Abstract
Natural antisense transcripts (NATs) can interfere with the expression of complementary sense transcripts with exquisite specificity. We have previously cloned NATs of Slc34a loci (encoding Na-phosphate transporters) from fish and mouse. Here we report the cloning of a human SLC34A1-related NAT that represents an alternatively spliced PFN3 transcript (Profilin3). The transcript is predominantly expressed in testis. Phylogenetic comparison suggests two distinct mechanisms producing Slc34a-related NATs: Alternative splicing of a transcript from a protein coding downstream gene (Pfn3, human/mouse) and transcription from the bi-directional promoter (Rbpja, zebrafish). Expression analysis suggested independent regulation of the complementary Slc34a mRNAs. Analysis of randomly selected bi-directionally transcribed human/mouse loci revealed limited phylogenetic conservation and independent regulation of NATs. They were reduced on X chromosomes and clustered in regions that escape inactivation. Locus structure and expression pattern suggest a NATs-associated regulatory mechanisms in testis unrelated to the physiological role of the sense transcript encoded protein.
Keywords: Natural antisense transcript, Evolution, Na-phosphate transport, Human, Mouse
Highlights
-
•
Some Slc34a genes encoding Na-phosphate transporters are transcribed in both directions.
-
•
We report the cloning of a human SLC34A1-related antisense transcript.
-
•
SLC34A1 sense and antisense transcripts show complementary expression in kidney and testis, respectively. This suggests that the antisense transcript is not directly involved in the physiological regulation of the Na-phosphate transporter.
-
•
We report a detailed comparison of natural antisense transcripts in human and mouse.
1. Introduction
Natural antisense transcripts (NATs) are long regulatory RNAs that are transcribed in the opposite direction of protein-coding transcripts and display extended sequence complementarity [1]. A regulatory cascade can unfold in cis at the bi-directionally transcribed locus or in trans via base-paring of the processed, complementary sense/antisense transcripts. Various regulatory mechanisms involving natural antisense transcripts have been identified, remarkably in genes that, apart from being bi-directionally transcribed, have not much in common. The genome-wide scale of antisense transcription became first apparent in large scale cDNA sequencing projects and bioinformatics approaches [2], [3], [4]. It is now recognized that a significant proportion of mammalian genes are transcribed in both directions; the estimates for a particular species depend crucially on sequencing depth and annotation quality of the relevant genomes and transcriptomes [5]. For both human and mouse there is evidence that 50% or more of all genomic loci are bi-directionally transcribed [2].
Bi-directional transcription is found in the genomes of most organisms, however, species specific regulatory strategies have evolved. In prokaryotes antisense transcripts act post-transcriptionally and interfere with secondary structures of complementary RNAs. These interactions ‘melt’ either inhibitory or stimulatory complexes leading to enhanced or repressed gene expression. An additional well documented regulatory mechanism involves so-called ‘kissing complexes’ with short RNA-RNA interactions [6]. Many bacterial NATs have been characterized in detail and are extensively reviewed [7].
In eukaryotes sense-antisense interactions involve extended RNA-RNA complementarity. The potential formation of double-stranded RNA (dsRNA) resembles viral structures and poses a deadly threat to the cell [8]. Therefore, co-expression of sense-and antisense transcripts may trigger a defensive response rather than a regulatory cascade. Specific strategies have evolved to mitigate this dilemma: Plants, for example have entered a veritable arms race with viruses involving components of the RNA interference pathway [9]. In addition, endogenous siRNAs originating from sense-antisense duplexes are abundant and readily detectable in plant transcriptome sequencing projects. Vertebrates, on the other hand, fight viral structures using a dsRNA-specific kinase that elicits an immune response [10]. Moreover, dsRNA is an essential intermediate in genome defence strategies against transposable elements. Both pathways are well described [11]. In contrast, the biological relevance of dsRNA formed between sense-and antisense transcripts in vertebrates is still poorly understood and only few examples have been studied in detail [12], [13], [14].
NATs in animals show development and tissue specific regulation with significant enrichment in testis and brain [15], [16]. Several lines of evidence confirm a co-expression of sense and antisense transcripts, though the NAT is generally expressed at a much lower level than the protein coding sense RNA. Whether the complementary transcripts form dsRNA is controversial: The evidence is either indirect or blurred through experimental complications (see Discussion). Nonetheless, it is plausible that sense-antisense dsRNA structures are being formed in animal cells, likely during small developmental windows, in specific cell types or in particular cellular compartments. An interesting genome-wide feature represents the significant under-representation of NATs on the mouse and human X chromosome, which may be related to the generally mono-allelic expression of X chromosomal genes [3], [4].
The Slc34a gene family encodes epithelial Na-phosphate transporters that are essential in maintaining homeostasis of inorganic phosphate in most animals [17]. Their expression is tightly controlled by hormones and metabolic factors. Many Slc34a genes are transcribed in both orientations resulting in natural antisense transcripts complementary to a few exons of the protein-coding sense transcript. The Slc34a-related NATs characterized so far are fully processed, however, different start sites and splicing strategies have evolved; the common denominator appears to be substantial exonic complementarity [18], [19], [20]. Here, we report the cloning of a human SLC34A1 antisense transcript. We use the publicly available databases to characterize the transcript and trace its evolutionary development. Moreover, we compare these findings to other well characterized sense/antisense pairs to gain insights into common patterns of gene regulation by NATs.
2. Material and methods
2.1. Molecular biology
Human testis RNA was purchased from Clontech/TaKaRa and used for 3′RACE (rapid amplification of cDNA ends) and RT-PCR without further purification. 3′RACE was performed using an oligo dT adaptor primer and 3 nested primers that all localize to the 5′ end of the PFN3 gene. The resulting fragment (780 bp) was cloned using the pGEM®-T vector system (Promega) and positive clones were identified by colony PCR. The sequence of the insert was determined by Sanger sequencing (Genevision, Newcastle, UK).
