Abstract
Aims
Somatic mutations in exon 2 of the MED12 gene have been identified in 60% of breast fibroadenomas (FAs). The aim of this study was to define whether phyllodes tumors (PTs) would harbor MED12 somatic mutations in a way akin to FAs.
Methods and results
A collection of 73 fibroepithelial tumors (including 26 FAs, 25 benign PTs, 9 borderline PTs and 13 malignant PTs) from 64 patients was retrieved from the authors' institution. Sections from FFPE blocks were microdissected to ensure an enrichment in neoplastic stromal elements of >70%. DNA samples extracted from tumor and matched normal tissues were subjected to Sanger sequencing of exon 2 of the MED12 gene. MED12 exon 2 somatic mutations, including 28 somatic single nucleotide variants and 19 insertions and deletions, were found in 65%, 88%, 78% and 8% of FAs, benign PTs, borderline PTs and malignant PTs, respectively. Malignant PTs significantly less frequently harbored MED12 exon 2 somatic mutations than FAs, benign and borderline PTs.
Conclusions
Although MED12 exon 2 somatic mutations likely constitute the driver genetic event of most FAs, benign and borderline PTs, our results suggest that the majority of malignant PTs may be driven by other genetic/epigenetic alterations.
Keywords: fibroepithelial tumors, breast, MED12, sequencing, somatic mutations
Introduction
Fibroadenomas (FAs) and phyllodes tumors (PTs) are fibroepithelial tumors of the breast composed of biphasic proliferations of both epithelial and stromal elements.1 FAs are benign and constitute the most frequent type of fibroepithelial tumors,2,3 being characterized by an admixture of epithelial and stromal components that can be arranged in two growth patterns: pericanalicular, characterized by a circumferential stromal proliferation around ductal structures, and intracanalicular, where the proliferating stromal cells compress the ductal structures into clefts.1 PTs are rarer than FAs, accounting for approximately 2.5% of all fibroepithelial tumors of the breast. Although in the majority of cases PTs develop de novo from intralobular or periductal stroma of the breast, there are reports of progression from FAs to PTs and of examples of FAs found in the proximity of PTs.1,4 Histologically, PTs have a cellular stroma and display an intracanalicular growth pattern, which is often more exuberant than that found in FAs and results in leaf-like projections protruding into the dilated luminal spaces of ductal structures.1 PTs are classified as benign, borderline or malignant based on specific histologic criteria,1 however these form a continuum, and grading these lesions is often challenging.5
The Mediator Complex Subunit 12 (MED12) gene (Xq13.1) encodes a member of the Multiprotein mediator complex. MED12 plays role in the regulation of transcription of all RNA polymerase II-dependent genes.6 The kinase/CDK8 module formed by MED12 together with CDK8/CDK19, Cyclin C, and MED13 is often associated with transcriptional repression and functions as a stimulus-specific positive coregulator within the p53 transcriptional program.7 Recent studies have demonstrated that up to 60% of FAs, 80% of benign and borderline PTs, and 40% of malignant PTs harbor somatic mutations in the exon 2 of the MED12 gene.8,9 Transcriptomic analysis of FAs provided additional evidence to support the contention that MED12 exon 2 somatic mutations likely constitute driver genetic alterations in FAs.8 In addition, MED12 somatic mutations have been reported in several tumor types, including hormone-associated lesions, such as uterine leiomyomas, uterine leiomyosarcomas, pelvic/retroperitoneal smooth muscle tumors, prostate cancer and adrenocortical cancer.10-14
Given the histologic similarities between FAs and PTs, and the evidence to suggest that at least a subset of PTs may originate from FAs,1,4 we sought to define whether PTs would harbor mutations affecting exon 2 of the MED12 gene in a way akin to FAs. Furthermore, as a hypothesis generating aim, we characterized the repertoire of MED12 exon 2 mutations in distinct fibroepithelial tumors from patients with multiple lesions to define their potential clonal relatedness.
Materials and Methods
Cases
The files of the Department of Pathology of Memorial Sloan Kettering Cancer Center (MSKCC) were searched for FAs and PTs diagnosed and surgically removed at our Institution between January 1996 and October 2014. The diagnostic slides and tissue blocks of 26 FAs, 25 benign PTs, 9 borderline PTs and 13 malignant PTs from 64 female patients were retrieved. Samples were anonymized prior to analysis, and the study was approved by the MSKCC Institutional Review Board. All cases were independently reviewed by three pathologists (MM, FLL and EB), and classified and graded according to the latest World Health Organization classification (WHO) criteria,1 which are detailed in the Supplementary Methods.
Microdissection and DNA extraction
Representative 8μm-thick sections from formalin-fixed paraffin-embedded histologic blocks of FAs and PTs were subjected to microdissection of tumor and matched normal tissue with a sterile needle under a stereomicroscope as previously described.15,16 Tumor areas were microdissected to enrich for neoplastic cells from the stromal elements (>70%) of the FAs and PTs analyzed. DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to manufacturer's guidelines.
