The ability of two three-dimensional bioscaffold systems to reverse the primary limitations of induced pluripotent stem cell (iPSC)-derived hepatocytes was compared. Proliferation and function of iPSC hepatocytes were significantly enhanced when cultured within scaffolds made from extracellular matrix (ECM). This ECM scaffold enhanced phenotypic maturation of iPSC hepatocytes compared with other platforms, likely owing to its biologically diverse makeup.
Keywords: Liver, Stem cell-microenvironment interactions, Stem/progenitor cell, Tissue regeneration, Induced pluripotent stem cells
Abstract
Induced pluripotent stem cells (iPSCs) are new diagnostic and potentially therapeutic tools to model disease and assess the toxicity of pharmaceutical medications. A common limitation of cell lineages derived from iPSCs is a blunted phenotype compared with fully developed, endogenous cells. We examined the influence of novel three-dimensional bioartificial microenvironments on function and maturation of hepatocyte-like cells differentiated from iPSCs and grown within an acellular, liver-derived extracellular matrix (ECM) scaffold. In parallel, we also compared a bioplotted poly-l-lactic acid (PLLA) scaffold that allows for cell growth in three dimensions and formation of cell-cell contacts but is infused with type I collagen (PLLA-collagen scaffold) alone as a “deconstructed” control scaffold with narrowed biological diversity. iPSC-derived hepatocytes cultured within both scaffolds remained viable, became polarized, and formed bile canaliculi-like structures; however, cells grown within ECM scaffolds had significantly higher P450 (CYP2C9, CYP3A4, CYP1A2) mRNA levels and metabolic enzyme activity compared with iPSC hepatocytes grown in either bioplotted PLLA collagen or Matrigel sandwich control culture. Additionally, the rate of albumin synthesis approached the level of primary cryopreserved hepatocytes with lower transcription of fetal-specific genes, α-fetoprotein and CYP3A7, compared with either PLLA-collagen scaffolds or sandwich culture. These studies show that two acellular, three-dimensional culture systems increase the function of iPSC-derived hepatocytes. However, scaffolds derived from ECM alone induced further hepatocyte maturation compared with bioplotted PLLA-collagen scaffolds. This effect is likely mediated by the complex composition of ECM scaffolds in contrast to bioplotted scaffolds, suggesting their utility for in vitro hepatocyte assays or drug discovery.
Significance
Through the use of novel technology to develop three-dimensional (3D) scaffolds, the present study demonstrated that hepatocyte-like cells derived via induced pluripotent stem cell (iPSC) technology mature on 3D extracellular matrix scaffolds as a result of 3D matrix structure and scaffold biology. The result is an improved hepatic phenotype with increased synthetic and catalytic potency, an improvement on the blunted phenotype of iPSC-derived hepatocytes, a critical limitation of iPSC technology. These findings provide insight into the influence of 3D microenvironments on the viability, proliferation, and function of iPSC hepatocytes to yield a more mature population of cells for cell toxicity studies and disease modeling.
Introduction
Induced pluripotent stem cell (iPSC) technology is a powerful strategy to develop multipotent cells from mature tissues with the ability to direct differentiation toward any tissue lineage to model disease [1–6], analyze drug pharmacokinetics [7], and potentially develop cells as therapies for a variety of diseases [8–11]. The development of patient-specific hepatocytes with intact synthetic xenobiotic catalytic activity would accelerate pharmaceutical testing and drug development. However, the ability of iPSC-derived hepatocytes to replicate the function of endogenous cells is limited by a blunted phenotype with reduced synthetic ability and metabolic enzyme activity.
Primary hepatocytes are in high demand as a cell source for pharmacology and toxicity testing owing to their synthetic and detoxification activities and because toxicity is a common failure mode in the development of new drugs. However, the use of primary cells has been limited by low proliferative capacity, loss of function in vitro, and tendency for dedifferentiation during prolonged culture [12]. Several strategies to develop hepatocyte-like cells from pluripotent stem cells (iPSCs and embryonic stem cells) have been established because of this important role in pharmaceutical testing and discovery [13–19]. The resulting hepatocyte-like cells express hepatic lineage markers (e.g., cytokeratin-7, 8, and 18, albumin, CYP7A1, hepatocyte nuclear factor-4α) and secrete liver-specific proteins (e.g., albumin, α1-antitrypsin). However, they consistently retain a mixed immature phenotype with expression of α-fetoprotein (AFP) and decreased overall cellular potency relative to primary hepatocytes, leading to the slower metabolism of toxins and small molecules by the cytochrome P450 system [17, 19–21]. Few synthetic three-dimensional (3D) cell culture systems, such as rudimentary collagen matrices [22] or spheroid culture [23], recreate cell-cell junctions and, thus, liver architecture, more effectively than traditional sandwich cultures and lead to enhanced iPSC hepatocyte function. Compared with these defined culture environments, the natural liver extracellular matrix (ECM) is biochemically complex, containing more active substrates with a diverse repertoire of proteins with biological function, carbohydrates, and growth factors. The natural liver ECM has recently been shown to maintain the phenotype of primary rat hepatocytes for a short duration in vitro [24]. In the present study, we investigated whether ECM might mimic the in vivo liver architecture to enhance proliferation, function, and maturation of iPSC hepatocytes during in vitro culture. Biomimetic coculture of human iPSC hepatocytes with rodent-derived, facilitating stroma cells leads to a more robust hepatic phenotype, as others have shown. However, the addition of a xenogeneic cell population limits the potential therapeutic application of the iPSC-derived population [21]. The objectives of our study were to develop a cell-free scaffold system that would enhance function of cultured iPSC hepatocytes and to determine the differential contribution of naturally occurring ECM-derived scaffolds compared with a biosynthetic 3D-printed scaffold. We developed a structurally complex 3D scaffold derived from a decellularized rat liver ECM (ECM scaffold) using perfusion decellularization [25–28], which retains the chemical composition of the liver matrix but is small in size (∼8 mm), allowing for rapid characterization of evolving cell phenotypes over multiple weeks. To control for the influence of the raw 3D structure on cell-cell interactions compared with the “outside-in” signaling capacity of the matrix niche of the ECM scaffold, we also used 3D bioplotting technology to print hybrid scaffolds composed of poly-l-lactic acid (PLLA) coated and infused with type I collagen only (PLLA-collagen scaffold) to represent a “deconstructed” 3D scaffold with structural homogeneity but limited chemical intricacy compared with the ECM scaffold. Both systems were further compared with the widely used reference standard control of hepatocyte sandwich culture. We found that prolonged culture within the ECM scaffolds enhanced maturation and function of iPSC hepatocytes compared with the growth in the PLLA-collagen scaffolds, although both environments allow for 3D growth and development of cell-cell contacts, together suggesting that discrete cell-matrix interactions are important for cellular maturation.
Materials and Methods
Development of ECM and Bioplotted Scaffolds
ECM Scaffolds
Male Sprague-Dawley rats weighing 220–250 g (Charles River Laboratories, Wilmington, MA, http://www.criver.com) were cared for in accordance with protocols approved by the institutional animal care and use committee of Northwestern University. After adequate induction of anesthesia, 2 × 103 U/kg body weight of heparin was injected intravenously. The liver was isolated and perfused with 20 ml of cold saline and decellularized by the perfusion of agents through a portal vein cannula: deionized water, followed by the decellularization solution (1% Triton X-100 and 0.1% NH4OH), and rinsed with deionized water [26]. To prepare the ECM scaffolds, decellularized liver lobes were embedded within optical cutting temperature compound, flash frozen, sectioned to a thickness of 500 µm, and biopsy punched into 8-mm-diameter disks (Integra Miltex, York, PA, http://www.miltex.com).
