loci associated with antidepressant response is low statistical power, especially in the GWAS that have been published to date [23–27]. For example, a recently published meta-analysis had a discovery sample of 865 MDD patients in which a GWAS was conducted, and the meta-analysis that also included the replication samples had a total sample size of 2394 MDD patients [27]. With such relatively small sample sizes, the identification of significant effects at the accepted GWAS significance threshold of p < 5 × 10-8 would require very large effect sizes on decrease in depressive symptoms over time. For this reason, larger collaborating efforts, as were also required to identify genetic loci in other phenotypes (e.g., [28,29]), are likely required to have well-powered studies to identify genetic loci with modest effects on the decrease in depressive symptoms [30]. Besides increasing the sample size, researchers have also applied other solutions to overcome the issues of the sample size, which includes studies of intermediate phenotypes. However, also this strategy has some important limitations that need to be considered. For example, a GWAS on plasma drug concentrations of citalopram and escitalopram, which was performed in 435 MDD patients, identified two genome-wide significant independent loci that included the CYP2C9 and CYP2D6 genes [31]. These genes are encoding for proteins that affect drug metabolism. However, studies as these are not able to identify genes affecting antidepressant pharmacodynamics [31]. Furthermore, although genetic variation in CYP2D6 is also associated with citalopram and nortriptyline in the GENDEP trial, no significant association was observed with drug response [20]. The clinical implication with respect to antidepressant drug response needs therefore to be elucidated.
An additional option for an intermediate phenotype could be, theoretically, the prescribed antidepressant dosage, as has also been done to identify loci involved in treatment resistance to coumarins [32–34]. However, coumarin treatment is precisely titrated by reference to the international normalized ratio (INR) while any antidepressant dose titration would rely on clinical response (unless blood levels are assessed on a regular basis). In the case of antidepressant drugs, participants at the upper end of the dosage phenotype distribution will include patients who were prescribed a higher prescribed antidepressant dosage because of a more severe type of depression as well as patients who were prescribed a higher antidepressant dosage because of nonresponse.
Of equal importance is the choice of the design to be able to identify genetic variants associated with antidepressant drug response. The study design most commonly used to date in genomic studies of antidepressant drug response is the ‘treated-only’ design, which is restricted to MDD patients who initiate antidepressant treatment and who are then followed over time to characterize treatment resistance. As was shown before in simulation models [35], genetic variants related with response in a treated-only design could theoretically reflect the main effect of the SNP on the indication of use. For example, it has been previously shown that antidepressant drugs have only a significant benefit over placebo treatment in patients with severe depressive symptoms [36]. When a study consists of a heterogeneous population of patients with mild and severe depressive symptoms, a larger drop in depressive symptoms during follow-up could theoretically be restricted to individuals with more severe depressive symptoms. In this scenario, genes associated with more severe depressive symptoms could thus be associated with heterogeneous antidepressant drug response. Although the effect of baseline severity of the depressive symptoms can be decreased by taking the percentage decrease in depressive symptoms, which is done in a limited number of the studies, this effect estimate can still be in part dependent on the severity of the depressive symptoms at baseline. Genes that have been previously published in relation with depressive symptoms or MDD as well as with antidepressant drug response could be examples of genes that only show an association with antidepressant drug response because of their association with a subset of severe depressive symptoms. Indeed, a systemic review and meta-analysis reported that the S variant of the HTTLPR polymorphism in the SLC6A4 gene, a variant that has been repeatedly associated with antidepressant drug response [12], was associated with an 11% higher risk of MDD [37]. In theory, the observed association between SLC6A4 and antidepressant drug response also could be influenced by the association between SLC6A4 and the risk of MDD, and the true effect of SLC6A4 on the therapeutic response of antidepressants could be in theory be smaller. Yang et al. observed that genetic variation in the APC gene was associated with both MDD as well as with the therapeutic response to antidepressants [38]. In addition to these examples, more genes have been shown to associate both to MDD as well as to the decrease in depressive symptoms after initiation of treatment (e.g., [18]). As no reference group was taken into account in the pharmacogenetic analysis, it is currently unknown whether or not these variants are truly associated with the therapeutic response of antidepressants in depression [38] or simply reflect genetic main effects.
