This study shows that exosomes secreted from umbilical mesenchymal stem cells (uMSCs) have potent antiviral activity against hepatitis C virus, especially by targeting viral replication. The high-throughput microRNA (miRNA) sequencing of uMSC exosomes and subsequent functional assay suggest that the antiviral process is mainly mediated by a series of miRNAs transported specifically through exosomes.
Keywords: Exosomes, Umbilical mesenchymal stem cell, Hepatitis C virus, Antiviral therapy, MicroRNA
Abstract
Hepatitis C virus (HCV) is a significant global public health problem, causing more than 350,000 deaths every year. Although the development of direct-acting antivirals has improved the sustained virological response rate in HCV patients, novel anti-HCV agents with higher efficacy as well as better tolerance and cheaper production costs are still urgently needed. Cell-based therapy, especially its unique and strong paracrine ability to transfer information to other cells via extracellular vesicles such as exosomes, has become one of the most popular therapeutic methods in recent years. In our study, exosomes secreted from umbilical mesenchymal stem cells (uMSCs), which are widely used in regenerative medicine, inhibited HCV infection in vitro, especially viral replication, with low cell toxicity. Our analysis revealed that microRNAs (miRNAs) from uMSC-derived exosomes (uMSC-Exo) had their unique expression profiles, and these functional miRNAs, mainly represented by let-7f, miR-145, miR-199a, and miR-221 released from uMSC-Exo, largely contributed to the suppression of HCV RNA replication. These four miRNAs possessed binding sites in HCV RNA as demonstrated by the target prediction algorithm. In addition, uMSC-Exo therapy showed synergistic effect when combined with U.S. Food and Drug Administration-approved interferon-α or telaprevir, enhancing their anti-HCV ability and thus improving the clinical significance of these regenerative substances for future application as optimal adjuvants of anti-HCV therapy.
Significance
This work reported, for the first time, the identification of stem cell-derived exosomes of antiviral activity. Umbilical mesenchymal stem cell-secreted exosomes inhibited hepatitis C virus infection through transporting a mixture of microRNAs complementing the viral genomes to the host cells. This finding provides insights and prospects for physiologically secreted substances for antiviral therapy.
Introduction
Hepatitis C virus (HCV) infects approximately 3% of the world population [1]. It is one of the major causes of chronic liver diseases, such as steatosis, cirrhosis, and hepatocellular carcinoma. Despite that, it is still difficult to develop effective vaccines against HCV. The traditional standard of care for HCV infection consists of pegylated interferon (IFN)-α and ribavirin. The recent advanced direct-acting antivirals, such as the protease inhibitor simeprevir and the nucleotide analog sofosbuvir, have optimized the therapeutic effect by increasing the sustained virological response (SVR) rate (the frequency of treated patients who achieve SVR) in HCV patients [2, 3]; however, there are still some unsolved problems, such as virus resistance, concomitant adverse reactions, and undeniably high medical expenses, related to their use in HCV treatment [4].
Mesenchymal stem cells (MSCs) are promising progenitor cells that harbor valuable therapeutic or biological capacities [5, 6]. Studies have revealed many functions mediated by this type of cell, such as reducing inflammation, modulating immune responses, promoting tissue regeneration, and inhibiting tumor progression [7–9]. In addition, MSC treatment has been the focus of more than 560 clinical trials that supply exogenous or activate endogenous MSCs [10] and has become the most effective source of cell-based therapy [11, 12]. Human umbilical cord MSCs (uMSCs) are a kind of MSC derived from umbilical cord through a noninvasive procedure, allowing robust in vitro culture expansion, and its physiological properties could be basically reserved under cryostorage [13–15]. They require relatively low cost to harvest and are promising candidates for regenerative medicine in the future.
Recent studies indicate that the functional mechanism of MSCs mainly relies on paracrine signaling, and exosomes are likely to be the key constituents of this process [16, 17]. Exosomes are membrane vesicles of 50–100 nm in size, which can be released from all kinds of cells [18, 19]. They can deliver specific functional RNAs (e.g., microRNAs [miRNAs]), proteins, and lipids from donor to recipient cells by direct fusion or active uptake [20, 21]. Because of this shuttling property, exosomes take part in many physiological or pathological processes, including intracellular communication, modulation of immune responses, and tumorigenesis [22–24]. Reports have also shown that exosomes could shuttle both infectious cargo and protective host molecules between cells [25–31], but the research of their roles in pathogen infection is still in an early stage. MSCs can produce a high amount of exosomes (MSC-Exo) compared with other cell types [32], and MSC-Exo exert an influence on various diseases similar to that of MSCs [17].
It was reported that the main form of functional RNA in exosomes is miRNA [33, 34]. The expression profile of miRNA has a signature of the origin parental cells, which could be determined by specific techniques, such as RNA sequencing. Increasing evidence suggests that exosome-packaged miRNAs are maintained in stable condition and play important regulatory roles in recipient cells [35]. miRNAs wrapped in MSC-Exo play a role in several diseases, such as promoting recovery in Parkinson’s disease and spine injury [36]. Hence, exosomal miRNAs may be valuable tools for treating certain diseases.
Herein we report that uMSC-derived exosomes (uMSC-Exo) were potent anti-HCV agents. By testing exosomes from multiple cell types, we found that uMSC-Exo were able to prevent viral replication and showed low cytotoxicity compared with other antiviral agents. Through a series of mechanism-related studies, we discovered that the antiviral mechanism of uMSC-Exo was mainly through transporting a series of antiviral exosomal miRNAs to target cells. Our study demonstrated for the first time that uMSC-Exo could repress HCV infection, thus adding a new functional role of uMSC-Exo and providing new insights and prospects for the development of optimal antiviral agents in the future.