PLLA-Collagen Scaffolds
A 3D bioplotter (EnvisionTEC GmbH, Gladbeck, Germany, http://www.envisiontec.com) was used to fabricate PLLA-collagen scaffolds (8-mm diameter, 500-µm thick). PLLA (Corbion, Amsterdam, The Netherlands, http://www.corbion.com) was melted at 220°C, 3D-printed via hot-melt extrusion onto a stage heated to 60°C, subsequently treated with 70% ethanol to reduce inherent hydrophobicity and induce surface hydrophilicity, and infused with 0.05 wt% type I bovine collagen solution in 50 mM acetic acid. The infused scaffolds were immediately frozen at −80°C for several hours before lyophilization.
Cells and Culture Conditions
Individual scaffolds were placed in a 48-well cell culture plate (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com), sterilized with 70% ethanol, and incubated at 37°C overnight in hepatocyte medium (Roswell Park Memorial Institute 1640 medium; Thermo Fisher Scientific Life Sciences) with B27 (Thermo Fisher Scientific Life Sciences), 20 ng/ml oncostatin M (R&D Systems, Minneapolis, MN, http://www.rndsystems.com), 0.1 μM dexamethasone (Thermo Fisher Scientific Life Sciences), and 25 μg/ml gentamicin (Thermo Fisher Scientific Life Sciences). The scaffolds were dried for 5 minutes before cell seeding.
Human iPSC hepatocytes (iCell Hepatocytes; Cellular Dynamics International, Madison, WI, http://www.cellulardynamics.com) were received fresh, in suspension, within a T-flask at room temperature. All cells were differentiated from a single-donor iPSC clone. The results from multiple production (differentiation) runs were found to yield consistent results. The cells were grown and handled according to the manufacturer’s recommendations. A 20-µl cell suspension containing 1 × 106 cells was pipetted onto each scaffold, and the cells were allowed to attach for 20 minutes. Next, 10 of each scaffold type with cells were transferred to a T25 flask, and 10 ml of hepatocyte medium (In Vitro Technologies Inc., Baltimore, MD, http://www.invitrotech.com) was added. The scaffolds were maintained under dynamic culture conditions at 37°C and 5% CO2 on an orbital shaker (Stovall Life Science, Greensboro, NC, http://www.ibisci.com) set at 1 rotation per second to prevent settling of the scaffolds on the bottom of the flask and minimizing foam formation on the surface of the media.
In the sandwich control group, 0.5 × 106 iPSC hepatocytes were seeded onto 24-well polystyrene plates coated with a type 1 collagen gel (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). Next, 0.5 ml of hepatocyte medium was added to maintain the same cell/media ratio used in microscaffold culture conditions. The following day, 0.25 mg/ml growth factor-reduced Matrigel Matrix (BD Biosciences) was placed over the cells.
Fresh primary human hepatocytes expanded in triple knockout Fah−/−/Rag2−/−/Il2rg−/− mice (Yecuris Inc., Tualatin, OR, http://www.yecuris.com) or cryopreserved primary human hepatocytes (male, age 56 years; In Vitro Technologies Inc.) were adopted as positive controls. Primary hepatocytes in hepatocyte medium were seeded onto the ECM, PLLA-collagen scaffolds, or type 1 collagen-coated polystyrene plates with growth factor-reduced Matrigel overlay using the same method described for the iPSC hepatocytes. The cell culture medium was changed every other day for all groups.
DNA Quantification and Quantitative Reverse Transcription-Polymerase Chain Reaction
The amount of DNA remaining after cell removal was used to evaluate the degree of decellularization in the acellular scaffolds compared with the normal (untreated) organ. The tissue was completely dehydrated and DNA extracted and subsequently purified using a standard kit (Qiagen, Gaithersburg, MD, http://www.qiagen.com). The DNA concentration was measured on a nanodrop machine at 260 nm and was represented as nanograms of DNA per milligram of dry weight of the sample.
To quantify the cell proliferation within the scaffolds after 1, 3, 7, and 14 days in culture, cell-laden scaffolds were digested using 0.1% Triton X-100 for 30 minutes, followed by 1 minute of ultrasonic treatment (50 Hz; Branson Ultrasonics Corp., Danbury, CT, http://www.emersonindustrial.com). DNA within the resulting lysates was measured by reading fluorescein at 485/535 nm using Quant-iT PicoGreen kit (Thermo Fisher Scientific Life Sciences). Known concentrations of iPSC hepatocytes and primary hepatocytes were used to construct a standard curve and correlate the DNA concentration to cell number.
A two-step quantitative real-time reverse transcription polymerase chain reaction was performed at day 14. Total RNA was isolated using the TRI Reagent solution (Thermo Fisher Scientific Life Sciences). Reverse transcription using the High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific Life Sciences) was performed. The mRNA expression of each target gene was normalized to cyclophilin expression. Primer sequences are shown in the supplemental online data. Quantitative polymerase chain reaction was performed with iQ SYBR Green and detected using the iQ5 Optical System (Bio-Rad Laboratories Inc., Des Plaines, IL, http://www.bio-rad.com). The thermal profile used was 50°C for 2 minutes, 95°C for 10 minutes, 40 amplification cycles of 95°C for 60 seconds, and 58°C for 60 seconds. The relative degree of gene amplification was calculated using 2^(Ct gene 2 – Ct Cyclophilin2) − (Ct gene 1 – Ct Cyclophilin 1). “Ct gene 1” represents the threshold cycle (Ct) of the target gene of the reference population and “Ct gene 2” the target gene in the sample of interest.
Immunohistochemical Characterization, Scanning Electron Microscopy, and Transmission Electron Microscopy
The ECM specimens were fixed in 4% paraformaldehyde and stained with hematoxylin and eosin (H&E). For immunohistochemistry staining, after rehydration and antigen retrieval, the sections were incubated at 4°C overnight with anti-laminin B2-γ1 antibody (1:100; Abcam, Cambridge, MA, http://www.abcam.com) or anti-fibronectin antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com). The secondary antibody (goat anti-mouse IgG horseradish peroxidase, 1:500; Abcam) was incubated at room temperature for 60 minutes. Visualization of the immunohistochemical reaction was performed with 3,3′-diaminobenzidine/H2O2 solution (Nichirei Bioscience, Tokyo, Japan, http://www.nichirei.co.jp) and counterstained with Mayer's hematoxylin. H&E and immunohistochemical characterization could not be performed on PLLA-collagen specimens owing to the sensitivity to histological solvents and the resulting inability of the sample to remain fixed to glass slides during the dehydration processes.