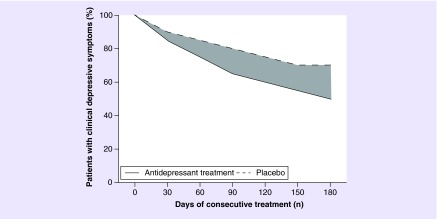
An additional challenge to successfully identify genetic variants associated with antidepressant drug response is the reduction in depressive symptoms that is unrelated to the use of antidepressant drugs themselves [39], which is fairly common following antidepressant drug treatment in MDD patients. When people with MDD are enrolled at the height of their symptoms, subsequent improvement may be spontaneous, the response to the nonpharmacological part of treatment, or the result of ‘regression to the mean’ [40]. This issue is visualized in Figure 1, in which a hypothetical placebo-controlled randomized trial on antidepressant response in depression is displayed. Here, both the group treated with antidepressants and the placebo-treated group showed significant reduction in depressive symptoms. However, one should take into account that only the area displayed in gray is attributable to the use of the antidepressant. When a treated-only design (with only depressive patients that are treated) is used to identify loci associated with antidepressant drug response in depression, the decrease in depressive symptoms will be in part be the result of the antidepressant drug treatment, but is also in part unrelated to the antidepressant drug treatment. In this case, both the depressive indication of antidepressants as well as the natural disease course of the depression can influence the results from a pharmacogenomic study on antidepressant drug response in depression. Again, taking into account an untreated reference population may be able to disentangle the reduction of depressive symptoms caused by the drug with the reduction of depressive symptoms caused by nonpharmacological reasons (e.g., if patients expect improvement because of receiving treatment) or by the natural disease course.
Figure 1. . Hypothetical randomized clinical trial on antidepressant response.
The dark gray area reflects the difference in response between depressive patients treated with antidepressant drugs depressive patients treated with placebo.
Taken together, studies using a treated-only design to identify genetic variants associated with antidepressant drug response in depression will not only identify loci that are associated with the therapeutic response of the antidepressant drug treatment, but also identify loci associated with the severity of the depressive indication and with the natural disease course of the depression. As we describe below, choosing a study design that includes a reference population with similar indication that is using placebo treatment will be able to disentangle the true antidepressant response related to the drug from the ‘response’ that is unrelated to the use of antidepressants.
Drug–gene interaction models in pharmacogenetic studies
In epidemiology, interaction terms are used to demonstrate whether the association between a determinant and outcome is modified by another factor. This way, patients at excessively high risk to develop a certain disease can be identified. For pharmacogenomics, this means that the association between the drug and outcome is modified by certain genetic factors, and that patients with excessively high risk to develop adverse drug reactions or treatment resistance can be identified. However, these so-called ‘drug–gene interaction studies’ to identify pharmacogenetic loci that are associated with response or adverse drug reactions are not often performed. Using collaborative efforts, the ‘Cohorts for Heart and Aging Research in Genomic Epidemiology’ consortium [41] aimed to identify genetic loci that interact with drug use on multiple outcomes of drug use. However, neither genome-wide drug–gene interaction studies of antihypertensive agents and cardiovascular disease outcomes [42] nor of predefined drug classes and QT interval [43] identified genetic loci at genome-wide significant thresholds. Although these results could mean that there are no genetic loci that substantially modify the association between the drug and study outcome (and thus have no or limited clinical importance), the statistical power of these studies could also be too limited. Notably, the statistical power of a genome-wide drug–gene interaction study is dependent on multiple factors [43]. First, the frequency of drug use in the study population should be sufficient, with less commonly used drugs necessitating larger study populations. Second, the effect size should be substantial at least for the interaction, otherwise larger sample sizes are required. And third, the effect size of the drug on the outcome of interest. In case of a fairly low effect size of the drug on the outcome (e.g., a rare adverse drug event or a relatively small intended drug effect), a larger study population is required. Because of the issue of a limited sample size, new statistical programs have been developed to further increase the statistical power by leveraging longitudinal drug and phenotype assessments often available in prospective cohort studies [44], which can considerably increase statistical power [43].
Despite indications of a better performance of studies that use drug–gene interaction analyses, studies analyzing the effect of a SNP on the change of the study outcome after initiation of drug treatment can still provide valuable results. For example, several loci were identified in a GWAS on cholesterol lowering in users of statin therapy [45]. Loci that were identified with the change in LDL cholesterol level after initiation of statin therapy include SORT1, SLCO1B1, APOE and LPA. Besides being related to change in LDL cholesterol level after initiation of statin therapy, three of these loci (notably SORT1, APOE and LPA) have also been identified in relation to LDL cholesterol level in the most recent GWAS [46]. Based on the simulation studies described earlier, these loci could, besides being related to the LDL cholesterol lowering response of statins, also be related to the main effect of the SNP on LDL cholesterol level [35], despite having adjusted for the baseline LDL cholesterol level [45]. Similar as discussed before, these genes do not necessarily reflect the biological mechanism how statins lower LDL cholesterol level despite showing an association with LDL cholesterol reduction in statin users. Different from antidepressant drug response in depression is that the response unrelated to the drug itself (e.g., placebo response or natural relief in depressive symptoms; Figure 1) is smaller in users of statins because LDL-cholesterol concentrations will not decrease spontaneously or as part of a disease course (e.g., [47]). This particular issue has been raised in a review by Hall et al. who showed that several genetic variants are associated with the placebo response [48]. In case of the pharmacogenetics of antidepressant drug response, several genes of particular interest to the field (e.g., TPH2, SLC6A4, HTR2A) are highlighted in this paper to be genes involved in placebo response. As we highlighted above that a relatively large placebo effect is a likely cause of the difficulties with antidepressant pharmacogenomics, it is not surprising that all genetic loci for LDL cholesterol reduction in statin users were replicated in a drug–gene interaction analysis [45].