Materials and Methods
Cell Culture
Human umbilical cords and skin were obtained from Changhai Hospital affiliated to Second Military Medical University. uMSCs and human dermal fibroblast (HF) cells were then isolated and cultured as previously described [37]. Human umbilical cords were donated from five healthy mothers who had natural childbirth, all of whom were negative for human pathogens, including HCV. Briefly, fresh umbilical cords and skin were first washed with 70% ethanol and then serum-free DMEM (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com) to remove excess blood. The tissues were then minced into small pieces (2–4 mm) and cultured in DMEM containing 10% fetal bovine serum (FBS) at 37°C. When the cells reached 70% confluence, they were trypsinized for subculture with 0.05% trypsin and 0.025% EDTA. uMSCs were cultured in DMEM with 10% FBS, 100 IU/ml streptomycin and penicillin, and 2 mM l-glutamine (Thermo Fisher Scientific Life Sciences). HF cells were cultured in high-glucose DMEM supplemented with 10% FBS, 100 IU/ml streptomycin and penicillin, and 2 mM l-glutamine. The fourth and fifth passages of the cells were used in the subsequent experiments. Human hepatoma (Huh) 7 cells and human embryonic kidney (HEK) 293T cells were cultured in DMEM containing 10% FBS, 1× nonessential amino acids, 100 IU/ml of streptomycin and penicillin, and 2 mM l-glutamine at 37°C with 5% CO2 and 90% humidity.
Viability assay was performed after 24 hours of treatment of indicated concentrations of uMSC-Exo using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H tetrazolium bromide (Beyotime Biotechnology, Haimen, China, http://www.beyotime.com) according to the manufacturer’s instructions [38]. For exosome-related microwell experiments, we used standard medium volume.
Exosomes Isolation and Identification
Exosomes were extracted and purified according to previous protocols [39, 40]. Briefly, uMSCs were cultured in DMEM with 10% FBS, which was pretreated by ultracentrifugation at 120,000 g overnight at 4°C to remove serum exosomes in T75 or T150 flasks. When cells reached 80% confluence (about 2–3 days), cell medium was harvested every other day. Cells in each flasks were continuously cultured for exosome collection for no more than 12 days. Cell supernatants were collected and centrifuged at 10,000 g for 30 minutes. The supernatants were then filtered through a 0.22-μm membrane and ultracentrifuged at 120,000 g for 70 minutes at 4°C. The supernatants were transferred to a new tube to undergo another ultracentrifugation at 120,000 g for 3 hours at 4°C to pellet the exosomes. The exosomes were resuspended in RNAase-free phosphate-buffered saline (PBS) and quantified by measuring their protein contents using the bicinchoninic acid protein assay kit (Thermo Fisher Scientific Life Sciences). uMSC-Exo generated from different donors were labeled individually and used in discrete experiments.
Isolated exosomes were subsequently identified by measuring the rate of Brownian motion with NTA NS300 (NanoSight, Malvern Instruments, Malvern, U.K., http://www.malvern.com/) equipped with fast video capture and particle-tracking software. The exosomal surface marker protein expression of CD81 and CD63 was detected using Western blotting.
PKH67 Analysis
Isolated exosomes were tested for the ability to enter cells. The uMSC-Exo were labeled using PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) according to the manufacturer’s protocol. The supernatant of ultracentrifugal exosomes was also labeled as a negative control. Labeled exosomes were then incubated with Huh7 cells for 6 hours at 37°C and then washed three times with PBS. The nuclei were stained with Hoechst 33342 (10 μg/ml) for 20 minutes before the cells were observed under a fluorescence microscope (Olympus, Tokyo, Japan, http://www.olympus-global.com).
Indirect Immunofluorescence Assay
Infected Huh7 cells were washed with PBS and fixed with cold methanol, and NS5A expression in the cells was detected using a primary antibody of NS5A monoclonal antibody 9E10 (at 1:200 dilution) and Alexa 488-conjugated goat anti-mouse IgG secondary antibody (at 1:500 dilution) (Thermo Fisher Scientific Life Sciences) to check the infection rate [41]. The nuclei were stained with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific Life Sciences) for 20 minutes at room temperature.
Isolation of RNA and qRT-PCR
Detailed information is given in the supplemental Materials and Methods. The primer sequences used are listed in supplemental online data file 1.
Analysis of RNA Sequencing Data and miRNA Target Prediction
For analysis of global uMSC-derived exosomal miRNAs, we downloaded the sequencing data from GEO dataset GSE69909 (the following link can be used to view the raw data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=svwvciucfzipvev&acc=GSE69909). The raw counts of miRNA reads were further normalized by transcripts per million values ([miRNA total reads/total clean reads] × 106).
For miRNA/HCV target prediction, we used the miRanda algorithm (http://www.mirbase.org), which evaluated binding possibility based on the duplex binding energy. The predicted results of exosomal miRNAs are listed in supplemental online data file 2.
miRNA Transfection
To assess the role of miR-125b, let-7f, miR-145, miR-1260a, miR-1260b, miR-199a, miR-221, and miR-222 derived from uMSC-Exo during HCV infection, we chemically synthesized the mimics of the nine miRNAs and transfected them into Huh7 cells by FuGENE HD transfection reagent (Thermo Fisher Scientific Life Sciences). The inhibitors of the four miRNAs (antagomir) with anti-HCV activity (let-7f, miR-145, miR-199a, miR-221) were purchased from GenePharma Corporation (Shanghai, China) and transfected to Huh7 cells or uMSCs by FuGENE HD transfection reagent (Promega, Madison, WI, http://www.promega.com/) according to the manufacturer's instructions.