To assess cell viability, both ECM and PLLA-collagen scaffolds were subjected to live-dead staining using a live/dead kit (Thermo Fisher Scientific Life Sciences) and observed with the Nikon C2 confocal laser scanning system (Nikon Instruments, Tokyo, Japan, http://www.nikoninstruments.com). For scanning electron microscopy (SEM), the samples were fixed in 5 wt% glutaraldehyde for 4 hours, dehydrated in a graded ethanol series, critically point dried, coated with osmium, and observed by SEM (LEO Gemini 1525; Leo Electron Microscopy, Inc., Cambridge, U.K., http://www.smt.zeiss.com). For transmission electron microscopy (TEM), the specimens were fixed with 2.5% glutaraldehyde, subsequently postfixed with 1% osmium tetroxide for 1 hour, with 1% uranyl acetate in maleate buffer for 1 hour, dehydrated with graduated concentrations of ethanol and propylene oxide, and then embedded in Epon (polymerized at 60°C for 48 hours). Ultrathin specimens were observed using the Hitachi HT-7700 Biological TEM (Hitachi, Tokyo, Japan, http://www.hitachi.com).
Measurement of CYP1A1, CYP2C9, and CYP3A4 Enzyme Activities
Cells seeded in sandwich control culture, ECM, and PLLA-collagen scaffolds treated with or without induction agents (5 µM 3-methylcholanthrene for CYP1A2 or 50 µM rifampicin for CYP2C9 and CYP3A4) for 48 hours were incubated with 100 µM Luc-ME (CYP1A2) for 0.5 hour, 100 µM Luc-H (CYP2C9) for 4 hours, or 3 µM Luc-IPA (CYP3A4) for 0.5 hour (Promega, Madison WI, http://www.promega.com). Next, 100 µl of this luciferin medium was added to a 96-well white opaque plate with 100 µl of luciferin detection reagent, and the luminescence signal was measured on a Cytation 3 Cell Imaging Multi-Mode Reader (BioTek Inc., Winooski, VT, http://www.biotek.com). After determining the activity, the amount of cells in each scaffold or sandwich culture was analyzed using the Quant-iT PicoGreen Kit (Thermo Fisher Scientific Life Sciences) and the described protocols.
Statistical Analysis
Experimental data are presented as the mean ± SD. One-way analysis of variance was used for statistical analysis, with the Holm-Sidak test used for post hoc pairwise comparisons and testing against the control group. The Student t test was applied for two-group comparisons using SPSS (IBM Corp., Armonk, NY, http://ibm.com) and Microsoft Excel software (Microsoft Corp., Redmond, WA, http://www.microsoft.com). Differences were considered statistically significant at p ≤ .05. Additional methods are available in the supplemental online data.
Results
Development and Characterization of 3D Liver ECM Bioscaffolds
Acellular ECM scaffolds were developed by sequential perfusion of weak detergents through the liver vasculature (supplemental online Fig. 1A). The resulting scaffold appeared opaque (supplemental online Fig. 1A), and quantitative DNA analysis revealed a 98.9% reduction in DNA content after decellularization (native liver, 6163.7 ± 1221.6 ng/mg; decellularized ECM, 67.9 ± 7.7 ng/mg; p < .01; n = 4 for each group). Despite the near absence of DNA, indicating removal of the cellular compartment, the growth factors remained immobilized to structural proteins of the ECM. The content of hepatocyte growth factor (HGF) was 41.61 ± 13.36 ng/g in the decellularized liver matrix and 86.89 ± 16.76 ng/g in the native, untreated liver (p < .01). The content of basic fibroblast growth factor (bFGF) was 21.80 ± 5.02 ng/g in the decellularized liver scaffold and 45.50 ± 12.36 ng/g in the native, untreated liver (p = .03; supplemental online Fig. 1B). These results indicate that ∼50% of HGF and bFGF were preserved after decellularization, similar to scaffolds developed using other cell-removal strategies [25]. Fibronectin, laminin, and type I collagen proteins were further detected in the liver ECM by Western blot (supplemental online Fig. 1C). SEM and hematoxylin and eosin staining of the decellularized liver matrix also revealed the acellularity of liver ECM with preservation of the 3D lacunae structure (supplemental online Fig. 1D). Immunohistochemical characterization of the liver ECM (supplemental online Fig. 1D) further confirmed the matrix content and found laminin and fibronectin to be more prevalent around the vessel remnants and Glisson’s capsule.
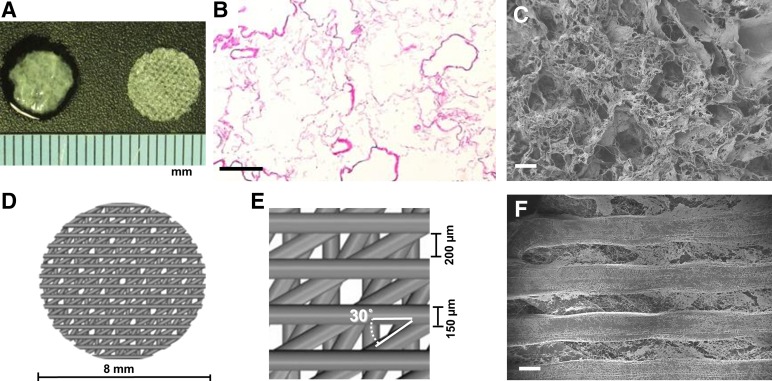
Individual ECM scaffolds (Fig. 1A, left), measuring 8 mm in diameter, were developed from the decellularized liver matrix, and preservation of the ECM porous microstructure was revealed by H&E staining (Fig. 1B) and SEM imaging (Fig. 1C). As a comparison matrix, we also developed a bio-hybrid PLLA-collagen scaffold (Fig. 1A, right) as a “deconstructed” 3D environment possessing an open pore structure and containing only type 1 collagen without growth factors (supplemental online Fig. 1B) or other structural proteins (supplemental online Fig. 1C). These scaffolds are composed of four 125-µm-thick layers (Fig. 1D). Each layer is composed of parallel PLLA struts measuring 150 µm in width and spaced 200 µm apart, with each successive layer oriented 30° with respect to the previous layer to mimic the porous nature of the liver ECM, albeit with a more homogenous architecture and thicker strut size afforded by the 3D bioprinting process (Fig. 1E). Infused collagen fibers were observed between the PLLA struts by SEM (Fig. 1F).
Figure 1.
Matrix architecture of three-dimensional extracellular matrix (ECM) and bioplotted poly-l-lactic acid (PLLA)-collagen scaffolds. (A): ECM scaffold (left) and porous PLLA-collagen scaffold (right). (B, C): Surface view of the ECM scaffold by hematoxylin and eosin staining (B) and scanning electron microscopy (SEM) imaging (C). (D, E): Computer-aided schematic diagram of the PLLA-collagen scaffold showing struts and pores. (F): SEM micrograph of the surface of PLLA-collagen scaffold depicting type 1 collagen infused between PLLA struts. Scale bars = 200 µm.