PCR amplification and Sanger sequencing
The entire MED12 exon 2 was amplified from 10ng of genomic DNA using the AmpliTaq 360 master mix (Life Technologies) as previously described.16 For this, a primer pair that amplifies a 373bp fragment encompassing exon 2 of the MED12 gene was employed (5′-TGTTCTACACGGAACCCTCCTC-3′ (forward) and 5′-CTGGGCAAATGCCAATGAGAT-3′ (reverse)) as described by Lim et al.8 (Supplementary Figure 1). Sequencing reactions were performed on an ABI3730 capillary sequencer using the ABI BigDye Terminator chemistry (v3.1, Life Technologies) according to manufacturer's instructions. To confirm that the MED12 mutations identified in FAs and PTs were somatic events, matched normal DNA was sequenced for all cases included in this study. Sequences of the forward and reverse strands were analyzed using MacVector software (MacVector Inc).16 All analyses were performed in duplicate. Insertions and deletion were manually annotated. Mutation function prediction was performed as previously described17 (Supplementary Methods).
Statistical analysis
The Chi-square test (χ2 test) and the Fisher's exact test were employed for the comparison of non-parametric variables. All statistical analyses were carried out using IBM SPSS Statistics v.20 (IBM). Two-tailed P values <0.05 were considered significant.
Results
Seventy-three tumors from 64 female patients were subjected to Sanger sequencing analysis for the presence of MED12 exon 2 somatic mutations. According to the WHO criteria,1 26 tumors were classified as FAs (36%), 25 benign PTs (34%), 9 borderline PTs (12%) and 13 malignant PTs (18%; Figure 1, Table 1 and Supplementary Tables 1 and 2).
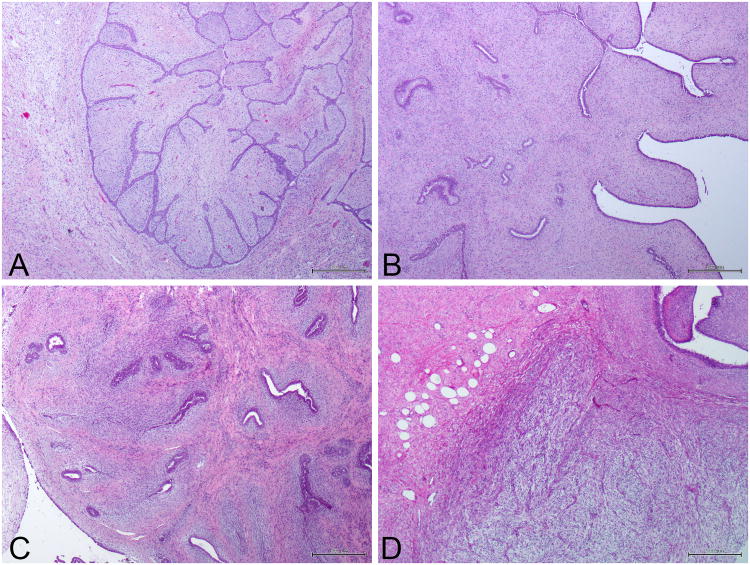
Figure 1. Representative micrographs of a fibroadenoma and phyllodes tumors of the breast included in this study.
(A) Fibroadenoma. (B) Benign phyllodes tumors of the breast. (C) Borderline phyllodes tumors of the breast. (D) Malignant phyllodes tumors of the breast. Scale bar, 500 μM.
Table 1. MED12 exon 2 mutations identified in 73 fibroepithelial tumors of the breast from 64 female patients.
| Patient ID | Sample ID | Ethnicity | Diagnosis | Laterality | Type of mutation | cDNA change | Protein change |
|---|---|---|---|---|---|---|---|
| Patient 01 | MD-01 | Black | Malignant PT | Left | wild-type | wild-type | wild-type |
| Patient 02 | MD-02 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 03 | MD-03 | Unknown | Borderline PT | Right | wild-type | wild-type | wild-type |
| Patient 04 | MD-04 | White | Malignant PT | Right | SNV | c.131G>A | p.G44D |
| MD-60a | White | Benign PT | Right | SNV | c.131G>T | p.G44V | |
| MD-60b | White | Fibroadenoma | Right | SNV | c.130G>T | p.G44C | |
| MD-60c | White | Fibroadenoma | Right | SNV | c.131G>T | p.G44V | |
| MD-60d | White | Fibroadenoma | Right | SNV | c.131G>A | p.G44D | |
| Patient 05 | MD-05 a | White | Malignant PT | Left | wild-type | wild-type | wild-type |
| Patient 06 | MD-06 | Asian | Borderline PT | Left | SNV | c.131G>A | p.G44D |
| Patient 07 | MD-07 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 08 | MD-08 | Asian | Malignant PT | Left | wild-type | wild-type | wild-type |
| Patient 09 | MD-09 a | White | Benign PT | Left | SNV | c.130G>A | p.G44S |
| MD-09 b | White | Benign PT | Left | SNV | c.130G>A | p.G44S | |
| MD-09 c | White | Benign PT | Left | SNV | c.131G>A | p.G44D | |
| MD-09 d | White | Benign PT | Left | SNV | c.131G>A | p.G44D | |
| Patient 10 | MD-10 | Asian | Borderline PT | Right | wild-type | wild-type | wild-type |
| Patient 11 | MD-11 | Black | Borderline PT | Left | Insertion (frameshift) | c.112_113insAC | p.L39_N40fs |
| Patient 12 | MD-12 | White | Borderline PT | Right | Deletion (in-frame) | c.115_141del27 | p.L39_N47del |
| Patient 13 | MD-13 | Black | Borderline PT | Left | SNV | c.131G>C | p.G44A |
| Patient 14 | MD-14 | White | Benign PT | Left | Deletion (in-frame) | c.122_139del18 | p.V41_N46del |
| Patient 15 | MD-15a | Asian | Borderline PT | Left | SNV | c.131G>T | p.G44V |
| Patient 16 | MD-16 | Asian | Borderline PT | Right | SNV | c.130G>A | p.G44S |
| Patient 17 | MD-17 | White | Borderline PT | Left | SNV | c.131G>T | p.G44V |
| Patient 18 | MD-18 | White | Benign PT | Left | Deletion (in-frame) | c.123_152del30 | p.K42_V51del |