HEK293 cells were a kind gift from Dr. Nicolas Watkins (Newcastle University) and grown according to standard protocols. Total RNA was extracted using Trizol (Invitrogen) and treated with DNaseI (Thermo Scientific). Approximately 0.5 μg of total RNA was used for reverse transcription using the Omniscript kit (Qiagen). RNA and random hexamers (2.5 μM) were denatured for 3 min at 70 °C and cooled to 37 °C. Polymerase, dNTPs, buffer and RNase inhibitor were added as detailed by the supplier. The final reaction was denatured for 2 min at 95 °C and stored at − 20 °C. Gene specific PrimeTime® qPCR primers and probes (Integrated DNA Technologies) were used to amplify a specific product from 0.5 μl of the RT reaction. 1 × Lightcycler 480 Probes Master mix (Roche) and with the cycling protocol: initial denaturation (95 °C for 10 min), 45 cycles of 95 °C for 5 s, 55 °C for 20 s and 72 °C for 1 s when fluorescence was determined. The sequence of all primers and probes are given as supplementary material (Supplementary Table 1).
2.2. Bioinformatics
For the phylogenetic analysis of human and mouse bi-directionally transcribed genes the Ensmbl genome browser was used with the Ensembl-Havana human GENCODE gene set (release 24) and mouse (release 83). The loci were selected by manually picking random areas (10 on each autosome) and selecting the closest pair of complementary transcripts. Protein coding and processed transcripts were included, pseudogenes and RNA genes excluded. Transcripts were deemed to be complementary when 20 nucleotides or more were in common, with the assumption that RNA duplexes of > 20 bp are likely to be stable under physiological conditions and biologically active. The loci were visually categorized with respect to overlap pattern (5′, 3′ or mid, Fig. 3). The EBI gene expression atlas was used to determine level and expression pattern of the individual transcripts (http://www.ebi.ac.uk/gxa/home). Peak expression as given by the expression atlas was used for the comparative analysis. (This choice was prior validated for 16 human and 8 mouse genes by normalizing individual data sets to the highest value (100%); the averaged expression levels were then used to calculate a scaling factor for comparison between sense and antisense expression. The expression ranking and pattern was identical for both approaches.) The expression pattern was manually assessed and grouped into ‘brain’, ‘testis’ and ‘others’. The two highest expressing tissues were selected (only one if the second highest expression was < 60% of the highest value) and colour-coded in an excel table (Fig. 4, Supplementary Table 2). To estimate expression patterns, a value of two was given to transcripts expressed in a single tissue and a value of one each if the transcript was found at high levels (> 60% of peak expression) in two or more tissues. This approach was validated using the scaled expression levels of 24 transcripts. All these data were compiled in spread sheets and excel functions were used to compare and sort the data according to the parameters indicated in the Results section. The names of genes that escape X chromosome inactivation were downloaded from Zhang et al. [21] and Berletch et al. [22].
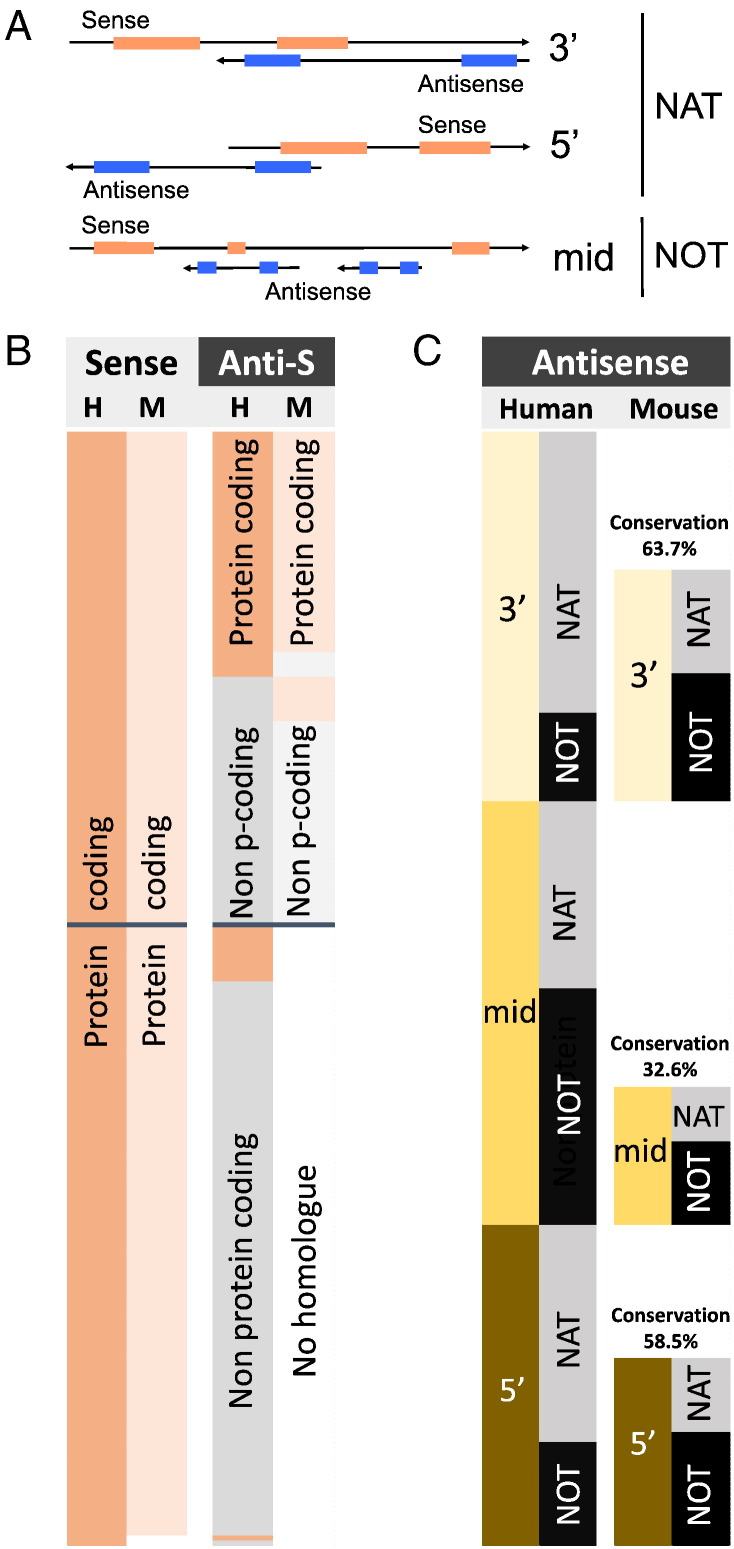
Fig. 3.