Cell Viability, Proliferation, and Migration
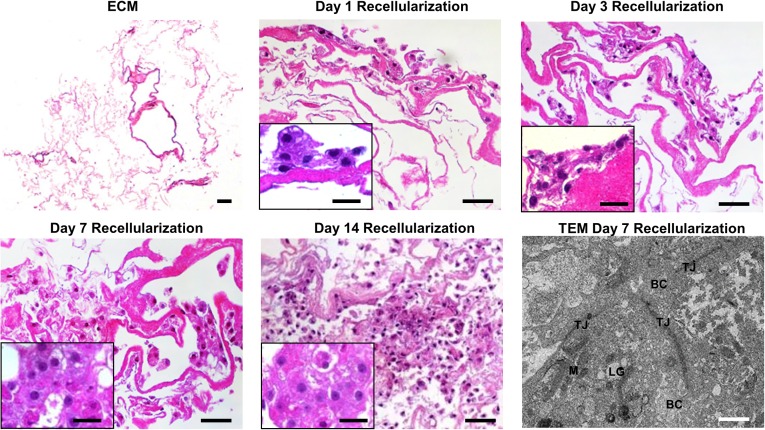
We performed initial phenotyping of commercially available, differentiated iPSC hepatocytes to establish a baseline and assess the initial hepatocyte-like characteristics compared with fresh primary hepatocytes. We found that hepatocyte-like cells differentiated from iPSCs express genes signifying hepatocyte lineage differentiation, albeit at a lower level (supplemental online Fig. 2A) with poorer synthetic function (albumin production; supplemental online Fig. 2B) and lower P450 enzyme activity (supplemental online Fig. 2C) compared with fresh primary human hepatocytes. The fetal hepatocyte markers, AFP and CYP3A7, were increased in iPSC hepatocytes compared with fresh primary hepatocytes. To assess the ability of our cell-free scaffolds to influence iPSC hepatocyte function and maturation, cells were seeded onto ECM or PLLA-collagen scaffolds or grown between a type 1 collagen monolayer and Matrigel overlay in a standardized control sandwich culture. SEM imaging of cell-laden scaffolds showed abundant iPSC hepatocytes within the interconnecting pore structure of the collagen network in the PLLA-collagen scaffolds and within the ECM scaffolds for 2 weeks (Fig. 2). Live/dead staining of iPSC hepatocytes within the scaffolds revealed that the cells remained viable and evenly distributed across the scaffolds after 14 days of culture (Fig. 2). H&E staining of ECM scaffolds in cross-section depicted cell attachment at day 1 and multilayered cell attachment after day 3, with penetration and migration into the matrix and formation of cell-cell connections on day 7, which continued through day 14 (Fig. 3). Bile canaliculi-like structures and formation of tight junctions between adjacent iPSC hepatocytes were observed on both ECM (Fig. 3; supplemental online Fig. 3) and PLLA-collagen (supplemental online Fig. 3) scaffolds by 7 days.
Figure 2.
Induced pluripotent stem cell (iPSC) hepatocytes survive and grow within ECM and PLLA-collagen scaffolds. SEM micrographs of recellularized ECM and PLLA-collagen scaffolds with iPSC hepatocytes over 14 days during an in vitro time course study. Red dashed lines delineate boundaries between PLLA bioprinted struts and infused type 1 collagen within three-dimensional bioprinted PLLA-collagen scaffolds. Staining of iPSC hepatocytes within ECM scaffolds or PLLA-collagen scaffolds showed live cells (green) with minimal dead cells (red) at day 14. Scale bars = 50 µm. Abbreviations: ECM, extracellular matrix; PLLA, poly-l-lactic acid; SEM, scanning electron microscopy.
Figure 3.
Induced pluripotent stem cell (iPSC) hepatocytes develop cell-cell contacts and biliary canaliculi within ECM scaffolds. Cross-sectional view of the ECM scaffold without cells and recellularized ECM scaffolds with iPSC hepatocytes after 1, 3, 7, and 14 days of in vitro culture by hematoxylin and eosin staining; scale bar = 50 µm. iPSC hepatocytes penetrated into the scaffold over time, indicating migration into the matrix scaffold. Insets: Higher magnification of iPSC hepatocytes within ECM scaffolds over time (×63; scale bars = 20 μm). TEM of iPSC hepatocytes in the ECM scaffold depicts formation of bile canaliculi-like structures flanked by tight junctions at day 7 (bottom right panel, scale bars = 1 μm; higher magnification shown in supplemental online Fig. 3). Abbreviations: BC, biliary canaliculi-like structures; ECM, extracellular matrix; LG, lipid globule; M, mitochondria; TEM, transmission electron microscopy; TJ, tight junction.
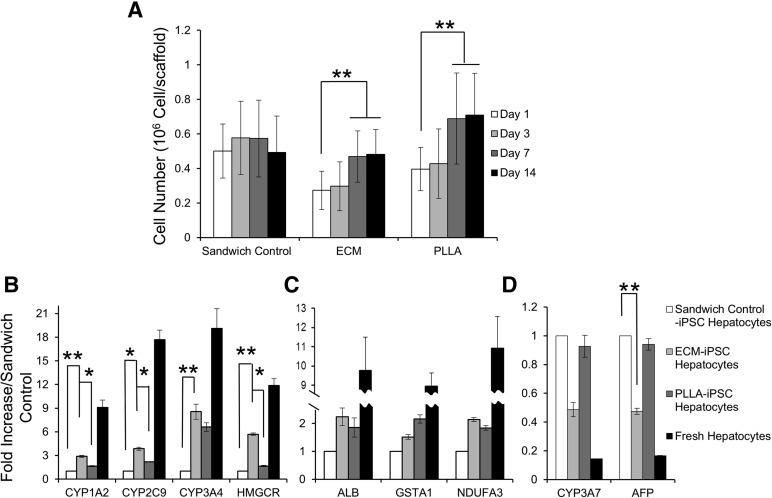
Total DNA content was used to determine the cell density and proliferation of iPSC hepatocytes in ECM, PLLA-collagen scaffold, and sandwich control groups at day 1, which was 0.27 ± 0.11 × 106, 0.39 ± 0.13 × 106, and 0.50 ± 0.15 × 106 cells per scaffold, respectively. The cell density in both engineered scaffolds increased during the first 7 days, with a 1.7-fold increase in iPSC hepatocytes grown in either the ECM scaffold or PLLA-collagen scaffold. The cell density of the sandwich control group did not show any significant change, indicating an absence of proliferation during the 14-day culture period (p = .54; Fig. 4A).
Figure 4.
iPSC hepatocyte culture within three-dimensional scaffolds leads to increased proliferation and expression of mature liver-specific markers. (A): Cell proliferation of iPSC hepatocytes in ECM scaffolds, PLLA-collagen scaffolds, and sandwich controls (n = 4 samples for each group). Quantitative reverse transcription-polymerase chain reaction of metabolic and synthetic genes in iPSC hepatocytes that either changed (B) or remained stable (C) in response to growth in ECM and PLLA-collagen scaffolds compared with growth in standard sandwich control culture at day 14. Fresh, human hepatocytes are shown for comparison. (D): Expression of fetal genes CYP3A7 and AFP in iPSC hepatocytes in ECM scaffolds or PLLA-collagen scaffolds compared with control sandwich culture at day 14 (∗, p < .05; ∗∗, p < .01; n = 4 for each group). Abbreviations: AFP, α-fetoprotein; ECM, extracellular matrix; HMGCR, human 3-hydroxy-3-methylglutaryl-coenzyme A reductase; iPSC, induced pluripotent stem cell; PLLA, poly-l-lactic acid.