| Patient 19 | MD-19 | Unknown | Benign PT | Left | SNV | c.130G>A | p.G44S |
| Patient 20 | MD-20 | White | Benign PT | Left | SNV | c.130G>A | p.G44S |
| Patient 21 | MD-21 | White | Benign PT | Right | SNV | c.130G>A | p.G44S |
| Patient 22 | MD-22 | White | Benign PT | Right | SNV | c.131G>T | p.G44V |
| Patient 23 | MD-23 | Black | Benign PT | Left | Deletion (in-frame) | c.141_167del27 | p.Q48_H56del |
| Patient 24 | MD-24 | White | Benign PT | Left | wild-type | wild-type | wild-type |
| Patient 25 | MD-25 | White | Benign PT | Left | Deletion (in-frame) | c.144__167del24 | p.P49_H56del |
| Patient 26 | MD-26 | White | Benign PT | Left | SNV | c.131G>A | p.G44D |
| Patient 27 | MD-27 | White | Benign PT | Right | SNV | c.130G>T | p.G44C |
| Patient 28 | MD-28 | White | Benign PT | Right | Deletion (in-frame) | c.100_129del30 | p.D34_Q43del |
| Patient 29 | MD-29 | White | Benign PT | Left | wild-type | wild-type | wild-type |
| Patient 30 | MD-30 | White | Benign PT | Right | SNV | c.130G>A | p.G44S |
| Patient 31 | MD-31 | White | Benign PT | Right | SNV | c.130G>A | p.G44S |
| Patient 32 | MD-32 | Asian | Benign PT | Left | Deletion (in-frame) | c.114_149del36 | p.L39_A50del |
| Patient 33 | MD-33 | White | Benign PT | Right | Deletion (in-frame) | c.121_138del18 | p.V41_N46del |
| Patient 34 | MD-34 | White | Benign PT | Right | Deletion (in-frame) | c.100_138del39 | p.D34_N46del |
| Patient 35 | MD-35 | Unknown | Benign PT | Right | Deletion (in-frame) | c.116_157del42 | p.L39_S52del |
| Patient 36 | MD-36 | White | Benign PT | Right | wild-type | wild-type | wild-type |
| Patient 37 | MD-37 | White | Fibroadenoma | Left | Deletion (in-frame) | c.121_135del15 | p. V41_F45del |
| Patient 38 | MD-38 | Unknown | Fibroadenoma | Right | Deletion (in-frame) | c.118_159del42 | p.N40_G53del |
| Patient 39 | MD-39 | White | Fibroadenoma | Right | Deletion (frameshift) | c.100_112del13 | p.D34_T37fs |
| Patient 40 | MD-40 | Unknown | Fibroadenoma | Left | wild-type | wild-type | wild-type |
| Patient 41 | MD-41 | White | Fibroadenoma | Right | SNV | c.130G>A | p.G44S |
| Patient 42 | MD-42 | White | Fibroadenoma | Right | Deletion (in-frame) | c.108_122del15 | p.T37_V41del |
| Patient 43 | MD-43 | White | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 44 | MD-44 | Black | Fibroadenoma | Left | Deletion (frameshift) | c.100_107del8 | p.D34_L36fs |
| Patient 45 | MD-45 | White | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 46 | MD-46 | Asian | Fibroadenoma | Right | SNV | c.131G>T | p.G44V |
| Patient 47 | MD-47 | Black | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 48 | MD-48 | White | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 49 | MD-49 | Unknown | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 50 | MD-50 | Unknown | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 51 | MD-51 | White | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| Patient 52 | MD-52 | White | Fibroadenoma | Left | Deletion (in-frame) | c.115_162del48 | p.L39_D54del |
| Patient 53 | MD-53 | White | Fibroadenoma | Left | Deletion (in-frame) | c.138_161del24 | p.N47_E55del |
| MD-54 | White | Fibroadenoma | Right | SNV | c.130G>A | p.G44S | |
| Patient 54 | MD-55 | White | Fibroadenoma | Right | SNV | c.131G>A | p.G44D |
| Patient 55 | MD-58 | White | Fibroadenoma | Left | SNV | c.130G>C | p.G44R |
| Patient 56 | MD-56 | White | Fibroadenoma | Right | wild-type | wild-type | wild-type |
| MD-57 | White | Fibroadenoma | Left | Deletion (in-frame) | c.117_152del36 | p.N40_V51del | |
| Patient 57 | MD-59 | Unknown | Fibroadenoma | Right | SNV | c.131G>A | p.G44D |
| Patient 58 | MD-61 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 59 | MD-62 | Unknown | Malignant PT | Left | wild-type | wild-type | wild-type |
| Patient 60 | MD-63 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 61 | MD-64 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 62 | MD-65 | White | Malignant PT | Left | wild-type | wild-type | wild-type |
| Patient 63 | MD-66 | White | Malignant PT | Right | wild-type | wild-type | wild-type |
| Patient 64 | MD-67 | Asian | Malignant PT | Right | wild-type | wild-type | wild-type |
PT, phyllodes tumor; SNV, single nucleotide variant.