Together with the simulation analyses [35], it is likely that the chance of successfully identifying genetic loci with the therapeutic response is dependent on the choice of the design. For drugs such as antidepressants, that have a relatively small ‘true therapeutic effect’ (drug treatment effect – placebo treatment effect) [36], taking into account a reference group could therefore have important implications.
Application & considerations of drug–gene interaction models in pharmacogenetic randomized clinical trials on antidepressant response
To the best of our knowledge, no studies have yet studied the association between genetic variants and antidepressant drug response in depression using drug–gene interaction models in a population containing a group of MDD patients treated with an antidepressants as well as a reference group (e.g., MDD patients treated with placebo). Nevertheless, a limited number of studies compared their results with a placebo-treated population [49], used a model-based approach to define the nonspecific response [50], or evaluated whether SNPs are specifically associated with response in nortriptyline or escitalopram [51]. In the case of a formal drug–gene interaction analysis, both the group of patients treated with an antidepressant drug and the group of patients treated with placebo would be followed over time and the depressive symptoms would be assessed at multiple time points. This procedure is exactly the same as what has been used to determine whether an antidepressant drug is more effective than a placebo [36]. Within this setting a drug–gene interaction model will determine whether a genetic variant is associated with the decrease in depressive symptoms in the group treated with an antidepressant relative to the group treated with placebo. The regression formulas for the statistical analyses for the treated-only and place-controlled pharmacogenomics study are as follows:
Regression formula for treated-only designs
Change in depressive symptoms = α + β1 × SNP + βs for covariates
Regression formula for drug–gene interaction models in randomized clinical trials
Change in depressive symptoms = α + β1 × SNP + β2 × (drug/placebo group) + β3 × SNP × (drug/placebo group) + βs for covariates
In these formulas, α is the intercept of the regression line and the βs reflect the effect size of the independent variable on the change in depressive symptoms over time. In the treated-only design, β1 measures the effect size of the SNP on the drug effect (e.g., reduction in depressive symptoms) in users. For the regression formula that is required in drug–gene interaction models in placebo-controlled studies, β3 reflects the effect size of the effect allele on the change in depressive symptoms over time in users of antidepressants relative to the placebo group. In case of the treated-only design β1 is the beta estimate of interest whereas in the placebo-controlled pharmacogenomic study β3 is the beta estimate of interest.
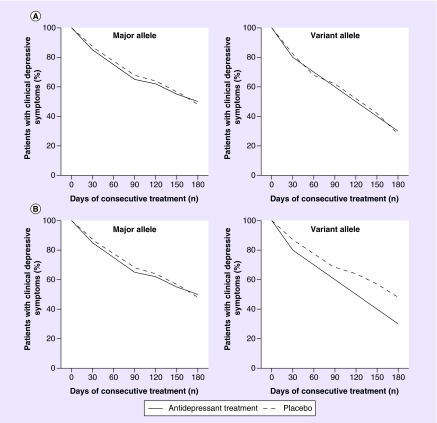
Hypothetical examples of two situations of a drug–gene interaction analysis in a placebo-controlled pharmacogenomic study are presented in Figure 2. According to the hypothetical situation depicted in the upper two graphs of the figure (Figure 2A), carriers of the variant allele (for simplicity reasons we combine heterozygous and homozygous variant allele carriers together in the examples) showed a larger reduction in the proportion of patients with clinical depressive symptoms over time than homozygous carriers of the major allele. However, the decrease in the proportion of patients with depressive symptoms over time is similar for users of antidepressant treatment (solid line) and for users of placebo treatment (dashed line). This indicates that, although associated with a faster relief in depressive symptoms, this allele is not modifying the antidepressant drug response. If we would have used a treated-only design, the conclusion for this specific hypothetical allele would be that it is associated with antidepressant drug response in depression. However, as the association of this genetic variant is similar in the placebo-treated group, this hypothetical allele is more likely to reflect associations with depressive indicators or natural disease course. In contrast, true drug–gene interactions are shown in Figure 2B. In this example, the variant allele has no effect on reducing the severity of the depressive symptoms in patients with depression treated with placebo, as the proportion of patients with clinical depressive symptoms at day 180 is similar to that observed among homozygous patients carrying the major allele. However, in patients treated with an antidepressant (solid line) who carry the variant allele, we observe a larger reduction in the proportion of patients with clinical depressive symptoms compared with homozygous patients carrying the major allele or patients carrying the variant allele, but who treated with placebo (dashed line). Therefore, the variant allele in this hypothetical situation demonstrates an allele that modifies therapeutic response to antidepressants in depression.