RNA Immunoprecipitation Assay
RNA immunoprecipitation (RIP) was performed using the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, http://www.emdmillipore.com/) according to the manufacturer’s protocol [42]. The direct binding of miRNA-mediated RNA-induced silencing complex (RISC) to HCV RNA was determined using anti-Ago2 antibody (Abcam, Cambridge, MA, http://www.abcam.com/) mediated RNA pulldown followed by quantitative polymerase chain reaction (qPCR) examine the amount of HCV RNA. In brief, 70%–80% confluence cells were washed twice with PBS and trypsinized. Cells were lysed and centrifuged at 12,000 rpm for 10 minutes at 4°C. Cell lysate was incubated with antibody/beads for 18 hours at 4°C. After incubation, beads were washed three times in NT2 buffer (50 mM Tris-HCl, 300 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40, RNase inhibitor), followed by treated with DNase I for 15 minutes at 37°C. RNAs were extracted by RNA purification kit (RNeasy MiniElute kit, Qiagen, Hilden, Germany, https://www.qiagen.com) and cDNA was synthesized using the SuperScript kit (Thermo Fisher Scientific Life Sciences) and the quality of cDNA was analyzed by qPCR.
Statistical Analysis
Values are shown as mean and standard deviation of at least three independent experiments. Statistical analysis was performed SPSS software, version 17.0 (IBM, Armonk, NY). Results were considered statistically significant at p < .05.
Results
uMSCs Inhibited HCV Infection Through Paracrine Signaling
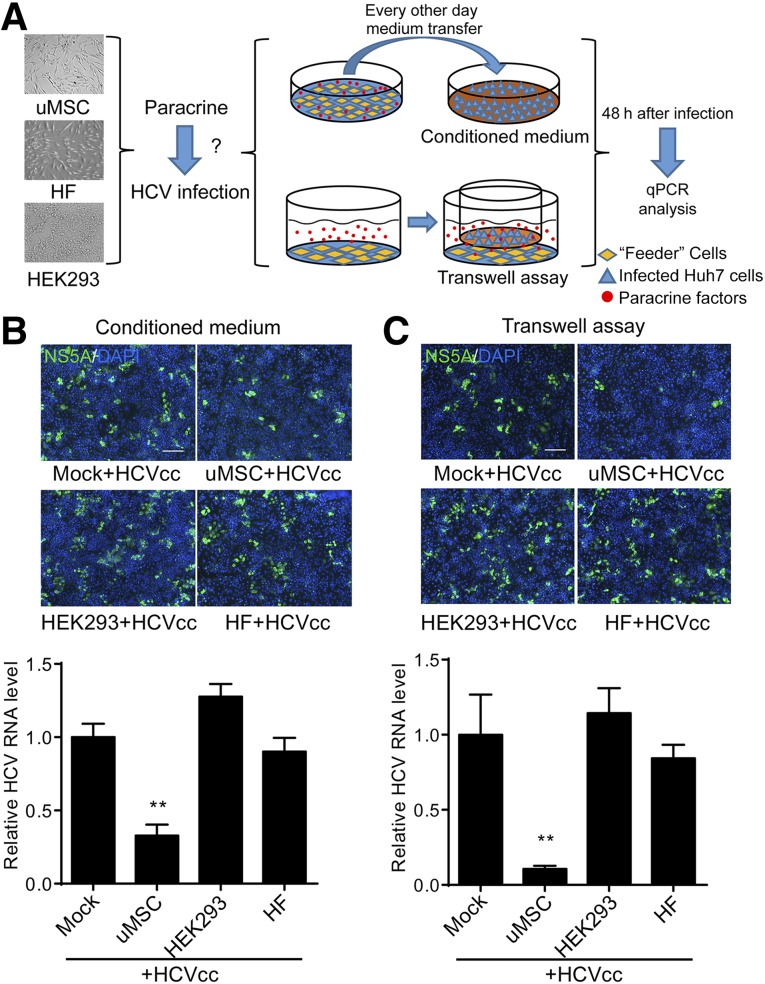
Isolated and subcultured uMSCs were identified through testing the expressions of CD29, CD90, CD44, CD105, CD73, CD34, CD14, CD45, CD19, and HLA-DR by flow cytometry. In accordance with previous studies [5, 43], the cells were positive for CD29, CD90, CD44, CD105, and CD73, while the surface markers of hematopoietic cells such as CD34, CD14, CD45, CD19, and HLA-DR were fairly weak to detect compared with the isotype control (supplemental online Fig. 1A). To evaluate the effect of uMSC paracrine signaling on HCV infection, we replaced the culture medium of cell-derived HCV (HCVcc, JFH1)-infected Huh7 cells with the uMSCs’ conditioned culture medium. Conditioned medium was collected and used to treat infected Huh7 cells every other day (Fig. 1A). The results showed that HCV infection rate of conditioned medium-treated Huh7 cells was significantly reduced compared with the cells treated with conditioned medium from HEK 293T cells or HF cells (Fig. 1B). To verify this finding, we performed a transwell-based assay to evaluate the paracrine function of uMSCs. The secreted components of uMSCs that were seeded in the lower chamber, passed through the membrane to the upper chamber that was seeded with Huh7 cells (Fig. 1A). The results showed that HCV infection on Huh7 cells was largely suppressed by coculture with uMSCs, while the secretion of HEK293 and HF cells did not exert any anti-HCV activity compared with the mock-treated group (Fig. 1C). The inhibitory effect of transwell-based coculture was consistent with the effect observed with conditioned medium but was even more potent, probably because of the persistent uMSCs’ paracrine signaling. However, the levels of IFN-α, IFN-β, and IFN-λ1, which are the major soluble anti-HCV factors, were barely detectable in the supernatant of uMSC (supplemental online Fig. 1B). Thus, our results suggested that uMSC-secreted components were capable of eliciting robust anti-HCV responses.
Figure 1.