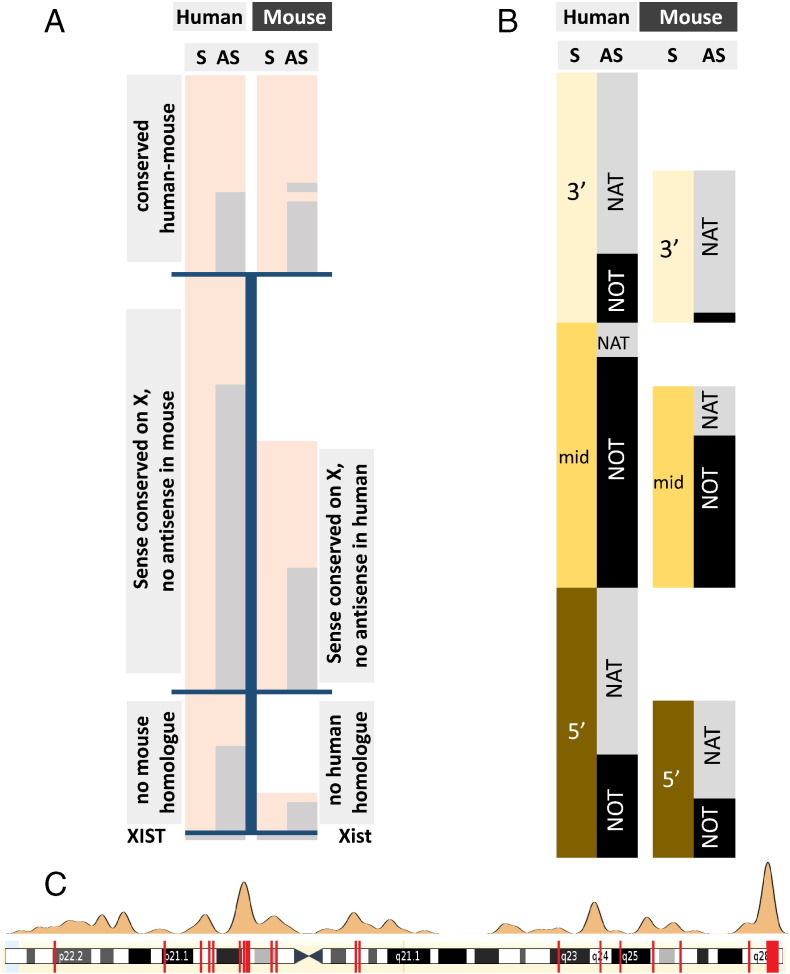
Natural antisense transcripts on autosomes of human (H) and mouse (M). A) Classification of sense-antisense overlapping structures including 3′ (tail-to-tail), mid (full overlap) and 5′ (head-to-head). Inclusion into the dataset required complementarity of > 20 nucleotides. B) Expression pattern of sense and antisense transcripts and their phylogenetic conservation. Peach and light peach indicate protein coding potential in human and mouse, respectively. Protein coding sense transcripts are well conserved between human and mouse (left two columns), the antisense transcripts to a much lesser extent (right columns) B) Comparison of the genomic arrangement of bi-directionally transcribed genes in human and mouse. 3′, mid and 5′ indicates the position of complementarity between sense and antisense transcripts. NAT stands for an antisense transcript with complementarity to the sense RNA; NOT indicates a non-overlapping configuration. S = sense, AS = antisense.
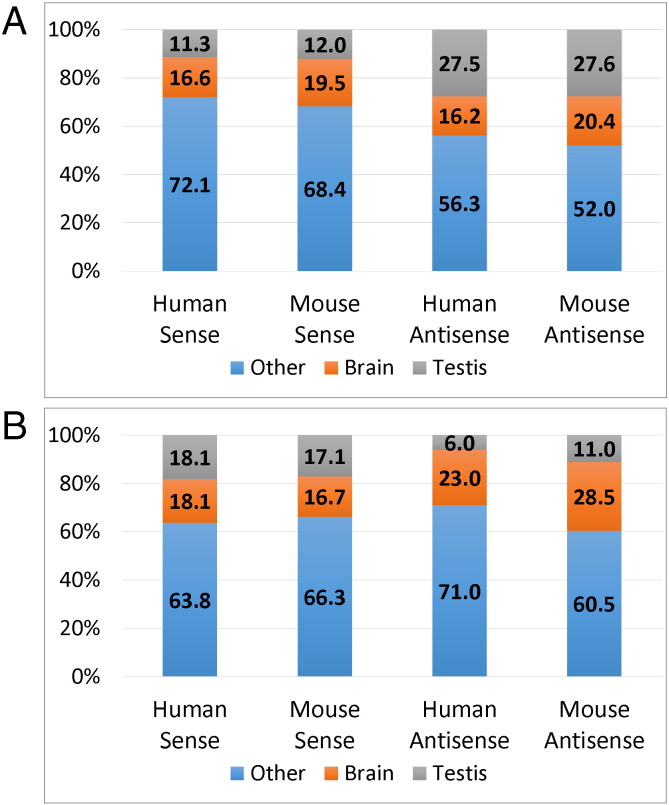
Fig. 4.
Schematic expression pattern of sense and antisense transcripts in human and mice. Expression data of individual genes was extracted from the gene expression atlas (http://www.ebi.ac.uk/gxa/home), scored and grouped into brain, testis and other tissues. A) Genes on autosomes including sense and antisense transcripts, B) Bi-directional X-chromosomal genes of human and mice.
3. Results
3.1. Cloning of the human SLC34A1 antisense transcript
To demonstrate the expression of an SLC34A1-related antisense transcript we used total RNA from human testis; we chose this tissue because NATs generally show highest expression levels in mammalian testis, including the mouse homologue of the candidate gene [18]. The fact that the mouse NAT represents an alternative splice product of a Pfn3 transcript further informed our search strategy (Fig. 1A). All cloned SLC34A-related NATs (from flounder, zebrafish and mouse [18], [19], [20]) are relatively short and alternatively spliced. We therefore applied3′ rapid amplification of cDNA ends (3′RACE) in combination with PFN3 specific primers to amplify fragments of the human SLC34A1 NAT. Three rounds of PCR with one specific primer each were necessary to amplify a specific fragment. Sanger sequencing revealed a transcript of 780 nucleotides containing 3 exons. The first one represents the start of PFN3 and the second and third one are complementary to exon 13 of SLC34A1 (Fig. 1A). A short open reading frame of 63 amino acids contains 47 N-terminal residues of Profilin 3 and 16 amino acids encoded by the noncoding 3′ end of the SLC34A1 transcript.