High frequency of MED12 somatic mutations in FAs, benign and borderline PTs but not in malignant PTs
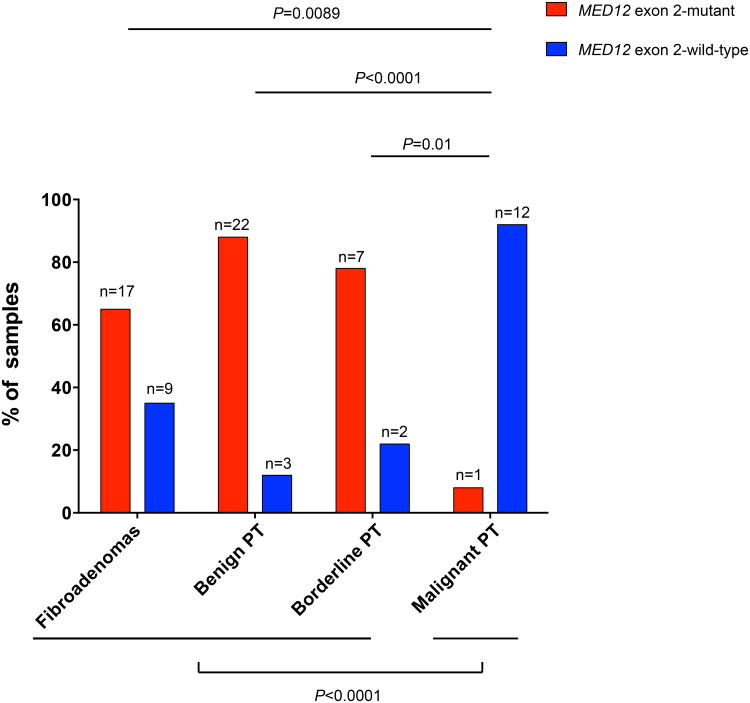
MED12 exon 2 somatic mutations were found in 65% of FAs (17/26), 88% of benign PTs (22/25), 78% (7/9) of borderline PTs and 8% (1/13) of malignant PTs. Six distinct somatic single nucleotide variants and nineteen distinct insertions and deletions affecting MED12 were identified in 28 and 19 cases, respectively (Figures 2 and 3, Table 1, Supplementary Table 1). All single nucleotide variants and insertions and deletions were predicted to be pathogenic by multiple mutation effect predictors (Supplementary Table 1). No nonsense single nucleotide variants were found. No associations between the presence of MED12 mutations and age of the patient or tumor size were observed (Student's t-test, P>0.1, Table 2).
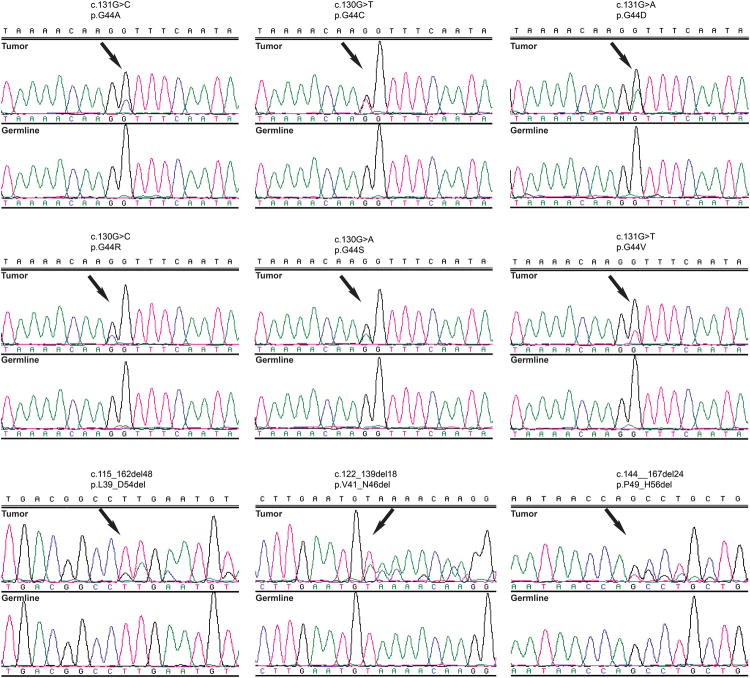
Figure 2. MED12 exon 2 mutations identified by Sanger sequencing analysis in fibroepithelial tumors.
Representative sequence electropherograms (tumor and matched germline) of the 6 single nucleotide variants, and of 3 of the 19 distinct insertions and deletions in exon 2 of MED12 identified by Sanger sequencing in our cohort of 73 fibroepithelial tumors.