Identification of uMSC and its anti-HCV activity by paracrine signaling. (A): Scheme demonstrating the manner in which conditioned medium treatment assay and transwell-based assay were carried out. (B): Huh7 cells were infected with HCVcc of JFH-1 (multiplicity of infections [MOI] = 1). After 6 hours of incubation, the supernatants were changed with conditioned culture medium of uMSCs. The medium was collected from uMSCs to infected Huh7 cells every other day. Infection of the cells was detected 48 hours after virus inoculation by immunofluorescence (IF) and real-time (RT) qPCR. Equal amounts of pure cell medium (mock) or conditioned medium from HEK 293 cells or HF cells were added as a control (2 × 105 copies/1 μg RNA for the mock treated group). (C): Confluent uMSCs were seeded in the lower chamber of the transwell plate, and 75% confluent Huh7 cells were seeded in the upper chamber and infected with HCVcc of JFH-1 (MOI = 1) for 6 hours. After 48-hour coculture between uMSCs and infected Huh7 cells, the upper chamber was taken out to test infection of the cells by IF and RT-qPCR. Pure cell medium (mock) or HEK 293 cells or HF cells were seeded in the lower chamber as a control (5 × 105 copies/1 μg RNA for mock treated group). Data are shown as mean ± SD. ∗∗, p < .01 compared with mock control. The scale bar represents 200 μm. Abbreviations: h, hours; HCV, hepatitis C virus; HCVcc, cell-derived hepatitis C virus; HEK, human embryonic kidney; HF, human dermal fibroblast; Huh, human hepatoma; qPCR, quantitative polymerase chain reaction; uMSC, umbilical mesenchymal stem cell.
uMSC-Exo Showed Antiviral Activities Against HCV Infection
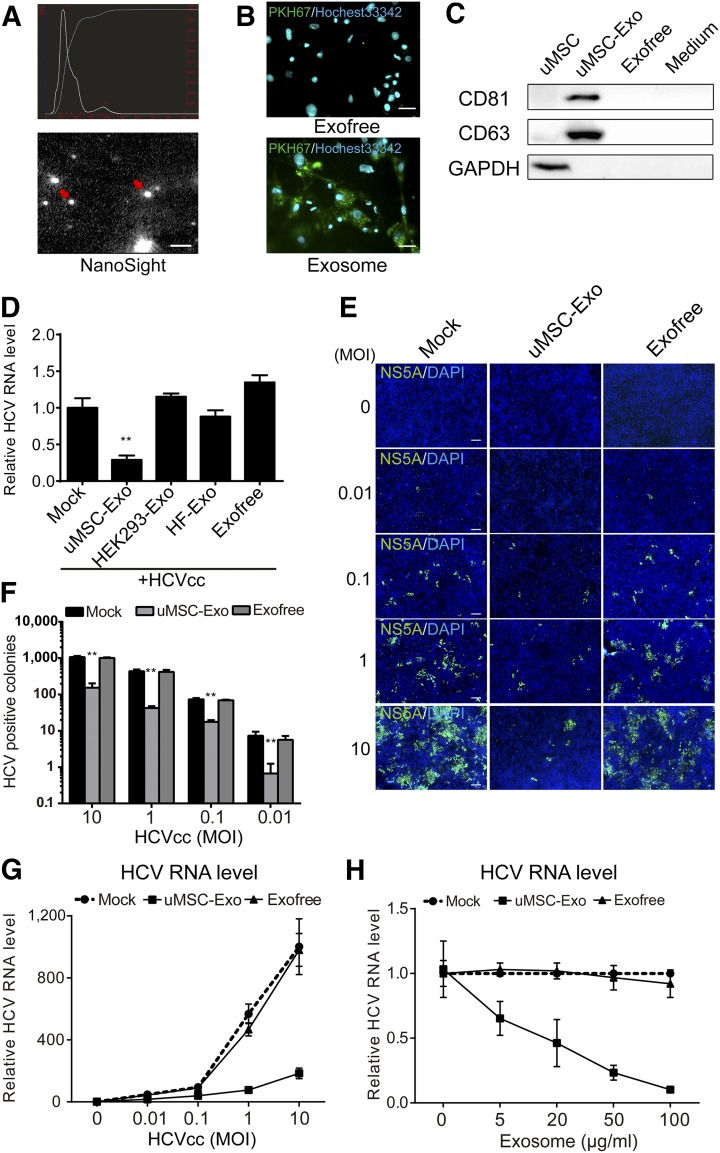
Because exosomes play a very important role in uMSC paracrine signaling, we investigated whether they were responsible for the anti-HCV activity. Exosomes were concentrated and purified from uMSC routine culture medium by ultracentrifugation, and their presence was further confirmed using the NanoSight instrument, which showed vesicles of approximately 100 nm size (Fig. 2A). Exosomes can enter into cells by fusion or active uptake. To confirm that our purified exosomes were competent to enter into cells, we used the membrane-specific dye PKH67 to monitor this process. After 6-hour incubation of PKH67-stained uMSC-Exo with Huh7 cells, fluorescent particles could be eminently observed in the cytoplasm of Huh7 cells, whereas no clear signals were observed using PKH67-stained exosome-free supernatant (Exofree) (Fig. 2B). Besides, exosomes surface markers were also detected by anti-CD81 and anti-CD63 Western blotting (Fig. 2C).
Figure 2.
Exosomes secreted from uMSCs inhibited HCV infection. (A): Purified uMSC-Exo were identified under NanoSight instrument. Particle size and image of the exosomes are shown. The red arrows indicate typical exosomes. The scale bar represents 200 nm. (B): uMSC-Exo were labeled by membrane-specific dye PKH67 and incubated with human dermal fibroblast 7 (Huh7) cells for 6 hours. Cell nuclei were stained with Hoechst33342. Huh7 cells were then observed under a fluorescent microscope after incubation. Exofree represents the supernatant free of exosomes. The scale bar represents 50 μm. (C): Expression of exosome-specific surface marker, such as CD81 or CD63, was analyzed by Western blotting in uMSC-Exo. Medium represents the culture medium of uMSCs. (D): Huh7 cells were infected with HCVcc of JFH-1 (MOI = 1) in the presence of exosomes derived from different cell types (50 μg/ml) for 6 hours, followed by media change with the same amount of exosomes. HCV RNA level was detected 48 hours after infection by real-time quantitative polymerase chain reaction (RT-qPCR) (2.5 × 105 copies/1 μg RNA for mock-treated group). ∗∗, p < .01 compared with mock control. Antiviral activity of uMSC-Exo was also tested under HCVcc infection of multiple MOIs. Infection of the cells was evaluated by both immunofluorescence of viral protein (E, F) and RT-qPCR of HCV RNA level (G). Cell nuclei was stained with DAPI. ∗∗, p < .01 compared with mock control. The scale bar represents 200 μm. (H): Huh7 cells were infected with HCVcc in the presence of indicated concentrations of uMSC-Exo for 6 hours before media change with the same amount of exosomes. HCV RNA level was tested 48 hours after virus inoculation (2.5 × 105 copies/1 μg RNA for mock-treated group with no exosomes). Data are shown as mean ± SD. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; Exo, exosomes; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCV, hepatitis C virus; HCVcc, cell-derived hepatitis C virus; HEK, human embryonic kidney; MOI, multiplicity of infections; uMSC, umbilical mesenchymal stem cell; uMSC-Exo, umbilical mesenchymal stem cell-derived exosomes.