Figure 2. . Hypothetical drug–gene interaction situations in a randomized clinical trial on antidepressant drug response in depression.
The trajectory of the depressive symptoms over time is depicted in antidepressant-treated patients as a solid line, and in placebo-treated patients as a dashed line. (A) Carriers of the variant allele (heterozygous and homozygous carriers combined for simplicity reasons) are associated with, on average, a larger decrease in depressive symptoms than homozygous carriers of the major allele, but the effect size is similar for users of antidepressant treatment and users of placebo treatment. (B) Carriers of the variant allele that use also antidepressant treatment have a steeper decline in depressive symptoms within the time period than homozygous carriers of the major allele and users of placebo treatment. This hypothetical genetic variant is therefore a modifier of antidepressant drug response.
Based on these hypothetical examples that are likely also influencing already published findings, the use of a reference population in studies that aim to identify genetic variants in antidepressant response will be able to disentangle the loci that are associated with the natural disease course or depressive indicators from the loci that truly modify antidepressant drug response in depression.
Drug–gene interaction analyses in population-based cohort studies
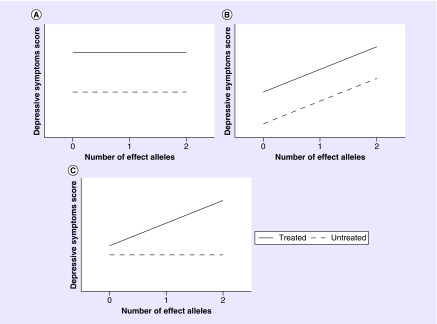
In addition to randomized controlled clinical trials, population-based cohort studies also can have valuable data available to identify genetic loci associated with response to antidepressants. For contributing in such studies, cohorts should have data available on drug use and have cross-sectional assessments of depressive symptoms collected concurrently [52]. Importantly, data on depressive symptoms should be collected for the total (unselected) study population. We defined genetic loci as being potential loci affecting antidepressant drug response in depression when genetic variants are associated with a different depressive symptoms score between users of antidepressants and nonusers of any antidepressant drug. The rationale of this theory is visualized in Figure 3, where interaction between the SNP and drug is present only in Figure 3C (the association between the genetic variant and depressive symptoms score is different between users and nonusers of antidepressants). In Figure 3B, the SNP is associated with depressive symptoms, but the association does not differ between treated and untreated individuals. Therefore, this locus is likely not associated with the therapeutic response, but instead with the depressive indication. In Figure 3A, there was interaction nor an association between the genetic variant and depressive symptoms. Likely, this variant has no role in depression nor with the therapeutic response to antidepressants in depression.
Figure 3. . Hypothetical drug–gene interaction situations in a (prospective) cohort study on antidepressant drug response in depression.
(A) No interaction and no association with cross-sectional assessments of depressive symptoms. (B) No interaction but association with cross-sectional assessments of depressive symptoms. (C) Interaction present, only association with cross-sectional assessments of depressive symptoms in treated participants.
To test the hypothesis that drug–gene interaction models can also be used to identify SNPs related to therapeutic response to antidepressants, we conducted a drug–gene interaction analysis in the prospective population-based Rotterdam Study cohort [53]. The Rotterdam Study, comparable to other population-based cohort studies, was originally designed to study the incidence of, and risk factors for, age-related diseases, and had no inclusion criteria other than being older than 55 years of age. During center visits, which took place every 4–5 years, data on depressive symptoms (center of epidemiological studies depression score or CES-D score) were collected, and data on use of drugs were continuously monitored [54]. Taking into account longitudinal data during which information on depressive symptoms was collected [44], we identified two potential interesting loci on a genome-wide level that interacted with the use of SSRIs on the depressive symptoms score (notably FSHR and HMGB4). Although a role in response to SSRIs has to be confirmed in future studies, mRNA expression levels were altered after stimulation with an SSRI in lower model organisms [55,56]. When we restricted our analysis to only genes that are involved in the serotonin signaling pathway [57,58], we identified genetic variation in the PLCB1 gene, which encodes for a postsynaptic signal transduction protein, to significantly interact with SSRI use on cross-sectional assessments of depressive symptoms. Although not earlier described in the literature in relation to antidepressant drug response in depression, common genetic variation in PLCB1 has been included in a patent of a pharmacogenetic testing kit for antidepressant drug response [59]. Furthermore, we observed that common genetic variation in the HTR2A gene interacted with use of SSRIs on depressive symptoms, which can be seen as additional confirmation of the role of HTR2A in the therapeutic response of SSRIs in depression [12,22].