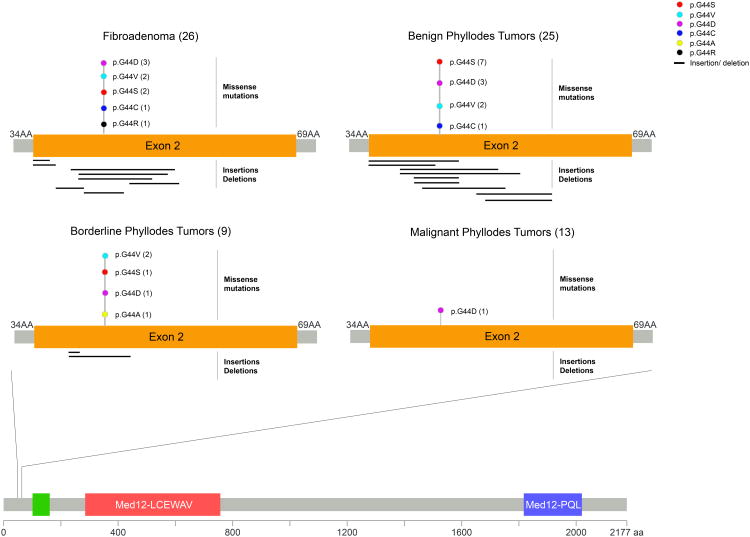
Figure 3. Distribution of MED12 mutations in exon 2.
Domain structure of the MED12 protein, and a close-up view of the amino acid residues in MED12 exon 2 affected by alterations in the fibroadenomas and phyllodes tumors analyzed in this study. The frequency of each alteration is denoted in parentheses. AA, amino acid.
Table 2. Correlation between MED12 exon 2 mutation status and patient age and tumor size in patients with fibroepithelial tumors.
| Age (Mean ± SD) MED12 mutant | n | Age (Mean ± SD) MED12 wild-type | n | P value | |
|---|---|---|---|---|---|
| Fibroadenoma | 31.8 ± 1.9 | 17 | 37.1 ± 3.7 | 9 | 0.158 |
| Benign PT | 42 ± 2.2 | 22 | 36.3 ± 8.1 | 3 | 0.404 |
| Borderline PT | 49.3 ± 4.5 | 7 | 44.5 ± 2.5 | 2 | 0.604 |
| Tumor size (cm) (Mean ± SD) MED12 mutant | n | Tumor size (cm) (Mean ± SD) MED12 wild-type | n | P value | |
| Fibroadenoma | 1.7 ± 0.2 | 17 | 2.2 ± 0.3 | 9 | 0.168 |
| Benign PT | 2.3 ± 0.2 | 22 | 2.4 ± 1.3 | 3 | 0.899 |
| Borderline PT | 5.8 ± 1.2 | 7 | 5.8 ± 3.1 | 2 | 0.986 |
Student's t-tests, two-tailed. PT, phyllodes tumor; SD, standard deviation. Malignant PTs were excluded from this comparison due to small sample size and the low frequency of MED12 mutations.
The prevalence of MED12 exon 2 somatic mutations in FAs (Table 1, Figure 3, Supplementary Table 1) was similar to that recently described by Lim et al.8 In FAs, MED12 exon 2 mutations were found to be similarly frequent in intracanalicular-type (8/8, 100%) and mixed-type FAs (3/3, 100%), but were significantly less prevalent in pericanalicular-type FAs (6/15, 40%) than in intracanalicular-type FAs or the group of intracanalicular-type or mixed-type FAs (Table 3). Of the 17 MED12-mutant FAs, 9 (53%) harbored hotspot point mutations in codon 44 (3 p.G44D, 2 p.G44V, 2 p.G44S, 1 p.G44R and 1 p.G44C). Eight (47%) MED12-mutant FAs displayed insertions or deletions, of which 2 were frameshift and 6 were in-frame (Figure 3, Table 1, Supplementary Table 1).
Table 3. MED12 exon 2 mutation status according to growth patterns in fibroadenomas.
| Growth pattern | n | MED12 wild-type n (%) | MED12 mutant n (%) | P value |
|---|---|---|---|---|
| Pericanalicular | 15 | 9 (60%) | 6 (40%) | 0.007 |
| Intracanalicular | 8 | 0 (0%) | 8 (100%) | |
| Intracanalicular | 8 | 0 (0%) | 8 (100%) | 1.000 |
| Mixed | 3 | 0 (0%) | 3 (100%) | |
| Pericanalicular | 15 | 9 (60%) | 6 (40%) | 0.205 |
| Mixed | 3 | 0 (0%) | 3(100%) | |
| Pericanalicular | 15 | 9 (60%) | 6 (40%) | 0.002 |
| Intracanalicular or mixed | 11 | 0 (0%) | 11 (100%) |
Twenty-two (88%) benign PTs and 7 (78%) borderline PTs harbored MED12 exon 2 somatic mutations. Of the 22 MED12-mutant benign PTs, 13 (59%) displayed hotspot point mutations in codon 44 (7 p.G44S, 3 p.G44D, 2 p.G44V and 1 p.G44C), 9 (41%) had in-frame insertions or deletions (Table 1, Figure 3, Supplementary Table 1). In the 7 MED12-mutant borderline PTs, point mutations in codon 44 (1 p.G44D, 1 p.G44S, 2 p.G44V and 1 p.G44A) were found in 5 cases (71%) and in-frame insertions or deletions were found in 2 cases (29%) (Table 1, Figure 3, Supplementary Table 1). By contrast, only 1 of the 13 malignant PTs analyzed (8%) was found to harbor a single nucleotide variant affecting codon 44 (p.G44D), which was considered as likely pathogenic by mutation function predictors.