Subsequently, we analyzed whether uMSC-Exo were capable of blocking HCV infection. Purified exosomes from uMSCs medium were incubated with Huh7 cells in which HCVcc was added afterward. Treatment of uMSC-Exo remarkably reduced the intracellular HCV RNA level, whereas HEK 293-Exo or HF-Exo showed no clear antiviral activity (Fig. 2D). This result was also consistent in Huh7.5.1 cells, which have a mutation in retinoic acid-inducible gene-I (RIG-I) of IFN pathway (supplemental online Fig. 1C). Moreover, we also found that uMSC-Exo suppressed HCVcc infection at multiple multiplicity of infections (MOIs) in both viral positive colonies (Fig. 2E, 2F) and RNA levels (Fig. 2G), inhibiting viral infection in a dose-dependent manner (Fig. 2H), whereas Exofree showed no effect, largely excluding the soluble factors in the supernatant of uMSC to be antiviral agents. Taken together, our results showed that uMSC-Exo were capable of inhibiting HCV infection.
uMSC-Exo Had No Effect on HCV Entry but Inhibited Viral Replication
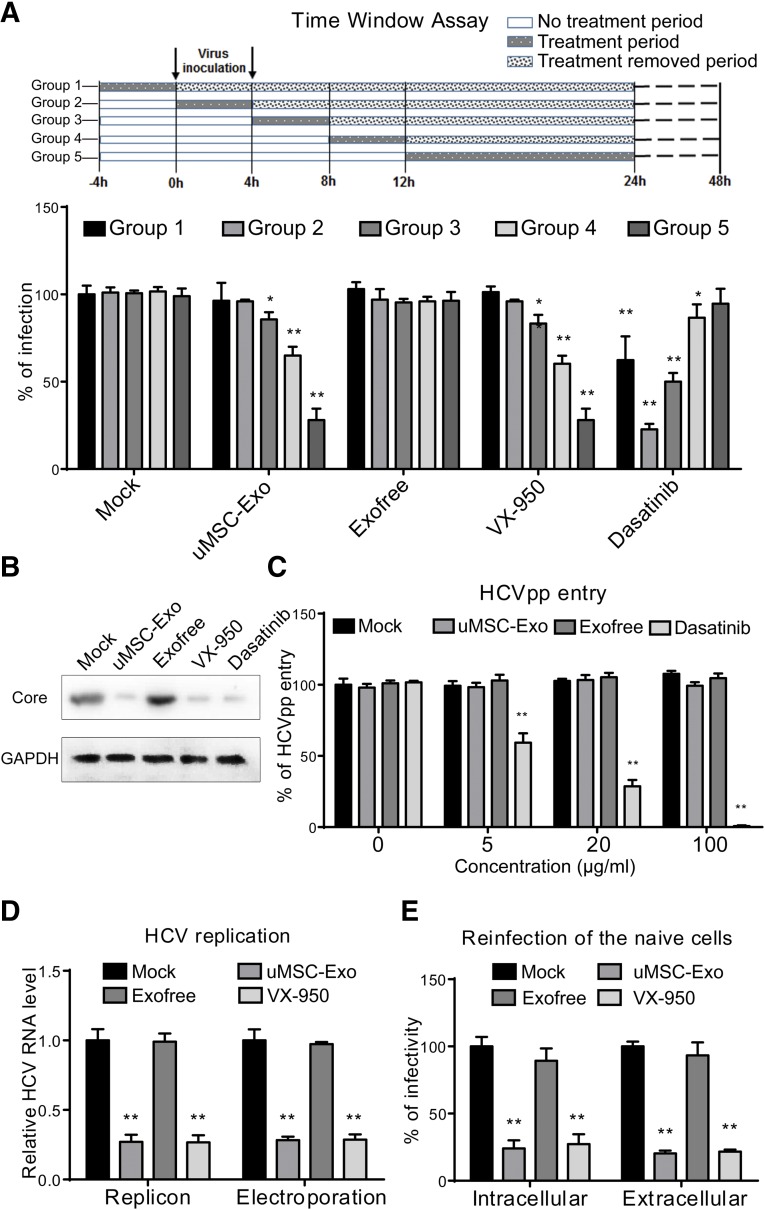
To reveal the stage in which uMSC-Exo exerted their inhibitory effect, Huh7 cells were infected with HCVcc together with relevant antiviral agents. As shown in Figure 3A, treatment of uMSC-Exo exhibited a similar inhibition pattern of VX-950, an NS3/4A protease inhibitor, also known as telaprevir, which prevents viral replication. uMSC-Exo did not manifest significant anti-HCV activity when added during the early phase of the HCV life cycle (−4 to 0 and 0–4 hours) but blocked more than 70% of viral infection 12 hours after virus inoculation. However, another drug, dasatinib, an inhibitor of HCV entry, showed potent inhibitory effect when added during the initial 8 hours of virus infection.
Figure 3.