Increased Expression of Hepatocyte-Specific Markers in ECM Scaffolds
mRNA transcripts encoding phase I and II xenobiotic enzymes (CYP1A2, CYP2C9, CYP3A4, and human 3-hydroxy-3-methylglutaryl-coenzyme A reductase [HMGCR]) assessed on day 14 were higher in iPSC hepatocytes grown in either ECM or PLLA-collagen scaffolds compared with the sandwich control group. Expression of CYP2C9 increased approximately threefold, CYP3A4 increased approximately eightfold, and HMGCR, a rate-limiting enzyme in the isoprenoid/cholesterol biosynthesis pathway and involved in cholesterol metabolism [29], increased approximately sixfold (Fig. 4B) compared with the sandwich control group. In addition, iPSC hepatocytes grown within ECM scaffolds had significantly higher mRNA levels compared with those in the less chemically intricate PLLA-collagen scaffolds: CYP1A2 (1.7-fold; p = .039), CYP2C9 (1.7-fold; p = .028), and HMGCR (3.3-fold; p = .02). However, mRNA synthesis of a mitochondrial membrane respiratory chain enzyme (NDUFA3; Fig. 4C; p = .11, ECM vs. sandwich control), a glutathione synthesis enzyme (GSTA1; Fig. 4C; p = .32, ECM vs. sandwich control), and albumin (Fig. 4C, p = .11, ECM vs. sandwich control) did not show significant differences between ECM scaffolds and either the PLLA-collagen scaffolds or sandwich control group.
To investigate potential phenotypic maturation of cells in ECM scaffolds, mRNA transcripts of fetal markers CYP3A7 and AFP in iPSC hepatocytes grown in ECM scaffolds displayed a decreasing trend at day 14 (CYP3A7, 49% of sandwich control [p = .16]; AFP, 47% of sandwich control [p = .041]). However, both CYP3A7 and AFP remained essentially unchanged in the PLLA-collagen scaffold (CYP3A7, 93% of sandwich control [p = .64]; AFP, 94% of sandwich control [p = .54]; Fig. 4D).
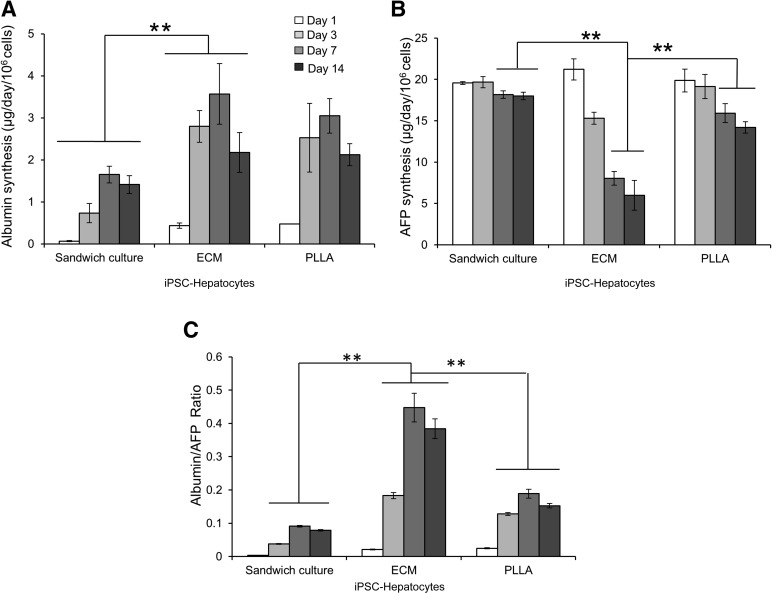
Synthesis of albumin by iPSC hepatocytes grown within the ECM was significantly higher than that in the sandwich control group throughout the 14-day culture period (p < .01; Fig. 5A) and was similar to that in the PLLA-collagen scaffolds (p = .58). Nonetheless, at day 14, albumin synthesis by iPSC hepatocytes grown within ECM scaffolds (2.18 ± 0.47 µg/day per 106 cells) was slightly lower than at day 7 (Fig. 5A) but was not significantly different from that of primary human hepatocytes grown within the ECM scaffolds (2.75 ± 0.31 µg/day per 106 cells; p = .057; supplemental online Fig. 4) over the same time course.
Figure 5.
iPSC hepatocytes express a higher degree of maturity and albumin synthesis within ECM scaffolds. Graphs show albumin production (A), AFP biosynthesis (B), and albumin/AFP protein synthesis ratio (C) in iPSC hepatocytes within ECM scaffolds, PLLA-collagen scaffolds, and sandwich controls throughout 14 days of culture (∗∗, p < .01; n = 4 for each group). Abbreviations: AFP, α-fetoprotein; ECM, extracellular matrix; iPSC, induced pluripotent stem cell; PLLA, poly-l-lactic acid.
AFP production in iPSC hepatocytes grown within ECM scaffolds consistently and significantly decreased from a baseline on day 1 (21.21 ± 1.27 µg/day per 106 cells) through day 7 (8.03 ± 0.82 µg/day per 106 cells; p < .01) to day 14 (5.98 ± 1.78 µg/day per 106 cells; p < .01; Fig. 5B). However, AFP synthesis in iPSC hepatocytes grown within the sandwich control environment did not show any significant decline during this interval (p = .87). Moreover, AFP production in PLLA-collagen scaffolds decreased slightly until day 14 (19.86 ± 1.39 µg/day per 106 cells on day 1; 14.19 ± 0.69 µg/day per 106 cells on day 14; p = .015), although it did not reach the low level of synthesis found in iPSC hepatocytes grown in ECM scaffolds. This finding suggests that the more complex biochemical milieu within the ECM scaffolds leads to the enhanced maturation of iPSC hepatocytes. Nonetheless, AFP synthesis in primary human hepatocytes remained low compared with that in iPSC hepatocytes grown in all environments (p < .01; supplemental online Fig. 4). Using the ratio of albumin to AFP synthesis as a gauge of cellular maturation, iPSC hepatocytes grown in ECM scaffolds consistently had a higher ratio compared with cells grown in either PLLA-collagen scaffolds or sandwich culture (Fig. 5C), clearly demonstrating cellular maturation in the ECM bioscaffold system.