Although a similarly high prevalence of MED12 exon 2 mutations was found in FAs, benign and borderline PTs (P>0.1, Fisher's exact test), malignant PTs were found to display MED12 exon 2 somatic mutations significantly less frequently than FAs, benign or borderline PTs (P<0.001, Fisher's exact test, Figure 4). These results suggest that whilst the majority of FAs, benign PTs and borderline PTs harbored MED12 exon 2 mutations, only a minority of malignant PTs display these mutations.
Figure 4. Frequencies of MED12 exon 2 mutations in different fibroepithelial tumors of the breast.
The percentages of cases with and without MED12 exon 2 somatic mutations are depicted. The MED12 exon 2 mutational frequencies in fibroadenomas, benign, borderline and malignant phyllodes tumors of the breast were compared using Fisher's exact tests. PT, phyllodes tumor.
MED12 mutational status in different tumors affecting the same patients
Four of the 64 women part of the study had multiple mammary fibroepithelial lesions (Table 1, Supplementary Table 1). We therefore compared the MED12 mutational status in multiple FAs and/or PTs from the same patient to define whether the lesions would be clonally related. In patient 04, the FAs displayed distinct MED12 exon 2 somatic mutations (p.G44V, p.G44D and p.G44C), the benign PT displayed a p.G44V mutation and the malignant PT harbored a p.G44D mutation. Interestingly, the malignant PT and one of the intracanalicular FAs harbored the p.G44D MED12 mutation, whereas the benign PT and another intracanalicular FA displayed the p.G44V MED12 mutation. In patient 09, two p.G44S and two p.G44D were found in the 4 benign PTs in the right breast, demonstrating that some of the PTs were likely not clonally related. Patients 53 and 56 presented with bilateral FAs; in patient 53, a p.G44S mutation was found in the FA of the right breast, whereas the left breast FA harbored a p.N47_E55del in-frame deletion. In patient 56, the left breast FA harbored a p.N40_V51del in-frame deletion, whereas the right breast FA was MED12 wild-type (Table 1, Supplementary Table 1). Taken together, these results demonstrate that fibroepithelial lesions arising in patients with multiple FAs and/or PTs do not necessarily harbor identical MED12 somatic mutations, even when the lesions are ipsilateral, suggesting that some of these lesions likely originated independently. Furthermore, the presence of a p.G44D in a FA and in a malignant PT affecting the same breast of patient 4 may be interpreted as supportive of a clonal relationship between these two lesions and raises the possibility that in this case the FA may have constituted the substrate for the development of the malignant PT.
Discussion
Somatic mutations in MED12 exon 2 have been recently reported in benign indolent types of stromal tumors, including the majority of leiomyomas,18-23 FAs8 and benign and borderline PTs9, and in a subset of malignant PTs.9 In FAs, which are biphasic tumors with epithelial components, these mutations have been found to be restricted to the stromal elements.8 Here we provide an independent validation of the results reported by Lim et al.8 and Cani et al.9 as we have also identified MED12 exon 2 mutations in 65%, 88% and 78% of FAs, benign and borderline PTs, respectively. In contrast to the high prevalence of MED12 somatic mutations found in FAs, benign and borderline PTs, MED12 exon 2 somatic mutations were found to be significantly less frequent in malignant PTs, being found in only in 8% (1/13) of cases. These observations are consistent with those reported by Cani et al.,9 who observed MED12 somatic mutations in 40% (2/5) of malignant PTs, but in 80% of benign (4/5) and borderline (4/5) PTs.
There is burgeoning evidence to demonstrate that MED12 exon 2 mutations likely constitute genomic drivers of FAs.8 The first 100 amino acids of MED12, which include its entire exon 2, have been shown to be essential for the interaction of MED12 with Cyclin C, and somatic mutations in MED12 exon 2 have been reported to be tumorigenic, decreasing the interaction between MED12 and Cyclin C-CDK8/CDK19 and resulting in loss of Mediator-associated CDK activity24 and in RAD51B overexpression.25 These mutations have also been shown to impact on estrogen signaling. Not only does the Mediator complex interact with estrogen receptors α and β,26 but also MED12 exon 2-mutated FAs have been shown to display dysregulated estrogen signaling.8 Given that breast and uterine stromal cells and uterine smooth muscle cells are estrogen responsive, it is plausible that the high frequency of MED12 exon 2 somatic mutations in these tumors and their relative rarity in stromal tumors originating in other non-estrogen responsive anatomical sites stems from the impact of MED12 mutations on estrogen signaling.8
MED12 somatic mutations appear to be less frequent in malignant than in benign tumors.13,20,27-29 We have observed that MED12 exon 2 somatic mutations were significantly more frequently found in FAs, benign and borderline PTs than in malignant PTs. Consistent with these observations, leiomyosarcomas have also been shown to harbor MED12 mutations at significantly lower frequencies than leiomyomas.29,30 These observations may be reflective of the fact that although some leiomyosarcomas and malignant PTs may originate from leiomyomas and FAs/benign PTs, respectively, which are estrogen dependent lesions, others may originate de novo through an estrogen independent pathway. Consistent with this notion, the expression both ERβ (in the stromal component) and ERα (in the epithelial component) of fibroepithelial lesions of the breast has been reported to be inversely correlated with their degree of aggressiveness.31,32 On the other hand, 60% of the malignant PTs analyzed by Cani et al. lacked MED12 somatic mutations,9 and these cases harbored TERT gene amplification. Further studies are required to define fully the mechanistic interactions between MED12 exon 2 mutations and estrogen signaling in MED12 mutant lesions and the alternative driver genetic alterations in MED12 wild-type malignant PTs.