Effect of uMSC-Exo on different stages of hepatitis C virus (HCV) infection. (A): Human dermal fibroblast 7 (Huh7) cells were infected with cell-derived HCV of JFH-1 for 4 hours before the medium change. uMSC-Exo (50 μg/ml) were added at different time points and allowed for indicated time window of treatment according to the above panel. At 48 hours after infection, immunofluorescence was performed to check virus infection. VX-950 (2 μM) and dasatinib (2 μM) were introduced as positive control for HCV replication and entry. ∗, p < .05; ∗∗, p < .01 compared with mock-treated group (620 colonies per well for mock-treated group 1). (B): Antiviral activity of uMSC-Exo (50 μg/ml) on viral infection by detection of HCV core expression. VX-950 (2 μM) and dasatinib (2 μM) were introduced as positive controls for HCV infection. (C): Huh7 cell were incubated with HCVpp of H77 for 6 hours in the presence of indicated concentrations of uMSC-Exo. At 72 hours after inoculation, entry rate was assayed by flow cytometry. Dasatinib (2 μM) was introduced as a positive control. ∗∗, p < .01 compared with mock-treated group (466 positive cells per well for mock-treated group). (D): uMSC-Exo (50 μg/ml) were incubated with HCV BB7 replicon or HCV RNA electroporated Huh7 cells. At 48 hours after infection, HCV RNA level was detected by real-time quantitative polymerase chain reaction. VX-950 (2 μM) was introduced as a positive control. ∗∗, p < .01 compared with mock-treated group. (E): Intracellular and extracellular virion infectivity was tested by reinfection of naïve Huh7 cells. At 48 hours after infection, immunofluorescence was performed to test virus infectivity. ∗∗, p < .01 compared with mock-treated group (293 and 388 colonies per well for intracellular and extracellular mock-treated group). Data are shown as mean ± SD. Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCVpp, hepatitis C virus pseudoparticles; uMSC-Exo, umbilical mesenchymal stem cell-derived exosomes.
Treatment of uMSC-Exo effectively suppressed viral infection, showing the same effect as VX-950 and dasatinib, as demonstrated by the viral core expressions (Fig. 3B). To exclude the possibility that uMSC-Exo affected viral entry, HCV pseudoparticles (HCVpp) of H77 were incubated with Huh7 cells in the presence of indicated concentrations of uMSC-Exo. As shown in Figure 3C, entry of HCVpp was not significantly altered even by the treatment of uMSC-Exo at 100 μg/ml, whereas dasatinib reduced their entrance.
Next, we evaluated the effect of uMSC-Exo on the viral postentry step. Replicon BB7 cells, which only support HCV replication, were treated with 50 μg/ml uMSC-Exo for 12 hours. The replication efficiency of HCV RNA levels was then detected by qPCR. The result showed that HCV RNA level decreased predominantly after uMSC-Exo as well as after VX-950 treatment, whereas Exofree showed no significant effect. This inhibitory effect was also consistent with the subsequent JFH-1 electroporated cells treated with the uMSC-Exo (Fig. 3D).
The supernatants of electroporated Huh7 cells subjected to different concentrations of uMSC-Exo treatments were collected to analyze extracellular infectivity. Intracellular virions were also acquired by repeated cycles of cell freezing and thawing. Intracellular and extracellular infectivity were analyzed by reinfection of naïve Huh7 cells, and our results indicated that both of them were significantly reduced by uMSC-Exo treatment at a similar rate. This phenomenon was probably due to the suppression of viral replication because incubation of VX-950 also caused a similar rate of reduction on intracellular and extracellular virion infectivity (Fig. 3E).
uMSC-Exo-Specific miRNAs Showed Inhibitory Effects in HCV Infection
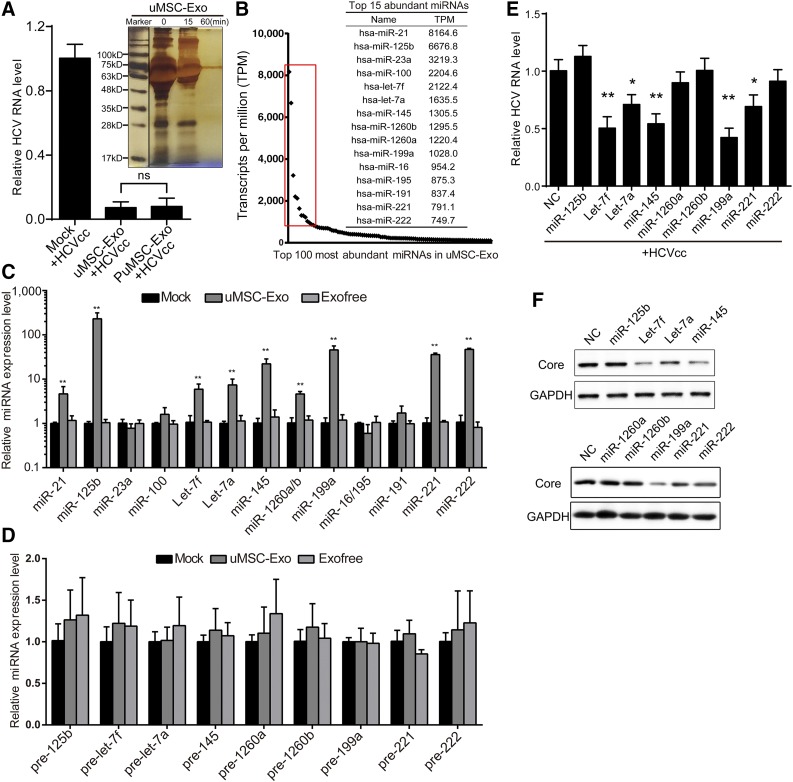
According to previous studies, exosomes mainly contain various proteins and RNAs. Therefore, we used the exclusion method to determine which factors in uMSC-Exo could efficiently inhibit HCV infection. Purified exosomes were subjected to proteinase K treatment (PK, supplemented with low concentration of detergents) at specific periods of time, and the digestion efficacy was determined by silver staining. After 60 minutes of PK incubation, the proteins contained in uMSC-Exo were barely detectable (Fig. 4A). However, these PK-treated uMSC-Exo still maintained exosomes integrity as detected, by the NanoSight instrument (supplemental online Fig. 1D), and they still possessed strong antiviral capacity, almost the same as the untreated uMSC-Exo (Fig. 4A). Therefore, the preceding result demonstrated that the RNAs contained in uMSC-Exo might be the functional anti-HCV factors instead of protein components.