Increased Liver-Specific Function in 3D ECM Scaffolds
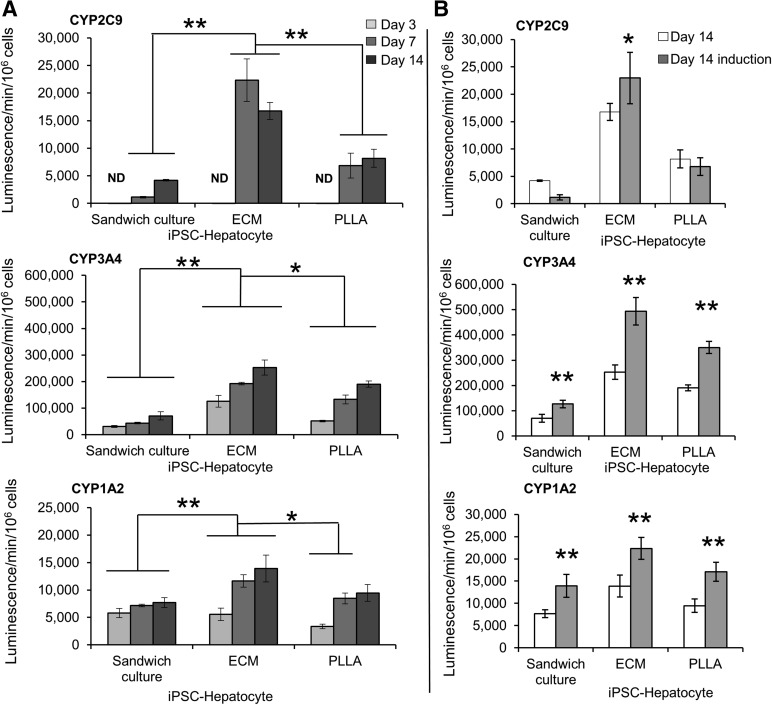
Notably, CYP2C9 activity was nearly undetectable in iPSC hepatocytes before growth within scaffolds (supplemental online Fig. 2C) or within the sandwich control environment, ECM, or PLLA-collagen scaffolds at day 3 (Fig. 6A). Nonetheless, on day 7, the CYP2C9 activity of iPSC hepatocytes in ECM scaffolds increased 20.2-fold (Fig. 6A) beyond that of iPSC hepatocytes grown in sandwich culture on the same day and was also significantly higher than that in iPSC hepatocytes in PLLA-collagen scaffolds (p = .02 for both; Fig. 6A). CYP3A4 activity in iPSC hepatocytes grown within ECM scaffolds at day 14 was 3.6-fold higher than that in iPSC hepatocytes in the sandwich control group (p < .01) and 1.3-fold higher than that in iPSC hepatocytes in the PLLA-collagen scaffold (p = .02). CYP1A2 activity of iPSC hepatocytes grown on ECM scaffolds at day 14 was 1.8-fold higher than that of iPSC hepatocytes in the sandwich control group (p = .01) and 1.5-fold higher than that of iPSC hepatocytes in PLLA-collagen scaffolds (p = .04; Fig. 6A). Rifampicin induced CYP3A4 activity by twofold in iPSC hepatocytes within ECM scaffolds, slightly more than that in iPSC hepatocytes within either the sandwich control or PLLA-collagen scaffold on day 14 (Fig. 6B). However, induction of CYP1A2 (by 3-methylcholanthrene) or CYP2C9 (by rifampicin) in iPSC hepatocytes was modest compared with that in primary hepatocytes. Taken together, the activity of CYP1A2, CYP2C9, and CYP3A4 in iPSC hepatocytes tended to increase in 3D culture, with ECM scaffolds conferring the most robust phenotype and primary hepatocytes generally losing activity in all three environments over 2 weeks (Fig. 7).
Figure 6.
ECM scaffolds confer increased P450 activity to iPSC hepatocytes. (A): CYP2C9, CYP3A4, and CYP1A2 activity in iPSC hepatocytes within ECM scaffolds, PLLA-collagen scaffolds, and sandwich control environments at 3, 7, and 14 days of culture. (B): Response of iPSC hepatocytes to treatment with P450 inducers, rifampicin (CYP2C9 and CYP3A4) and 3-methylcholanthrene (CYP1A2), within ECM scaffolds, PLLA-collagen scaffolds, and sandwich controls at day 14 (∗, p < .05; ∗∗, p < .01; n = 3 for each group at each time point). Abbreviations: ECM, extracellular matrix; iPSC, induced pluripotent stem cell; ND, not detected; PLLA, poly-l-lactic acid.
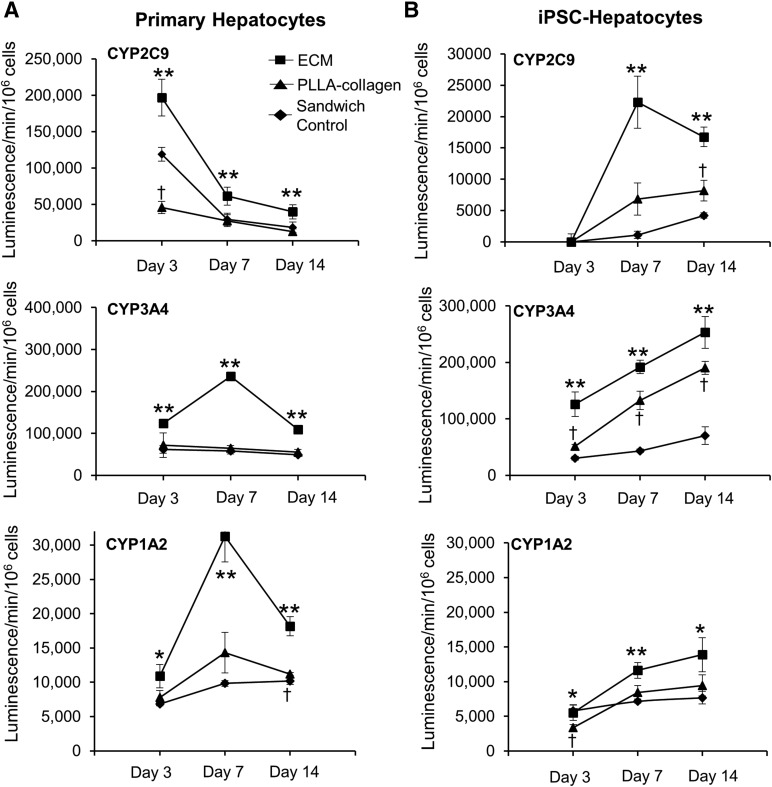
Figure 7.
P450 activity showed a trend toward higher in iPSC hepatocytes over time. CYP2C9, CYP3A4, and CYP1A2 activity showed a trend in primary cryopreserved hepatocytes (A) and iPSC hepatocytes (B) within ECM scaffolds, PLLA-collagen scaffolds, and sandwich control groups after 3, 7, and 14 days of culture. CYP activity of primary hepatocytes decreased and CYP activity of iPSC hepatocytes increased over time, with ECM scaffolds conferring the most robust phenotype for both cell types. Data are presented as mean ± SD (n = 3 for each group at each time point). Squares indicate ECM scaffold; triangles, PLLA-collagen scaffold; diamonds, sandwich control. ∗, Significant difference between ECM scaffolds versus PLLA-collagen scaffolds or sandwich control groups; ∗∗, significant difference between ECM scaffolds versus both PLLA-collagen scaffolds and sandwich control groups; †, significant difference between PLLA-collagen scaffolds and sandwich control groups. Abbreviations: ECM, extracellular matrix; iPSC, induced pluripotent stem cell; PLLA, poly-l-lactic acid.
Discussion
iPSCs can be engineered from patient tissue and then differentiated along defined developmental pathways toward maturity. Thus, iPSC-derived cells have the same genetic makeup as the donor patient, making them an ideal cell source for disease modeling, pharmacokinetics, and hepatotoxicity testing. We hypothesized that the 3D microenvironment could enhance the growth, function, and maturation of iPSC hepatocytes without the need for coculture with a stromal cell population. To evaluate this, we compared two cell-free scaffold systems, the first using natural ECM scaffolds, developed from decellularized rat livers that contain the original pore architecture and overall structure of the whole organ matrix, and a second, biosynthetic hybrid PLLA-collagen scaffold developed using 3D bioplotting technology, an advanced rapid-prototyping technique to produce geometrically tunable micron-scale scaffolds. This new technology allows us to create a “deconstructed” 3D control environment to investigate the mechanism of action by defining the incremental contribution of both the 3D architecture and the additive matrix function by infusing collagen into the bioplotted scaffold. Moreover, the small scale of both scaffolds provides an easy and rapid platform for studies analyzing hepatocyte development and drug efficacy and toxicity.