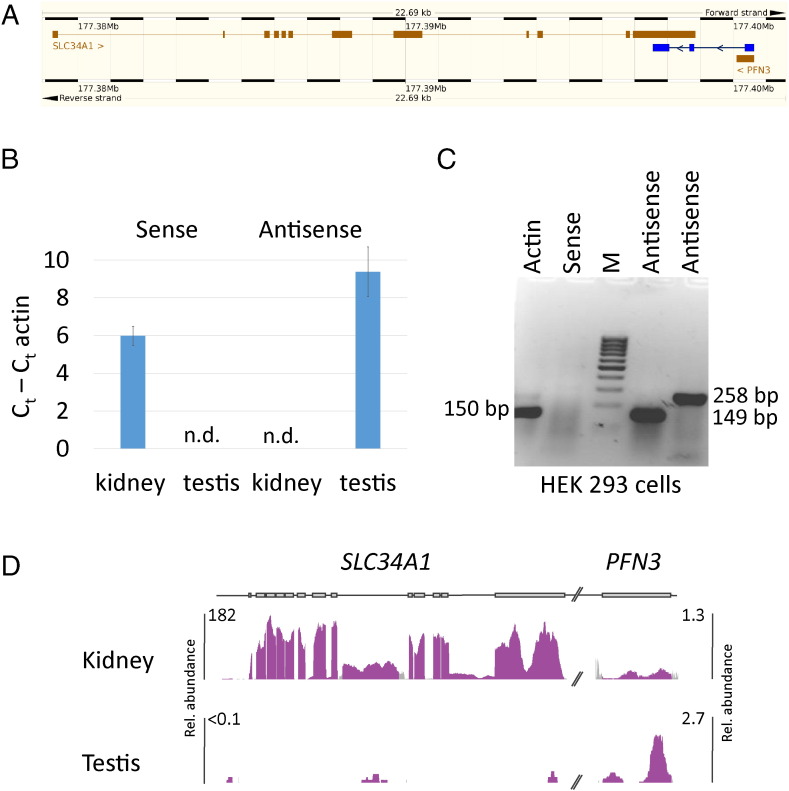
Fig. 1.
The natural antisense transcript of SLC34A1. A) Genomic arrangement of the SLC34A1 antisense transcript, representing an alternative splice form of PFN3 (in blue). B) RT-qPCR of SLC34A1 sense and antisense transcripts. C) RT-PCR with SLC34A1 specific primers using HEK293 total RNA. Two different primer pairs for the antisense transcript were used (Supplementary Table 1). D) Representation of RNA-Seq data from human protein atlas. (For interpretation of the references to colour in this figure, the reader is referred to the web version of this article.)
The natural antisense transcript of SLC34A1. A) Genomic arrangement of the SLC34A1 antisense transcript, representing an alternative splice form of PFN3 (in blue). B) RT-qPCR of SLC34A1 sense and antisense transcripts. C) RT-PCR with SLC34A1 specific primers using HEK293 total RNA. Two different primer pairs for the antisense transcript were used (Supplementary Table 1). D) Representation of RNA-Seq data from human protein atlas.
RT-qPCR, RT-PCR and publicly available expression data (http://www.proteinatlas.org) were used to investigate the expression pattern of both SLC34A1-related sense and antisense transcripts (Figs. 1B–D). Both transcripts showed a very restricted expression pattern with the expected dominant expression of the sense transcript in kidney, traces were found in testis but none in other tissues. The antisense transcript was found in testis at a low and in kidney at a hardly detectable level. Interestingly, the antisense transcript was also expressed in HEK293 cells without detectable levels of the protein coding sense RNA. We were unable to localize the SLC34A1 antisense transcript on human kidney sections by in situ hybridization (not shown).
3.2. Phylogeny of Slc34a-related antisense transcripts
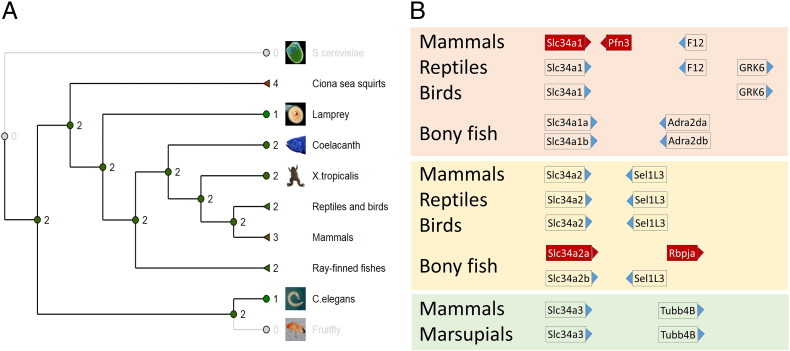
Antisense transcripts have been identified and cloned from different Slc34a isoforms. We used the comparative genomics tools from Ensembl to build a phylogenetic tree of Slc34a (Fig. 2A) and to visualize the region downstream where potential antisense transcripts may originate (Fig. 2B). The Slc34a gene family initially comprised two members in the animal kingdom, denoted Slc34a1 and Slc34a2. Through contractions and gene duplications lineage specific genotypes have evolved. For example, in mammals a third isoforms is expressed in kidney, Slc34a3, and the protein shows distinct functional properties. On the other hand, a documented gene duplication in zebrafish has resulted in two Slc34a1 and two Slc34a2 isoforms. The gene arrangements downstream of the Slc34a loci predict that the Slc34a-linked antisense transcripts cloned and analyed to date (Slc34a1 in mammals and Slc34a2 in fish) have different origins: The Slc34a1 related NATs derive from the Pfn3 promoter and this gene is only found in mammals and marsupials. Antisense transcripts complementary to Slc34a2 from zebrafish are thought to be driven by the bi-directional Rbpja promoter. The NAT-relevant promoter in winter flounder has not been defined [19]. The arrangement of genes around flounder Slc34a2 suggests that the antisense transcript is either an alternatively spliced Sel1L3 product or derives from an undefined promoter between Slc34a2 and Sel1L3. The apparently rather random emergence of the Slc34a antisense transcripts in contrast to the well-conserved sense transcript and the established physiological role of the related transport proteins implies that the sense and antisense transcripts follow distinct evolutionary agendas. Specifically, phosphate is essential for growth and its levels are tightly regulated, in vertebrates by hormonally controlled expression of Slc34a encoded proteins. At any point, testis is neither directly nor indirectly involved in maintaining phosphate homeostasis. This finding raises two questions; first, if the antisense transcripts is unlikely to modulate the physiological functioning of the sense encoded protein, what else does it do; and second, to which extend is the situation in Slc34a relevant to other bi-directionally transcribed genes?