However, although being a promising model for future studies on the genetic basis of antidepressant drug response in depression, there are some important limitations that need to be taken into account. For example, it should be noted that antidepressants were prescribed for indication other than depression. In case of The Netherlands, SSRIs are most often prescribed for depressive indications [6], but participants with an SSRI prescribed for another indication might negatively affect the study results. Furthermore, this approach does also not take into account factors associated with a GP's decision whether or not to initiate antidepressant drug treatment in case of a diagnosed MDD. As long as the genetic variant is unrelated to the indication ‘depression’ as well as with factors related to the choice of treatment, this will result in increased heterogeneity and variation in the outcome, meaning that the observed effect sizes are likely underestimated. In any case, larger sample sizes are required. Furthermore, as already highlighted earlier, it should be taken into account that for drug–gene interaction analyses in cohort studies using cross-sectional assessments of depressive symptoms requires large sample sizes, especially to capture genetic variants with a relatively low effect size and relatively low allele frequency. Nevertheless, future studies that are done with larger sample sizes will likely provide additional insights in the underlying biology of antidepressant treatment resistance in depression. However, significant interactions from these type of studies should be replicated in placebo-controlled trials, as in theory the scenario depicted in Figure 3C can also be the other way around when the SNP is associated with depressive symptoms in untreated individuals, but not in treated individuals. Visualization, as done in our study in case of the HTR2A findings [53], is one way to understand whether the observed interaction is driven by the treated or untreated individuals.
Conclusion & future perspective
With the increased possibilities to study genetic determinants in relation to certain study outcomes, researchers all over the world recognized the possibilities to increase the accuracy to predict disease outcomes and investigated the possibilities of precision medicine. Despite many initiatives, only a handful of genetic loci have been repeatedly associated with antidepressant drug response in depression [12]. These loci, however, could theoretically also be associated with aspects of decrease in depressive symptoms that are unrelated to the use of an antidepressant drug, including the depressive indication as well as the natural disease course of the depression. Furthermore, studies aiming to identify genetic loci associated with antidepressant drug response in depression are often performed with a small number of depressive patients, including the GWAS which have been done to identify new genetic variants associated with antidepressant drug response. For example, the largest GWAS to date on lipid lowering during statin therapy required more than 18,000 users of statin therapy to identify four independent genetic variants associated with LDL cholesterol lowering after initiation of statin treatment [45]. Still, these identified loci explained only approximately 5% of the total variation in LDL cholesterol response in statin users. Although successful in identifying genetic markers in relation to cholesterol-lowering response in statin users, there is still a long way to accurately predict before initiation of statin treatment whether a patient will develop sufficient lowering in cholesterol level or not. A meta-analysis of the GENDEP, MARS and STAR*D trials on the association between genetic variants and antidepressant drug response in depression only comprised 2256 treated MDD patients, and no loci were identified that reached the level of statistical significance [26]. Therefore, also with respect to other pharmacogenomic studies on other phenotypes, this indicates that the sample size is one major limitation [60]. Drug–gene interaction studies on antidepressant drug response in depression should also be aware of this, as such studies normally require larger sample sizes.
In summary, the search for genetic markers associated with antidepressant drug response in depression is difficult and only moderately successful to date. Although some genetic loci associated with antidepressant treatment have been replicated by others, generally the heterogeneity is large between studies, possible because of the lack of a reference population in the design of the study. Drug–gene interaction models, which additionally take into account the association between the genetic variant and the reduction in depression symptoms that is unrelated to the use of the antidepressant, will probably increase the ability to identify new loci and to confirm some of the already identified loci. This will also mean that loci previously associated with antidepressant drug response in depression in a treated-only design could be left unconfirmed in these newer drug–gene interaction studies. We expect that drug–gene interaction models will reduce the heterogeneity between studies and will increase the list of loci affecting the therapeutic response of antidepressants. When ethical issues prevent the use of a placebo-reference population especially in studies to be conducted in severe cases of MDD, studies can selected different reference population in their study design, including a reference population on psychotherapy or on different antidepressant drug treatment. Nevertheless, studies should be performed in larger (collaborative) settings to increase the statistical power whatever the composition of the reference population is. Likely, the use of drug–gene interaction models will increase our understanding of the genetic basis of antidepressant response.
Executive summary.
Although many studies have been conducted aiming to identify genetic variants in relation to antidepressant drug response in depression, only few genes have been repeatedly and robustly identified, notably CYP2D6, ABCB1, SLC6A4 and HTR2A.
Important limitations of studies that have been conducted include the used sample size (and thus statistical power), but also the study design itself: the so-called ‘treated-only design’ in which only treated depressive patients are followed in time.
The reduction in depressive symptoms that is observed in a study with a treated-only design can have different origins, including the indication of use, the natural disease course of the depressive indication (independent of drug use) and the true response of the antidepressant drug treatment.
The inclusion of a reference group, such as a placebo-treated population, could disentangle the mix of origins why depressive patients show a decrease in depressive symptoms over time.