We demonstrated that decellularized rat ECM is cell-free by histologic and electron microscopy analysis with ∼1% residual DNA remaining compared with the native liver, similar to other published decellularization strategies for tissues and organs [25, 30]. Immunohistochemical and growth factor content analyses of the decellularized ECM further demonstrated preservation of fibronectin and laminin and ∼50% HGF and FGF. Moreover, the ECM scaffolds allowed iPSC hepatocyte survival, proliferation, and migration within this natural matrix.
One universal limitation of hepatocyte-like cells differentiated from iPSCs is their decreased phenotypic maturity with lower liver-specific enzyme function compared with that of primary human hepatocytes [31]. However, iPSC hepatocytes grown within our ECM scaffolds exhibited increased P450 function. Not only was the expression of CYP1A2 (acetaminophen metabolism), CYP2C9 (warfarin metabolism), CYP3A4 (metabolism of ∼50% of medications by the P450 system), and HMGCR transcripts higher in iPSC hepatocytes grown on ECM scaffolds, but this also translated into consistently higher P450 enzyme activity in iPSC hepatocytes on a day-by-day basis in ECM scaffolds compared with both “deconstructed” 3D controls (e.g., PLLA-collagen) and standard sandwich cultures. We found that iPSC hepatocytes display extremely low, and at times undetectable, CYP2C9 activity at initial (day 0) and early time points (∼3 days) in all environments, and this continued throughout the 14-day culture period in standard sandwich cultures. CYP2C9 metabolizes warfarin and phenytoin at varying rates based on the genetic polymorphism present within each individual. The cause of low CYP2C9 activity observed within the starting iPSC hepatocyte population is unclear, although it might result from the known genetic heterogeneity at the CYP2C9 locus conferring decreased activity or altered transcriptional regulation. Nonetheless, the use of ECM scaffolds led to a 20-fold increase beyond baseline in CYP2C9 activity at day 7. Although it is challenging to compare hepatocyte characteristics between studies using microscaffolding techniques owing to differences in controls, data analysis, and normalization methods, other microscaffolding systems have been developed to culture stem cell-derived hepatocytes; however, none showed as robust or as early induction of CYP2C9 [21, 23]. However, other liver ECM-based biomatrix scaffolds have been developed by delipidation using a separate strategy (phospholipase A2, high salt wash, and nuclease treatment) to obtain a scaffold that was >95% DNA free with retention of matrix-bound growth factors [32]. In that study, Wang et al. [32] demonstrated a 1.3-fold increase in CYP3A4 activity in cryopreserved human hepatocytes grown on these scaffolds compared with the same cells grown on type 1 collagen but did not assess CYP2C9 activity. In the present study, our ECM scaffold was produced with a distinct strategy (1% Triton X-100 and 0.1% NH4OH) that likewise produced a cell-free environment with bound growth factors and structural proteins. As a model to promote cellular maturation in iPSC-derived cells, we tested our 3D environments to enhance the proliferation and phenotypic properties of iPSC-derived hepatocytes, a relatively new cell type, and showed that growth on ECM scaffolds leads to a population closer to primary human hepatocytes with decreased AFP synthesis, an increased albumin/AFP ratio, and a trend toward lower expression of fetal-specific markers, AFP and CYP3A7. Together with the increasing enzyme function of CYP2C9, CYP3A4, and CYP1A2, this trend provides evidence that growth within ECM scaffolds helps to overcome a primary obstacle of iPSC hepatocytes by conferring increasing maturity and potency to hepatocyte-like cells derived from iPSCs, which could be used by others to better model disease or analyze the metabolism of pharmaceutical compounds.
Our use of bioprinting nanotechnology further allowed us to develop 3D comparison environments to evaluate cell phenotypes within a multidimensional, but chemically simplified, environment to indicate the potential mechanisms leading to changes in cell function observed in ECM scaffolds. Although clearly distinct from natural liver architecture, 3D bioprinting technology can be used to develop uniform, tunable scaffolds to evaluate cell growth within a defined matrix composition. We have shown that bioplotted PLLA-collagen scaffolds provide a 3D environment that permits iPSC hepatocyte proliferation with increased expression and activity of liver-specific enzymes relative to standard sandwich culture. However, the levels of each of these indexes were lower in 3D-printed scaffolds than in the cells grown in ECM-derived scaffolds. As such, the results from experiments using PLLA-collagen printed scaffolds suggest that growth in 3D scaffolds and the presence of type 1 collagen encourage hepatocyte function, albeit to a limited degree. A reduction in fetal genes CYP3A7 and AFP was not observed in iPSC hepatocytes grown on PLLA-collagen scaffolds, indicating that both multidimensional growth and appropriate matrix biochemical complexity (e.g., in chemically diverse ECM scaffolds) is a requirement for maturation of iPSC hepatocytes.
Our use of 3D-printed scaffolds allowed a degree of comparison with other collagen scaffolding systems such as the Real Architecture for 3D Tissues [33] system (TAP Biosystems, Royston, U.K., http://www.tapbiosystems.com), which uses type 1 collagen as a substrate [22]. Using this system, Gieseck et al. [22] found albumin secretion to be nearly constant at ∼1.0 μg/day per 106 cells 10–20 days after iPSC differentiation to a common hepatocyte progenitor cell. In contrast, our study showed that PLLA-collagen scaffolds led to an albumin synthesis rate of ∼3 μg/day per 106 cells and ECM scaffolds to an albumin synthesis rate of ∼3–4 μg/day per 106 cells between days 7 and 14.
Although iPSC hepatocytes in all our three culture systems remained immature compared with fresh human primary hepatocytes, we showed a specific effect of ECM composition on the biochemical function of iPSC hepatocytes compared with PLLA-collagen scaffolds. ECM scaffolds contain multiple structural proteins, including laminin and fibronectin, and growth factors that are likely contributory on a multifactorial level through induction of hepatic nuclear factors and gene networks. Further metabolic maturation of iPSC hepatocytes might be possible in both systems through coculture with liver stroma cells, which have been shown to increase the catalytic function of iPSC hepatocytes in biomimetic liver systems [21]. Our findings also suggest that proliferation, maturation, and hepatic function of iPSC hepatocytes might be further enhanced by producing highly controlled, fully porous, custom synthetic scaffolds through 3D bioprinting with increased matrix function by “adding back” individual specific ECM components to develop fully synthetic scaffolds that retain the individual, structural building blocks that maximize cell maturation. ECM scaffolds contain FGF, HGF, laminin, fibronectin, and other matrix components that are absent from 3D bioplotted PLLA-collagen scaffolds but might influence maturation of iPSC hepatocytes in our system, as a recent study revealed that a fragment of the ECM protein vitronectin can replace Matrigel to support differentiation of iPSCs into hepatocyte-like cells [34]. The development of tunable inks for 3D bioprinting [35, 36] to permit the incorporation of specific structural, growth factor, and mitogenic components can eventually lead to the development of fully synthetic scaffolds that confer a maturation advantage to iPSC hepatocytes similar to that of natural ECM scaffolds. Ongoing work entails altering the synthetic, bioplotted scaffold to better emulate the mechanical and bioactive properties of native liver tissue to determine direct influence on iPSC hepatocyte function.