Fig. 2.
Phylogenetic tree of Slc34A genes of Na-phosphate transporters. A) Phylogenetic tree with the number of isoforms indicated. B) The tree isoforms of mammalian Slc34A, Slc34A1 (peach), Slc34A2 (yellow) and Slc34A3 (green), and the genes downstream that are relevant to potential antisense transcription. Slc34A isoforms including the downstream genes with confirmed expression of antisense transcripts are in red.
3.3. Conservation of bi-directionally transcribed loci
The expression pattern as well as mechanistic considerations can provide clues to identify putative biological functions of sense and antisense transcript pairs. There are a number of mechanisms relating to natural antisense transcripts and particularly double-stranded RNA which have been discussed extensively and reviewed in detail [23], [24]. RNA editing, RNA interference and RNA masking involve dsRNA intermediates whereas transcriptional interference and chromatin changes may not require RNA-RNA pairing. We selected 10 bi-directionally transcribed loci that were previously characterized in detail and collected a number of coordinates from public databases including coding potential of the transcripts, expression patterns and phylogenetic conservation (Table 1). The selection is rather small because investigations into the mechanism of gene regulation by NATs in animals are limited. Nevertheless, the data confirm published findings that NATs are generally expressed at a low level poorly conserved. The observation that only one of 14 antisense transcripts is structurally conserved (intron/exon structure, not just antisense transcription per se) suggests that the overall structure of these transcripts is not essential for a potential biological function. All the reports that included mechanistic studies found evidence of dsRNA formation between sense and antisense transcripts; those who did not, found either no exonic complementarity within the locus (RUNX2/SUPT3H) or did not investigate the question. Moreover, the expression profile of the various transcripts suggested an accumulation of lowly expressed NATs in testis. The importance of a dsRNA intermediates is corroborated by a recent report that detected significant levels of sense-antisense hybrids in human cells [12].
Table 1.
Compilation of well-studied bi-directionally transcribed genes. Peach-coloured transcripts are protein coding, yellow indicates a mode of action that involves dsRNA formation. Expression levels and patterns are deduced from the EBI expression atlas (http://www.ebi.ac.uk/gxa/home). The proposed mechanism is informed by the indicated papers.
| Gene name | Expression | Antisense gene name | Expression | Overlap | Structure | Expression AS/S | Expression pattern sense | Expression pattern antisense | Proposed mechanism | Synteny human-mouse, sense | Synteny human-mouse, antisense | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BACE1 (8) | 47 | BACE1-AS | 12 | Mid | Multiple exons | NAT | 0.26 | Brain, pancreas | Brain, multiple | miRNA masking | Conserved | nd |
| BACE1 | 47 | RNF214 | 14 | 5′ | 1st exon | NAT | 0.30 | Brain, pancreas | Testis, multiple | Not determined | Conserved | Not conserved |
| WDR83 | 16 | WDR83-OS | 259 | Mid | Multiple exons | NAT | 16.19 | Brain, testis, multiple | Multiple | Not determined | Conserved | Not conserved |
| WDR83 (27) | 16 | DHPS | 118 | 5′ | 1st exon | NAT | 7.38 | Brain, testis, multiple | Brain, multiple | dsRNA + stability | Conserved | Not conserved |
| DHRS4 (17) | 49 | DHRS4-AS1 | 22 | Mid, 5′ | 1st exon | NAT | 0.45 | Multiple | Multiple | dsRNA, Me/histone | nd | nd |
| HAS2 (20, 30) | 12 | HAS2-AS | 3 | 5′ | 1st exon | NAT | 0.25 | Digestive tract, testis | Testis, digestive tract | dsRNA, + stability/chromatin | Conserved | nd |
| HIF1a (29) | 184 | HIF1a-AS1 | 4 | 5′ | 1st exon | NAT | 0.02 | Multiple | Multiple | + stability | Conserved | nd |
| TP53 (23) | 63 | WRAP53 | 5 | 5′ | 1st exon | NAT | 0.08 | Multiple | Testis, multiple | dsRNA, + stability | Conserved | Not conserved |
| PINK1 (24, 25) | 31 | PINK1-AS | 3 | 3′ | Multiple exons | NAT | 0.10 | Testis, multiple | Multiple | Concordant regulation | Conserved (?) | nd |
| RUNX2 (1) | 7 | SUPT3H | 62 | 5′ | Intronic | NOT | 8.86 | Immune system, epithelia | Brain, multiple | Promoter interaction | Conserved | conserved |
| RUNX2 | 7 | RP1-244F24 | 3 | Mid | Intronic | NOT | 0.43 | Immune system, epithelia | Multiple, glands | Not determined | Conserved | nd |
| SCN1A | 14 | AC010127.3 | 1 | Mid | Multiple exons | NAT | 0.07 | Brain | Testis | Conserved | nd | |
| SCN9A (16) | 4 | AC010127.3 | 1 | Mid | Multiple exons | NAT | 0.25 | Brain | Testis | Discordant | Conserved | nd |
| SLC34A1 (5) | 219 | PFN3 | 39 | 3′ | Multiple exons | NAT | 0.19 | Kidney | Testis | siRNA, discordant | Conserved | Not conserved\ |
In order to test the importance of dsRNA formation at a medium scale we compiled a random list of 226 human bi-directionally transcribed genes. The only requirement was complementarity of 20 or more base pairs of the primary transcripts. These transcripts were then catalogued with respect to their genomic arrangement, exonic complementarity and expression pattern (Supplementary Table 2). As demonstrated in Fig. 3B, natural antisense transcripts were found to be predominantly non-protein-coding (72.6%, compare column 1 and column 3). The genomic arrangement showed approximately equal proportions of 3′, mid and 5′ overlapping antisense transcripts (Fig. 3C). The proportion of antisense transcripts with exonic complementarity (NATs) to purely intronic transcripts (NOTs) was about 2:1 (62%/38%). Complementarity between sense and antisense transcripts was predominantly found at the 5′ and the 3′ end of genes (first and last exon, respectively), whereas antisense transcripts embedded within the sense encoding gene tend to be non-overlapping (NOTs) (Fig. 3C, compare columns 1 and 2). Our findings are in line with previously published large scale assessments of the human antisense transcriptome [3].