The drug–gene interaction model in a placebo-controlled trial investigates what genetic variants associate with antidepressant drug response in antidepressant-treated depressive patients beyond the association that is observed in placebo-treated depressive patients.
Genetic variants associated with ‘antidepressant response’ in both antidepressant-treated depressive patients and placebo-treated depressive patients are unlikely variants involved in antidepressant drug response in depression.
Drug–gene interaction models in prospective cohort studies, in which genetic variants are differently associated with depressive symptoms in antidepressant-treated participants and untreated participants, might be an alternative strategy to identify genetic variants of interest for antidepressant drug response in depression.
Future studies might benefit to investigate drug–gene interactions instead of main effects to identify genes for antidepressant drug response in depression.
Footnotes
Disclaimer
The funding agencies had no role in the preparation, review or approval of the manuscript.
Financial & competing interests disclosure
CL Avery and BH Stricker acknowledges funding from R01HL103612 (NIH). CL Avery also acknowledges funding from 15GPSPG239 (American Heart Association). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Hasin DS, Goodwin RD, Stinson FS, Grant BF. Epidemiology of major depressive disorder: results from the National Epidemiologic Survey on Alcoholism and Related Conditions. Arch. Gen. Psychiatry. 2005;62(10):1097–1106. doi: 10.1001/archpsyc.62.10.1097. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289(23):3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 3.Patten SB, Wang JL, Williams JV, et al. Descriptive epidemiology of major depression in Canada. Can. J. Psychiatry. 2006;51(2):84–90. doi: 10.1177/070674370605100204. [DOI] [PubMed] [Google Scholar]
- 4.Angst J. The epidemiology of depressive disorders. Eur. Neuropsychopharmacol. 1995;5(Suppl.):95–98. doi: 10.1016/0924-977x(95)00025-k. [DOI] [PubMed] [Google Scholar]
- 5.Luijendijk HJ, Van Den Berg JF, Dekker MJ, et al. Incidence and recurrence of late-life depression. Arch. Gen. Psychiatry. 2008;65(12):1394–1401. doi: 10.1001/archpsyc.65.12.1394. [DOI] [PubMed] [Google Scholar]
- 6.Noordam R, Aarts N, Verhamme KM, Sturkenboom MC, Stricker BH, Visser LE. Prescription and indication trends of antidepressant drugs in the Netherlands between 1996 and 2012: a dynamic population-based study. Eur. J. Clin. Pharmacol. 2015;71(3):369–375. doi: 10.1007/s00228-014-1803-x. [DOI] [PubMed] [Google Scholar]
- 7.Trivedi MH, Rush AJ, Wisniewski SR, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am. J. Psychiatry. 2006;163(1):28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 8.Franchini L, Serretti A, Gasperini M, Smeraldi E. Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J. Psychiatr. Res. 1998;32(5):255–259. doi: 10.1016/S0022-3956(98)00004-1. [DOI] [PubMed] [Google Scholar]
- 9.Tansey KE, Guipponi M, Hu X, et al. Contribution of common genetic variants to antidepressant response. Biol. Psychiatry. 2013;73(7):679–682. doi: 10.1016/j.biopsych.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 10.O'Reilly RL, Bogue L, Singh SM. Pharmacogenetic response to antidepressants in a multicase family with affective disorder. Biol. Psychiatry. 1994;36(7):467–471. doi: 10.1016/0006-3223(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 11.Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. Pharmacol. Ther. 2009;124(1):57–73. doi: 10.1016/j.pharmthera.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol. Psychiatry. 2010;15(5):473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- 13.Arias B, Fabbri C, Gressier F, et al. TPH1, MAOA sero-tonin receptor 2A and 2C genes in citalopram response: possible effect in melancholic and psychotic depression. Neuropsychobiology. 2013;67(1):41–47. doi: 10.1159/000343388. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Huang Y, Li J, et al. Association between the 5-HT1A receptor gene polymorphism (rs6295) and antidepressants: a meta-analysis. Int. Clin. Psychopharmacol. 2012;27(6):314–320. doi: 10.1097/YIC.0b013e32835818bf. [DOI] [PubMed] [Google Scholar]
- 15.Noordam R, Aarts N, Hofman A, Van Schaik RH, Stricker BH, Visser LE. Association between genetic variation in the ABCB1 gene and switching, discontinuation, and dosage of antidepressant therapy: results from the Rotterdam Study. J. Clin. Psychopharmacol. 2013;33(4):546–550. doi: 10.1097/JCP.0b013e318291c07b. [DOI] [PubMed] [Google Scholar]
- 16.Bijl MJ, Visser LE, Hofman A, et al. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br. J. Clin. Pharmacol. 2008;65(4):558–564. doi: 10.1111/j.1365-2125.2007.03052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bet PM, Verbeek EC, Milaneschi Y, et al. A common polymorphism in the ABCB1 gene is associated with side effects of PGP-dependent antidepressants in a large naturalistic Dutch cohort. Pharmacogenomics J. 2016;16(2):202–208. doi: 10.1038/tpj.2015.38. [DOI] [PubMed] [Google Scholar]
- 18.Lin KM, Chiu YF, Tsai IJ, et al. ABCB1 gene polymor-phisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet. Genomics. 2011;21(4):163–170. doi: 10.1097/FPC.0b013e32833db216. [DOI] [PubMed] [Google Scholar]
- 19.De Luca V, Mundo E, Trakalo J, Wong GW, Kennedy JL. Investigation of polymorphism in the MDR1 gene and antidepressant-induced mania. Pharmacogenomics J. 2003;3(5):297–299. doi: 10.1038/sj.tpj.6500196. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson K, Tansey K, Dernovsek MZ, et al. Genetic differences in cytochrome P450 enzymes and antidepressant treatment response. J. Psychopharmacol. 2014;28(2):133–141. doi: 10.1177/0269881113512041. [DOI] [PubMed] [Google Scholar]
- 21.Peters EJ, Slager SL, Kraft JB, et al. Pharmacokinetic genes do not influence response or tolerance to citalopram in the STAR*D sample. PLoS ONE. 2008;3(4):e1872. doi: 10.1371/journal.pone.0001872. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Meta-analysis of the most commonly studied genetic markers for antidepressant drug response.