Conclusion
In the present study, we compared the effect of two 3D bioscaffold systems to reverse the primary limitation of iPSC hepatocytes. We found that the cellular phenotypic properties of proliferation, function, and maturation are influenced to differing degrees within (a) biologically heterogeneous, naturally occurring ECM scaffolds derived from decellularized liver tissue and (b) biosynthetic, collagen-infused 3D-printed porous PLLA scaffolds. iPSC hepatocyte proliferation and function were significantly enhanced when cultured within ECM scaffolds compared with standard sandwich culture controls. In addition, although culture within the biosynthetic PLLA scaffold improved some important aspects of iPSC hepatocyte proliferation and function, of the two 3D-cultured systems, only the ECM scaffold enhanced phenotypic maturation of iPSC hepatocytes compared with the other platforms, likely because of its biologically diverse makeup, including both its 3D configuration and biological components, such as growth factors and structural proteins that regulate cell function and intracellular signaling via transmembrane receptors. Taken together, the small size of both 3D scaffolds allows for rapid, careful analysis of key growth conditions, allowing matrices such as these to be used as high-throughput platforms to influence the development of iPSC hepatocytes to ultimately model disease or evaluate hepatotoxicity to small molecular pharmaceutics.
Supplementary Material
Acknowledgments
We thank the Zell Family Foundation for support. We acknowledge the Liver Scholar Award from the American Association for the Study of Liver Diseases and the American Liver Foundation to J.A.W. We recognize support from the Northwestern Memorial Foundation Dixon Translational Research Grants Initiative and the NIH (Grants K01DK099454 and K08DK101757). We recognize the generous support of Frank A. Krumlovsky, a career development award from the Society for Surgery of the Alimentary Tract, and the Robert R. McCormick Foundation. We acknowledge the ARAID Foundation support to P.M.B. We acknowledge the support from the North Carolina General Assembly for the research performed by S.S. and P.M.B. We thank Cellular Dynamics International for supplying the iCell Hepatocytes. We thank Dr. David Mann for critical review of the manuscript. We acknowledge the use of the following facilities at Northwestern University: Northwestern University Microsurgery Core, Cell Imaging Facility, generously supported by National Cancer Institute Grant CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center; Electron Probe Instrumentation Center, which has received support from National Science Foundation Grant DMR-0520513 and Engineering Center Award EEC-0118025/003; and the Equipment Core Facilities at the Simpson Querrey Institute for BioNanotechnology at Northwestern University, developed by support from the U.S. Army Research Office, the U.S. Army Medical Research and Material Command, and Northwestern University.
Author Contributions
B.W.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, critical revisions for important intellectual content, final approval of manuscript; A.E.J.: collection and/or assembly of data, data analysis and interpretation, critical revisions for important intellectual content, final approval of manuscript; P.M.B., S.S., A.S.-G., M.M.A., and R.N.S.: data analysis and interpretation, critical revisions for important intellectual content, final approval of manuscript; J.A.W.: conception and design, data analysis and interpretation, manuscript writing, critical revisions for important intellectual content, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.M.A. has compensated stock options in Transplant Genomics, Inc. J.A.W. has nonfinancial research support from Cellular Dynamics International. The iPSC hepatocytes were provided free or at a discount; this material was not consumed by or used on patients. The authors indicated no potential conflicts of interest.
References
- 1.Cayo MA, Cai J, DeLaForest A, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz RE, Trehan K, Andrus L, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebert AD, Yu J, Rose FF, Jr, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye L, Chang JC, Lin C, et al. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raya A, Rodríguez-Pizà I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee G, Papapetrou EP, Kim H, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirenko O, Cromwell EF, Crittenden C, et al. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol Appl Pharmacol. 2013;273:500–507. doi: 10.1016/j.taap.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S, Rezvani M, Harbell J, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamao H, Mandai M, Okamoto S, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid ST, Corbineau S, Hannan N, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Chen S, Li W, et al. Rescue of ATP7B function in hepatocyte-like cells from Wilson’s disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176–3187. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- 12.Adams RM, Wang M, Crane AM, et al. Effective cryopreservation and long-term storage of primary human hepatocytes with recovery of viability, differentiation, and replicative potential. Cell Transplant. 1995;4:579–586. doi: 10.1177/096368979500400607. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Zhao Y, Liu Y, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 14.Duan Y, Catana A, Meng Y, et al. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25:3058–3068. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- 15.Hay DC, Fletcher J, Payne C, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires activin A and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basma H, Soto-Gutiérrez A, Yannam GR, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo A, Kim YH, Irion S, et al. The homeobox gene Hex regulates hepatocyte differentiation from embryonic stem cell-derived endoderm. Hepatology. 2010;51:633–641. doi: 10.1002/hep.23293. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan GJ, Hay DC, Park IH, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancho-Bru P, Roelandt P, Narain N, et al. Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol. 2011;54:98–107. doi: 10.1016/j.jhep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Berger DR, Ware BR, Davidson MD, et al. Enhancing the functional maturity of iPSC-derived human hepatocytes via controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 22.Gieseck RL, III, Hannan NR, Bort R, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayama K, Kawabata K, Nagamoto Y, et al. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 24.Loneker AE, Faulk DM, Hussey GS, et al. Solubilized liver extracellular matrix maintains primary rat hepatocyte phenotype in-vitro. J Biomed Mater Res A. 2016;104:957–965. doi: 10.1002/jbm.a.35636. [DOI] [PubMed] [Google Scholar]
- 25.Soto-Gutierrez A, Zhang L, Medberry C, et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng Part C Methods. 2011;17:677–686. doi: 10.1089/ten.tec.2010.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baptista PM, Siddiqui MM, Lozier G, et al. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–617. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 27.Kajbafzadeh AM, Javan-Farazmand N, Monajemzadeh M, et al. Determining the optimal decellularization and sterilization protocol for preparing a tissue scaffold of a human-sized liver tissue. Tissue Eng Part C Methods. 2013;19:642–651. doi: 10.1089/ten.TEC.2012.0334. [DOI] [PubMed] [Google Scholar]
- 28.Wertheim JA, Baptista PM, Soto-Gutierrez A. Cellular therapy and bioartificial approaches to liver replacement. Curr Opin Organ Transplant. 2012;17:235–240. doi: 10.1097/MOT.0b013e3283534ec9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 30.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Z, Cai J, Liu Y, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Cui CB, Yamauchi M, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2011;53:293–305. doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 33.Nash RA, Hutton GJ, Racke MK, et al. High-dose immunosuppressive therapy and autologous hematopoietic cell transplantation for relapsing-remitting multiple sclerosis (HALT-MS): A 3-year interim report. JAMA Neurol. 2015;72:159–169. doi: 10.1001/jamaneurol.2014.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaoka M, Kobayashi M, Kawai C, et al. Design of a vitronectin-based recombinant protein as a defined substrate for differentiation of human pluripotent stem cells into hepatocyte-like cells. PLoS One. 2015;10:e0136350. doi: 10.1371/journal.pone.0136350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakus AE, Rutz AL, Shah RN. Advancing the field of 3D biomaterial printing. Biomed Mater. 2016;11:014102. doi: 10.1088/1748-6041/11/1/014102. [DOI] [PubMed] [Google Scholar]
- 36.Rutz AL, Hyland KE, Jakus AE, et al. A multimaterial bioink method for 3D printing tunable, cell-compatible hydrogels. Adv Mater. 2015;27:1607–1614. doi: 10.1002/adma.201405076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.