Figure 4.
The specific miRNAs from uMSC-Exo contributed to the antiviral activity on HCV infection. (A): Purified uMSC-Exo were subjected to PuMSC-Exo for indicated time durations (15 minutes or 60 minutes), and the digestion efficacy was determined by silver staining. Human dermal fibroblast 7 were infected with HCVcc of JFH-1 in the presence of uMSC-Exo or PuMSC-Exo (60 minutes) or the solvent control (mock). At 48 hours after infection, HCV RNA level was detected by real-time quantitative polymerase chain reaction (RT-qPCR). (B): High-throughput small RNA sequencing of uMSC-Exo. The red rectangle frames the top 15 abundant miRNAs in uMSC-Exo. (C): Huh7 cells were treated with uMSC-Exo (50 μg/ml) for 24 hours before the expression of the 15 miRNAs was detected by RT-qPCR. ∗∗, p < .01 compared with mock-treated group. (D): Expression of precursors of the nine upregulated miRNAs was assayed in Huh7 cells by RT-qPCR after 24-hour treatment of uMSC-Exo (50 μg/ml). (E): Huh7 cells were transfected with chemically synthesized mimics of the nine miRNAs and infected with HCVcc of JFH-1. At 48 hours after virus inoculation, HCV infection was evaluated by both HCV RNA level by RT-qPCR (3 × 105 copies/1 μg RNA for mock-treated group with no exosomes) and viral core protein expression by Western blotting. ∗, p < .05; ∗∗, p < .01 compared with NC. Data are shown as mean ± SD. Abbreviations: Exo, exosomes; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCV, hepatitis C virus; HCVcc, cell-derived hepatitis C virus; miR and miRNA, microRNA; NC, negative control; ns, not significant compared with mock control; PuMSC, proteinase K-treated umbilical mesenchymal stem cell-derived exosomes; uMSC-Exo, umbilical mesenchymal stem cell-derived exosomes.
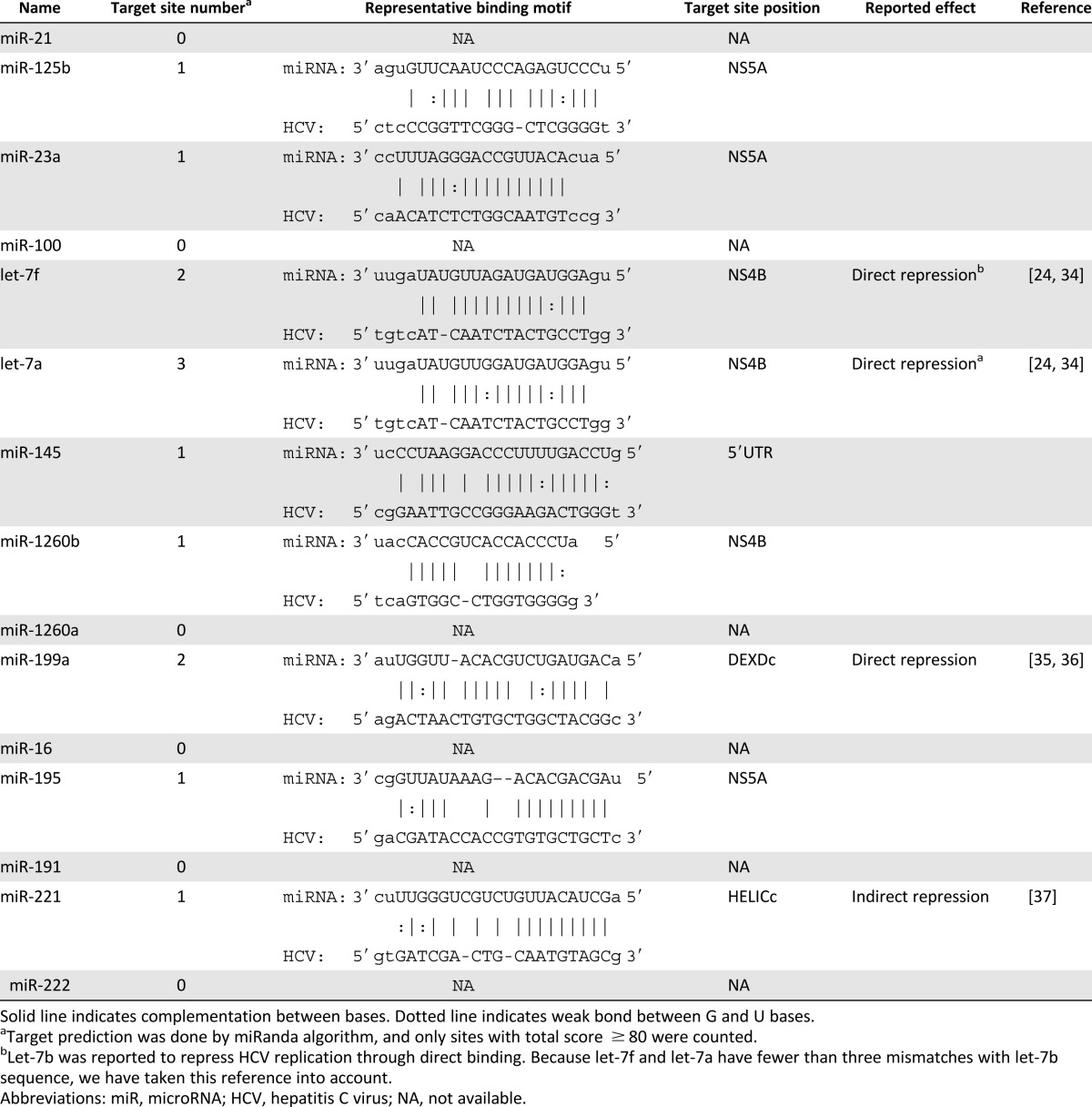
Various studies suggest that HCV infection can be regulated by some specific miRNAs. Therefore, we first validated the presence and abundance of exosomal miRNA using gel electrophoresis of extracted exosomal RNA. Our results showed that exosomal RNAs were mainly small RNAs less than 50 base pairs in length (supplemental online Fig. 2A). We thus took the advantage of high-throughput small RNA sequencing to analyze the global miRNAs in uMSC-Exo. As shown in Figure 4B, only a few types of miRNAs were highly abundant in the uMSC-Exo, as framed out by the red square (top 15 abundant miRNAs). To gain more insights regarding these high abundant miRNAs, we used a target prediction algorithm search to predict their binding sites in HCV RNA. The results suggested that most of them can complement the viral genomes (Table 1). Moreover, among the 15 miRNAs, some have already been reported to influence HCV infection [44–47].