The genotyping of multiple fibroepithelial lesions occurring in 4 patients constitutes a unique and intriguing aspect of our study. The identification of ipsilateral fibroepithelial lesions harboring distinct MED12 mutations is consistent with the notion that multiple coexisting ipsilateral fibroepithelial tumors can arise independently. The analysis of multiple ipsilateral FAs, benign and malignant PTs also revealed that in one case, identical MED12 mutations were found in a FA and in a malignant PT in the same breast, an observation that is consistent with a clonal relationship between the two lesions and highlights the possibility that, in this case, the FA may have constituted the substrate for the development of the malignant PT.
This study has several limitations. First, the sample size is small, in particular, the cohort of borderline PTs comprises only 9 tumors; despite the small sample size, we have documented a significantly lower prevalence of MED12 exon 2 mutations in malignant PTs than in FAs and benign and borderline PTs. This observation, however, should be considered as hypothesis-generating and warrants the analysis of independent cohorts of malignant PTs. Second, we have deliberately focused on somatic mutations affecting exon 2 of the MED12 gene. Although previous findings demonstrate that the vast majority of MED12 somatic mutations in FAs affect exon 2, we cannot rule out the possibility of somatic mutations affecting other coding sequences of the MED12 gene may be present in breast fibroepithelial lesions. Finally, we employed Sanger sequencing for the detection of MED12 somatic mutations; given that all lesions included in this study were microdissected and considering the sensitivity of Sanger sequencing, subclonal MED12 mutations present in <20% of the clones would not be accurately detected. It should be noted, however, that given the evidence that MED12 somatic mutations are early driver events in fibroepithelial lesions, these mutations would likely be present in the modal populations of MED12 mutant FAs and PTs.
In conclusion, our study demonstrates that somatic mutations in MED12 exon 2 likely constitute driver genetic events of not only FAs but also of benign and borderline PTs. MED12 exon 2 somatic mutations were found to be significantly less frequent in malignant PTs, indicating that genetic and/or epigenetic alterations other than MED12 exon 2 mutations may constitute the driving genetic events in the pathogenesis of malignant PTs.
Supplementary Material
Supplementary Figure 1: Schematic representation of the MED12 somatic mutations reported by Lim et al.8 and Cani et al.9 in relation to the primers employed for Sanger sequencing in this study.
Supplementary Table 1: Clinicopathologic characteristics of the fibroepithelial lesions included in this study, their MED12 exon 2 somatic mutation status, and the predicted pathogenicity of the mutations identified.
Supplementary Table 2: Histologic characteristics of the malignant phyllodes tumors included in this study.
Acknowledgments
SP is funded by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348); CM is funded by the Italian Association of Cancer Research (AIRC, MFAG13310). AMS is funded by a stipend from the German Cancer Aid (Dr. Mildred Scheel Stiftung).
BW, EB and JSR-F conceived the study. MM and EB provided the samples. MM, FLL and EB performed the pathologic review. SP, NF, AMS, CM, LGM and MA carried out experiments and analyzed data. SP, BW and JSR-F wrote the first draft of the manuscript. All authors interpreted the data, and reviewed and approved the final version of the manuscript.