We then assessed the phylogenetic conversation of the human loci in mouse and compiled the same set of parameters (genomic arrangement and expression pattern). Synteny was found for all but one locus (MMP26) and most of the (sense) transcripts show a similar expression levels in both human and mouse (Fig. 3A, r = 0.851 in Supplementary Fig. 1). In contrast, antisense transcript expression was neither correlated between human and mice nor in relation to the cognate sense transcripts (Supplementary Fig. 1). Only 50% (113) of the syntenic mouse loci showed evidence of bi-directional transcription (Fig. 3B, columns 3 and 4). As expected, protein coding antisense transcripts were more likely to be conserved (91.9% protein coding versus 34.1% non-protein coding, respectively, Fig. 3B, columns 3 and 4), particularly at the 5′ and 3′ end of genes: 58.5 and 63.7% versus 32.6% mid gene (Fig. 3C, columns 3 and 4). When the genomic organisation was taken into account, only 37 (33%) antisense transcripts displayed comparable features (exonic complementarity/location of the antisense transcript relative to the sense transcript) between mouse and human. The summarized expression pattern in brain/testis/other tissues of the transcripts was comparable between mouse and human and, for both species antisense transcripts were enriched in testis (Fig. 4A).
The significant expression of antisense transcripts in testis points to a biological function of – at least some – antisense transcripts that is not directly related to the sense encoded protein function. Our results also confirm previous reports that NATs are structurally not well conserved between species [25].
3.4. Antisense transcripts on the X chromosome
In an evolutionary context, bi-directionally transcribed genes on human and murine X chromosomes are of particular interest: Transposable elements, which drive the evolution of new genetic elements and are also linked to the emergence of antisense transcripts [26] are significantly enriched on mammalian sex chromosomes [27]. Despite the high transposon density the mammalian X chromosome show a significant reduction in NATs as compared to the density on autosomes [3], [4]. We argued that evolutionary conservation of a complementary gene arrangement on the X chromosome (where antisense transcripts are negatively selected) would provide support for a regulatory impact of the antisense transcript on the sense-encoded protein function. We catalogued bi-directionally transcribed loci on both human and mouse X chromosome as described above. We found 133 (16.1% of X-linked genes) bi-directionally transcribed loci in human and 93 (9.9% of X-linked genes) in mouse, respectively. These findings confirm the previously reported bias against complementary transcripts on mammalian X chromosomes. Interestingly, the expression pattern of sense transcripts was comparable to autosomal genes in both human and mice; X-encoded antisense transcripts, however, showed a tendency against expression in testis (contrary to autosomal encoded NATs, Fig. 4B). Synteny was detected for about 90% of the bi-directionally transcribed loci in both human-mouse (83.5%) and mouse-human direction (91.4%; Fig. 5A, compare the sum of the upper two panels with the bottom panel), which is lower than the almost complete (> 99.5%) synteny of autosomal genes. Bi-allelic transcription was conserved in 42 loci (Fig. 5A, top) 68 loci are specific to human, 43 to mouse (Fig. 5A, middle panel). In human, the proportion of non-protein coding to coding antisense transcripts was 35.8%, in mouse 51.4% respectively (sum of grey bar areas in Fig. 5A). Analysis of the genomic arrangement revealed a clear bias of complementarity between sense and antisense at the 3′ and the 5′ end of genes whereas non-complementary arrangements were enriched in the middle (Fig. 5B). The observation was made in both human and mouse.
Fig. 5.
Natural antisense transcripts on X chromosomes of human and mouse. A) Phylogenetic conservation of sense and antisense transcripts. Top panel, conserved bi-directional locus between human and mouse; middle panel, locus conserved but no antisense transcript in the other species; homologue in the other species. Light peach indicates protein coding potential, S = sense, AS = antisense. The nature of synteny is described in the grey boxes. B) Comparison of the genomic arrangement of bi-directionally transcribed genes on the X chromosome of human and mouse. 3′, mid and 5′ indicates the position of complementarity between sense and antisense transcript. NAT stands for an antisense transcript with complementarity to the sense RNA; NOT indicates a non-overlapping configuration. C) Natural antisense transcripts in regions that escape X chromosome inactivation. Upper panel indicates areas with relaxed inactivation, the red bars on the lower panel depict the bi-directionally transcribed human genes.
X chromosomal NATs were reported to be clustered in areas that escape X chromosome inactivation [28]. Since this original observation, more comprehensive lists of escapee genes have been published [21], [22]. In human, we could establish a significant overlap between bi-directionally transcribed loci (113, 13.7%) and our gene list: 25 genes or 22.1% of bi-directionally transcribed loci escape and these genes tend to be better conserved (Fig. 5C). In mouse, less genes escape X chromosome inactivation (37, 3.9%) and only 4 were found to be bi-directional and escapee, too few to warrant conclusions.
In summary, our findings point to selective benefits to retain antisense transcripts but also frequent novel incidences of bi-directional transcription in human and mouse lineages [29]. Moreover, the reduced number of bi-directionally transcribed genes on X chromosomes show lesser conservation than autosomal loci (31.6% vs. 50%).
4. Discussion
The scope of natural antisense transcripts is still largely speculative and often indirect evidence such as phylogenetic conservation is used to underpin biological relevance. The task of investigating functionality of NATs is also hampered by the fact that experimental up- or down regulation of either transcript of a sense-antisense pair will almost inevitably interfere with the expression of the counterpart. Therefore, such experiments need to be very carefully controlled and interpreted.
The cloning of a human antisense transcript of the SLC34A1/PFN3 locus that we report in this paper highlights another difficulty in the ‘antisense’ field: Despite ever increasing sequencing depth, the annotation of the antisense transcriptome is still incomplete. The generally low expression of antisense transcripts certainly exacerbates the problem. The material used for RNA extraction is another important factor to be considered; antisense transcripts are predominantly expressed in testis and systematic assessment of bi-directional genes should involve this tissue [30]. Moreover, unless the experimental strategy is aimed at maintaining strand specificity the bioinformatic assessment of RNA-seq data will have a bias against antisense transcripts because antisense reads are undiscernible from reverse transcription artefacts and DNA contamination.