- 22.Lin JY, Jiang MY, Kan ZM, Chu Y. Influence of 5-HTR2A genetic polymorphisms on the efficacy of antidepressants in the treatment of major depressive disorder: a meta-analysis. J. Affect Disord. 2014;168:430–438. doi: 10.1016/j.jad.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Ising M, Lucae S, Binder EB, et al. A genome-wide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch. Gen. Psychiatry. 2009;66(9):966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uher R, Perroud N, Ng MY, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am. J. Psychiatry. 2010;167(5):555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 25.Garriock HA, Kraft JB, Shyn SI, et al. A genomewide association study of citalopram response in major depressive disorder. Biol. Psychiatry. 2010;67(2):133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendep Investigators, Mars Investigators, STAR*D. Investigators. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am. J. Psychiatry. 2013;170(2):207–217. doi: 10.1176/appi.ajp.2012.12020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biernacka JM, Sangkuhl K, Jenkins G, et al. The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl. Psychiatry. 2015;5:e553. doi: 10.1038/tp.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan A, Go MJ, Zhang W, et al. Genome-wide transancestry meta-analysis provides insight into the genetic architecture of Type 2 diabetes susceptibility. Nat. Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17(R2):R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y, Schaid DJ, Desta Z, et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br. J. Clin. Pharmacol. 2014;78(2):373–383. doi: 10.1111/bcp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teichert M, Eijgelsheim M, Rivadeneira F, et al. A genome-wide association study of acenocoumarol maintenance dosage. Hum. Mol. Genet. 2009;18(19):3758–3768. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- 33.Teichert M, Eijgelsheim M, Uitterlinden AG, et al. Dependency of phen-procoumon dosage on polymorphisms in the VKORC1, CYP2C9 and CYP4F2 genes. Pharmacogenet. Genomics. 2011;21(1):26–34. doi: 10.1097/FPC.0b013e32834154fb. [DOI] [PubMed] [Google Scholar]
- 34.Giri AK, Khan NM, Grover S, et al. Genetic epidemiology of pharmacogenetic variations in CYP2C9, CYP4F2 and VKORC1 genes associated with warfarin dosage in the Indian population. Pharmacogenomics. 2014;15(10):1337–1354. doi: 10.2217/pgs.14.88. [DOI] [PubMed] [Google Scholar]
- 35.Avery CL, Der JS, Whitsel EA, Sturmer T. Comparison of study designs used to detect and characterize pharmacogenomic interactions in nonexperimental studies: a simulation study. Pharmacogenet. Genomics. 2014;24(3):146–155. doi: 10.1097/FPC.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Simulation study of pharmacogenetic studies with the treated-only design and overall population design.
- 36.Fournier JC, Derubeis RJ, Hollon SD, et al. Antidepressant drug effects and depression severity: a patient-level meta-analysis. JAMA. 2010;303(1):47–53. doi: 10.1001/jama.2009.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Meta-analysis describing that only depressive patients with a severe indication benefit from antidepressant treatment.