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare
References
- 1.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. Who classification of tumours of the breast. 4th. Lyon: IARC press; 2012. [Google Scholar]
- 2.Demetrakopoulos NJ. Three-dimensional reconstruction of a human mammary fibroadenoma. Q Bull Northwest Univ Med Sch. 1958;32:221–228. [PMC free article] [PubMed] [Google Scholar]
- 3.Foster ME, Garrahan N, Williams S. Fibroadenoma of the breast: A clinical and pathological study. J R Coll Surg Edinb. 1988;33:16–19. [PubMed] [Google Scholar]
- 4.Noguchi S, Yokouchi H, Aihara T, et al. Progression of fibroadenoma to phyllodes tumor demonstrated by clonal analysis. Cancer. 1995;76:1779–1785. doi: 10.1002/1097-0142(19951115)76:10<1779::aid-cncr2820761015>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SP, Chang YC, Liu TP, Lee JJ, Tzen CY, Liu CL. Phyllodes tumor of the breast: The challenge persists. World J Surg. 2006;30:1414–1421. doi: 10.1007/s00268-005-0786-2. [DOI] [PubMed] [Google Scholar]
- 6.Borggrefe T, Yue X. Interactions between subunits of the mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Donner AJ, Szostek S, Hoover JM, Espinosa JM. Cdk8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim WK, Ong CK, Tan J, et al. Exome sequencing identifies highly recurrent med12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46:877–880. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 9.Cani AK, Hovelson DH, McDaniel AS, et al. Next-gen sequencing exposes frequent med12 mutations and actionable therapeutic targets in phyllodes tumors. Mol Cancer Res. 2015 doi: 10.1158/1541-7786.MCR-14-0578. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makinen N, Heinonen HR, Sjoberg J, Taipale J, Vahteristo P, Aaltonen LA. Mutation analysis of components of the mediator kinase module in med12 mutation-negative uterine leiomyomas. Br J Cancer. 2014;110:2246–2249. doi: 10.1038/bjc.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertsch E, Qiang W, Zhang Q, et al. Med12 and hmga2 mutations: Two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod Pathol. 2014;27:1144–1153. doi: 10.1038/modpathol.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwetye KE, Pfeifer JD, Duncavage EJ. Med12 exon 2 mutations in uterine and extrauterine smooth muscle tumors. Hum Pathol. 2014;45:65–70. doi: 10.1016/j.humpath.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent spop, foxa1 and med12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assie G, Letouze E, Fassnacht M, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- 15.Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232:553–565. doi: 10.1002/path.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating prkd1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martelotto LG, Ng C, De Filippo MR, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makinen N, Mehine M, Tolvanen J, et al. Med12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 19.Makinen N, Heinonen HR, Moore S, Tomlinson IP, van der Spuy ZM, Aaltonen LA. Med12 exon 2 mutations are common in uterine leiomyomas from south african patients. Oncotarget. 2011;2:966–969. doi: 10.18632/oncotarget.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Je EM, Kim MR, Min KO, Yoo NJ, Lee SH. Mutational analysis of med12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer. 2012;131:E1044–1047. doi: 10.1002/ijc.27610. [DOI] [PubMed] [Google Scholar]
- 21.Markowski DN, Huhle S, Nimzyk R, Stenman G, Loning T, Bullerdiek J. Med12 mutations occurring in benign and malignant mammalian smooth muscle tumors. Genes Chromosomes Cancer. 2013;52:297–304. doi: 10.1002/gcc.22029. [DOI] [PubMed] [Google Scholar]
- 22.Matsubara A, Sekine S, Yoshida M, et al. Prevalence of med12 mutations in uterine and extrauterine smooth muscle tumours. Histopathology. 2013;62:657–661. doi: 10.1111/his.12039. [DOI] [PubMed] [Google Scholar]
- 23.McGuire MM, Yatsenko A, Hoffner L, Jones M, Surti U, Rajkovic A. Whole exome sequencing in a random sample of north american women with leiomyomas identifies med12 mutations in majority of uterine leiomyomas. PLoS One. 2012;7:e33251. doi: 10.1371/journal.pone.0033251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turunen M, Spaeth JM, Keskitalo S, et al. Uterine leiomyoma-linked med12 mutations disrupt mediator-associated cdk activity. Cell Rep. 2014;7:654–660. doi: 10.1016/j.celrep.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampjarvi K, Park MJ, Mehine M, et al. Mutations in exon 1 highlight the role of med12 in uterine leiomyomas. Hum Mutat. 2014;35:1136–1141. doi: 10.1002/humu.22612. [DOI] [PubMed] [Google Scholar]
- 26.Kang YK, Guermah M, Yuan CX, Roeder RG. The trap/mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the trap220 subunit and directly enhances estrogen receptor function in vitro. Proc Natl Acad Sci U S A. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assie G, Letouze E, Fassnacht M, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampjarvi K, Makinen N, Kilpivaara O, et al. Somatic med12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107:1761–1765. doi: 10.1038/bjc.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravegnini G, Marino-Enriquez A, Slater J, et al. Med12 mutations in leiomyosarcoma and extrauterine leiomyoma. Mod Pathol. 2013;26:743–749. doi: 10.1038/modpathol.2012.203. [DOI] [PubMed] [Google Scholar]
- 31.Sapino A, Bosco M, Cassoni P, et al. Estrogen receptor-beta is expressed in stromal cells of fibroadenoma and phyllodes tumors of the breast. Mod Pathol. 2006;19:599–606. doi: 10.1038/modpathol.3800574. [DOI] [PubMed] [Google Scholar]
- 32.Tse GM, Lee CS, Kung FY, et al. Hormonal receptors expression in epithelial cells of mammary phyllodes tumors correlates with pathologic grade of the tumor: A multicenter study of 143 cases. Am J Clin Pathol. 2002;118:522–526. doi: 10.1309/D206-DLF8-WDNC-XJ8K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Schematic representation of the MED12 somatic mutations reported by Lim et al.8 and Cani et al.9 in relation to the primers employed for Sanger sequencing in this study.
Supplementary Table 1: Clinicopathologic characteristics of the fibroepithelial lesions included in this study, their MED12 exon 2 somatic mutation status, and the predicted pathogenicity of the mutations identified.
Supplementary Table 2: Histologic characteristics of the malignant phyllodes tumors included in this study.