We report the cloning of an alternative splice product of PFN3 that is complementary to the last exon of SLC34A1. Since the two transcripts show a non-overlapping expression pattern, the sense transcript is almost exclusively expressed in kidney whereas the antisense transcript is found in testis, it is unlikely that the latter contributes to the physiological regulation of the Na-phosphate transporter and hence helps maintaining acute phosphate balance. It could, however, be involved in setting chromatin marks in kidney but also during sperm development. We have recently proposed a mechanism based on double stranded RNA formation and endo-siRNA processing in a perspective paper that may explain how epigenetic modifications can be targeted either to the sense or the antisense strand [31]. In that respect it is intriguing, that the kidney cell line HEK293 expresses the antisense transcript but lacks the message for the transporter.
SLC34A-encoded transporters play a central role in maintaining phosphate homeostasis in most animals as demonstrated in various animal models and knock-out experiments [32]. Accordingly, SLC34A isoforms from various species have been functionally assessed and shown to have adapted to specific physiologic/environmental constraints [33]. Bi-directional transcription has been detected in some isoforms but not in others; moreover, there is heterogeneity with respect to the phylogeny of SLC34A-related antisense transcripts: In fish they are driven by a bi-directional (Rbpja) promoter and in mammals they represent a read-through transcript of Pfn3. The unrelated occurrence of NATs in two different SLC34A isoforms suggests that the transcripts are unlikely to contribute to the acute regulation of Na-phosphate transporters. In general, the tissue expression of homologous (protein-coding) sense transcripts is well conserved between human and mouse but not so much the expression pattern of the cognate antisense transcripts (Fig. 3A. Supplementary Fig. 1, Supplementary Table 2). This observation provides additional support to the hypothesis that many antisense transcripts are not involved in the acute physiological regulation of the sense transcript.
The phylogenetic conservation of bi-directional loci between human and mouse has been investigated by others with comparable results [34], [35]. The figures crucially depend on the specific parameters to identify and categorize natural antisense transcripts but also on sequencing depth and annotation. The current deficit of mechanistic insights further complicates a comparative evaluation of antisense transcription. We focused on transcripts with at least 20 nucleotides of complementarity and only included protein coding and non-protein coding processed transcripts but excluded pseudogenes. Bi-directional transcription was conserved between human and mouse in about half of all investigated genes, in good agreement with other reports. Likewise, 3′ and 5′ complementary transcripts were better conserved than intronic, non-overlapping transcripts (NOTs). The slightly enhanced phylogenetic stability of complementary sense-antisense arrangements suggest that double-stranded RNA formation may represent an essential intermediate in an antisense mediated regulatory mechanism, a hypothesis that receives support from recently published mechanistic studies (Table 1).
The under-representation of antisense transcripts and overlapping gene arrangements becomes immediately apparent upon visual inspection of an annotated X chromosome in a genome browser [3], [4]. Comprehensive analysis revealed a slightly higher percentage of bi-directional loci on the human than the mouse X chromosome. The conservation of X-encoded antisense loci was slightly lower as compared to genes on autosomes (human → mouse 32.6%, mouse → human 45.7% versus 50% human → mouse on autosomes), this may reflect a slightly accelerated evolution of the X chromosome as compared to autosomes [36].
X chromosome inactivation in human and mouse show significant mechanistic differences that also result in a significantly higher number of human genes that escape silencing as compared to mouse [37]. Interestingly, we found a significant overlap between X inactivation escape and complementary antisense transcripts in human (potentially forming sense-antisense RNA hybrids). In mouse, numbers were too low to draw significant conclusions. These findings suggest that antisense transcription carries the potential of inducing transcriptional silencing which is potentially detrimental for already monoallelically expressed (X chromosomal) gene [38].
5. Conclusions
We have identified a novel antisense transcript complementary to the human SLC34A1 gene. Expression pattern of the sense and antisense transcripts argue against an involvement of the antisense RNA in the acute regulation of the sense encoded Na-phosphate transporter. Comparative studies between human and mouse bi-directionally transcribed loci indicate generally limited conservation of antisense transcripts. Moreover, one of potential mechanisms involves dsRNA formation that may lead to the establishment of an epigenetic signature and potentially transcriptional gene silencing [38], [39].
Comparative studies between human and mouse have revealed distinct expression signatures of (autosomal and X chromosomal) bi-directionally transcribed loci in testis, indicating that the expression pattern established for Slc34a1 transcripts is not unique. The particular importance of testis in the context of antisense-mediated gene regulation is also supported by expression differences of RNA interference enzymes in testis and somatic tissues [40], [41]. Our results did not identify a bias in human-mouse conservation of overlapping versus non-overlapping sense-antisense transcripts; hence the comparative findings do not strengthen the argument for a mechanism involving dsRNA. The various hallmarks in genomics and expression of antisense transcripts, however, suggest tissue specific biological roles that do not necessarily relate to the sense encoded protein function.
The following are the supplementary data related to this article.
Sequence information of primers and probes.
Compilation of bi-directional genes on autosomes and X-chromosomes.
Comparison of sense and antisense transcripts of human and mice.
Acknowledgment
This work was funded by The Dunhill Medical Trust (SA10/0210).
Footnotes
Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank Data Libraries (Submission 6 May 2016, No KX215110).