- 37.Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, et al. Meta-analyses of genetic studies on major depressive disorder. Mol. Psychiatry. 2008;13(8):772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Ma X, Wang Y, et al. Association of APC and REEP5 gene polymorphisms with major depression disorder and treatment response to antidepressants in a Han Chinese population. Gen. Hosp. Psychiatry. 2012;34(5):571–577. doi: 10.1016/j.genhosppsych.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Khan A, Detke M, Khan SR, Mallinckrodt C. Placebo response and antidepressant clinical trial outcome. J. Nerv. Ment. Dis. 2003;191(4):211–218. doi: 10.1097/01.NMD.0000061144.16176.38. [DOI] [PubMed] [Google Scholar]
- 40.Davis CE. The effect of regression to the mean in epidemiologic and clinical studies. Am. J. Epidemiol. 1976;104(5):493–498. doi: 10.1093/oxfordjournals.aje.a112321. [DOI] [PubMed] [Google Scholar]; • Describes the concept of regression to the mean when studying the pharmacological response of a drug.
- 41.Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circulation. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bis JC, Sitlani C, Irvin R, et al. Drug–gene interactions of anti-hypertensive medications and risk of incident cardiovascular disease: a pharmacogenomics study from the CHARGE consortium. PLoS ONE. 2015;10(10):e0140496. doi: 10.1371/journal.pone.0140496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avery CL, Sitlani CM, Arking DE, et al. Drug–gene interactions and the search for missing heritability: a cross-sectional pharmacogenomics study of the QT interval. Pharmacogenomics J. 2014;14(1):6–13. doi: 10.1038/tpj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitlani CM, Rice KM, Lumley T, et al. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat Med. 2015;34(1):118–130. doi: 10.1002/sim.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a statistical method to conduct a genome-wide drug–gene interaction study with repeated measures.
- 45.Postmus I, Trompet S, Deshmukh HA, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat. Comm. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a pharmacogenetic study on LDL cholesterol lower with statins using a treated-only design, but replicated results with a drug–gene interaction model.
- 46.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepherd J, Blauw GJ, Murphy MB, et al. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360(9346):1623–1630. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 48.Hall KT, Loscalzo J, Kaptchuk TJ. Genetics and the placebo effect: the placebome. Trends Mol. Med. 2015;21(5):285–294. doi: 10.1016/j.molmed.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durham LK, Webb SM, Milos PM, Clary CM, Seymour AB. The serotonin transporter polymorphism, 5HTTLPR is associated with a faster response time to sertraline in an elderly population with major depressive disorder. Psychopharmacology. 2004;174(4):525–529. doi: 10.1007/s00213-003-1562-3. [DOI] [PubMed] [Google Scholar]
- 50.Peters EJ, Slager SL, Mcgrath PJ, Knowles JA, Hamilton SP. Investigation of serotonin-related genes in antidepressant response. Mol. Psychiatry. 2004;9(9):879–889. doi: 10.1038/sj.mp.4001502. [DOI] [PubMed] [Google Scholar]
- 51.Uher R, Huezo-Diaz P, Perroud N, et al. Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J. 2009;9(4):225–233. doi: 10.1038/tpj.2009.12. [DOI] [PubMed] [Google Scholar]
- 52.Hek K, Demirkan A, Lahti J, et al. A genome-wide association study of depressive symptoms. Biol. Psychiatry. 2013;73(7):667–678. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noordam R, Direk N, Sitlani CM, et al. Identifying genetic loci associated with antidepressant drug response with drug–gene interaction models in a population-based study. J. Psychiatr. Res. 2015;62:31–37. doi: 10.1016/j.jpsychires.2015.01.005. [DOI] [PubMed] [Google Scholar]; •• First antidepressant–gene interaction study in a cohort study.
- 54.Hofman A, Brusselle GG, Darwish Murad S, et al. The Rotterdam Study: 2016 objectives and design update. Eur. J. Epidemiol. 2015;30(8):661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lister A, Regan C, Van Zwol J, Van Der Kraak G. Inhibition of egg production in zebrafish by fluoxetine and municipal effluents: a mechanistic evaluation. Aquat. Toxicol. 2009;95(4):320–329. doi: 10.1016/j.aquatox.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Malki K, Uher R, Paya-Cano J, et al. Convergent animal and human evidence suggests a role of PPM1A gene in response to antidepressants. Biol. Psychiatry. 2011;69(4):360–365. doi: 10.1016/j.biopsych.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Helton SG, Lohoff FW. Serotonin pathway polymorphisms and the treatment of major depressive disorder and anxiety disorders. Pharmacogenomics. 2015;16(5):541–553. doi: 10.2217/pgs.15.15. [DOI] [PubMed] [Google Scholar]
- 58.Sangkuhl K, Klein TE, Altman RB. Selective serotonin reuptake inhibitors pathway. Pharmacogenet. Genomics. 2009;19(11):907–909. doi: 10.1097/FPC.0b013e32833132cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Athanasiou M, Holick K, Reed C, Salisbury B, Serretti A, Zou W. Genetic markers associated with reponse to antidepressants (US 20090233942 A1) 2009. Patent.
- 60.Laje G, Mcmahon FJ. Genome-wide association studies of antidepressant outcome: a brief review. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35(7):1553–1557. doi: 10.1016/j.pnpbp.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]