References
- 1.Beiter T., Reich E., Williams R.W., Simon P. Antisense transcription: a critical look in both directions. Cell. Mol. Life Sci. 2008;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Suzuki H., Carninci P., Hayashizaki Y., Wells C., Frith M., Ravasi T., Pang K.C., Hallinan J., Mattick J., Hume D.A., Lipovich L., Batalov S., Engstrom P.G., Mizuno Y., Faghihi M.A., Sandelin A., Chalk A.M., Mottagui-Tabar S., Liang Z., Lenhard B., Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. (New York, N.Y) [DOI] [PubMed] [Google Scholar]
- 3.Chen J., Sun M., Kent W.J., Huang X., Xie H., Wang W., Zhou G., Shi R.Z., Rowley J.D. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiyosawa H., Yamanaka I., Osato N., Kondo S., Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yelin R., Dahary D., Sorek R., Levanon E.Y., Goldstein O., Shoshan A., Diber A., Biton S., Tamir Y., Khosravi R., Nemzer S., Pinner E., Walach S., Bernstein J., Savitsky K., Rotman G. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 6.Wagner E.G., Brantl S. Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem. Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner E.G., Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Carmichael G.G. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding S.W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Luo L.F., Hou C.C., Yang W.X. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene. 2016;578:141–157. doi: 10.1016/j.gene.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Portal M.M., Pavet V., Erb C., Gronemeyer H. Human cells contain natural double-stranded RNAs with potential regulatory functions. 2015;22:89–97. doi: 10.1038/nsmb.2934. [DOI] [PubMed] [Google Scholar]
- 13.Werner A., Cockell S., Falconer J., Carlile M., Alnumeir S., Robinson J. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genomics. 2014;15:19. doi: 10.1186/1471-2164-15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling K.H., Brautigan P.J., Moore S., Fraser R., Cheah P.S., Raison J.M., Babic M., Lee Y.K., Daish T., Mattiske D.M., Mann J.R., Adelson D.L., Thomas P.Q., Hahn C.N., Scott H.S. Derivation of an endogenous small RNA from double-stranded Sox4 sense and natural antisense transcripts in the mouse brain. Genomics. 2016;107:88–99. doi: 10.1016/j.ygeno.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Ge X., Rubinstein W.S., Jung Y.C., Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics. 2008;9:27. doi: 10.1186/1471-2164-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner A., Schmutzler G., Carlile M., Miles C.G., Peters H. Expression profiling of antisense transcripts on DNA arrays. Physiol. Genomics. 2007;28:294–300. doi: 10.1152/physiolgenomics.00127.2006. [DOI] [PubMed] [Google Scholar]
- 17.Werner A., Kinne R.K. Evolution of the Na-P(i) cotransport systems. Am. J. Phys. Regul. Integr. Comp. Phys. 2001;280:R301–R312. doi: 10.1152/ajpregu.2001.280.2.R301. [DOI] [PubMed] [Google Scholar]
- 18.Carlile M., Swan D., Jackson K., Preston-Fayers K., Ballester B., Flicek P., Werner A. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res. 2009;37:2274–2282. doi: 10.1093/nar/gkp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huelseweh B., Kohl B., Hentschel H., Kinne R.K., Werner A. Translated anti-sense product of the Na/phosphate co-transporter (NaPi-II) Biochem. J. 1998;332(Pt 2):483–489. doi: 10.1042/bj3320483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nalbant P., Boehmer C., Dehmelt L., Wehner F., Werner A. Functional characterization of a Na +-phosphate cotransporter (NaPi-II) from zebrafish and identification of related transcripts. J. Physiol. 1999;520(Pt 1):79–89. doi: 10.1111/j.1469-7793.1999.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Castillo-Morales A., Jiang M., Zhu Y., Hu L., Urrutia A.O., Kong X., Hurst L.D. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol. Biol. Evol. 2016;33:302. doi: 10.1093/molbev/msv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berletch J.B., Ma W., Yang F., Shendure J., Noble W.S., Disteche C.M., Deng X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magistri M., Faghihi M.A., St Laurent G., 3rd, Wahlestedt C. 2012. Regulation of Chromatin Structure by Long Noncoding RNAs: Focus on Natural Antisense Transcripts. (Trends Genet) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner A., Carlile M., Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 25.Veeramachaneni V., Makalowski W., Galdzicki M., Sood R., Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayarpadikannan S., Lee H.E., Han K., Kim H.S. Transposable element-driven transcript diversification and its relevance to genetic disorders. Gene. 2015;558:187–194. doi: 10.1016/j.gene.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 27.Costa F.F. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Kiyosawa H., Abe K. Speculations on the role of natural antisense transcripts in mammalian X chromosome evolution. Cytogenet. Genome Res. 2002;99:151–156. doi: 10.1159/000071587. [DOI] [PubMed] [Google Scholar]
- 29.Kvikstad E.M., Makova K.D. The (r)evolution of SINE versus LINE distributions in primate genomes: sex chromosomes are important. Genome Res. 2010;20:600–613. doi: 10.1101/gr.099044.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthes P., Kokkinaki M., Nef S., Gnirke A., Dym M., de Massy B., Mikkelsen T.S., Kaessmann H. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 31.Werner A., Piatek M.J., Mattick J.S. Transpositional shuffling and quality control in male germ cells to enhance evolution of complex organisms. Ann. N. Y. Acad. Sci. 2015;1341:156–163. doi: 10.1111/nyas.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biber J., Hernando N., Forster I. Phosphate transporters and their function. Annu. Rev. Physiol. 2013;75:535–550. doi: 10.1146/annurev-physiol-030212-183748. [DOI] [PubMed] [Google Scholar]
- 33.Forster I.C., Hernando N., Biber J., Murer H. Phosphate transporters of the SLC20 and SLC34 families. Mol. Asp. Med. 2013;34:386–395. doi: 10.1016/j.mam.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Wood E.J., Chin-Inmanu K., Jia H., Lipovich L. Sense-antisense gene pairs: sequence, transcription, and structure are not conserved between human and mouse. Front. Genet. 2013;4:183. doi: 10.3389/fgene.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galante P.A., Vidal D.O., de Souza J.E., Camargo A.A., de Souza S.J. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner J.M. Meiotic silencing in mammals. Annu. Rev. Genet. 2015;49:395–412. doi: 10.1146/annurev-genet-112414-055145. [DOI] [PubMed] [Google Scholar]
- 37.Gendrel A.V., Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu. Rev. Cell Dev. Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- 38.Tufarelli C., Stanley J.A., Garrick D., Sharpe J.A., Ayyub H., Wood W.G., Higgs D.R. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 39.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Gonzalez E., Lopez-Casas P.P., del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim. Biophys. Acta. 2008;1779:306–311. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Modzelewski A.J., Holmes R.J., Hilz S., Grimson A., Cohen P.E. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell. 2012;23:251–264. doi: 10.1016/j.devcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence information of primers and probes.
Compilation of bi-directional genes on autosomes and X-chromosomes.
Comparison of sense and antisense transcripts of human and mice.