Table 1.
Target site prediction summary of top 15 abundant microRNAs in umbilical mesenchymal stem cell-derived exosomes
To examine the function of these exosomal miRNAs, we first tested their expression levels in Huh7 cells under indicated treatments. Compared with the Exofree and mock-treated group, uMSC-Exo-treated cells expressed high levels of miR-125b, let-7f, let-7a, miR-145, miR-1260a, miR-1260b, miR-199a, miR-221, and miR-222 (Fig. 4C). However, the expression levels of relevant miRNA precursors was not significantly modified (Fig. 4D), indicating that the enhancement of specific miRNAs in Huh7 cells under uMSC-Exo treatment was caused by direct increasing of mature miRNAs, not by increasing endogenous production.
To further evaluate the antiviral effect of these nine highly upregulated miRNAs in HCV infection, we transfected the host cells with chemically synthesized miRNAs mimics. As shown in Figure 4E and 4F, overexpression of let-7f, miR-145, miR-199a, and miR-221 made Huh7 cells less susceptible to HCV infection, displaying a significant reduction of both HCV RNA level and viral core protein expression. However, we found that miR-221 overexpression in Huh7.5.1 cells did not cause a significant reduction of HCV RNA level (supplemental online Fig. 2B), which is consistent with our previous findings [47].
Let-7f, miR-145, miR-199a, and miR-221 Played Vital Roles in the Antiviral Effect of uMSC-Exo on HCV Infection
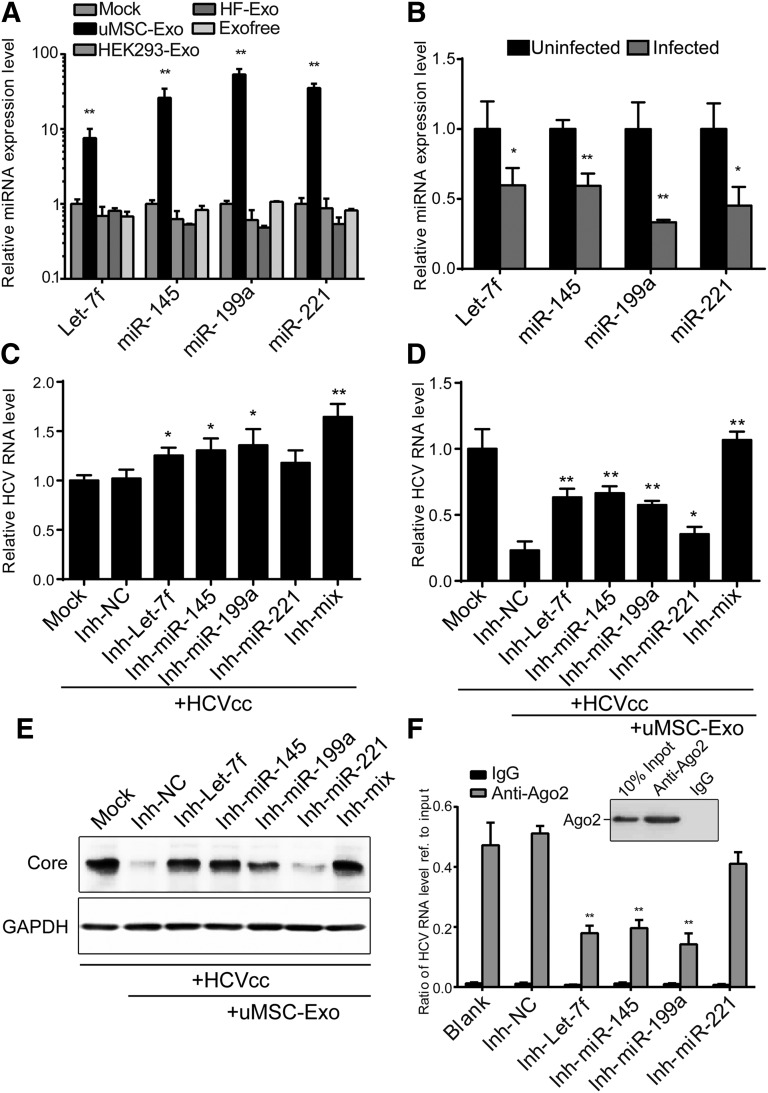
To confirm that these identified antiviral miRNAs were uMSC-Exo-specific, we used exosomes derived from several other cell types, including HEK 293-Exo and HF-Exo, to incubate them with the host cells. We then tested the expression level of these miRNAs. The results suggested that only the uMSC-Exo-treated group showed a clear increase in miRNAs expression (Fig. 5A). Moreover, the expression level of these miRNAs before and after HCV infection was also determined. As shown in Figure 5B, let-7f, miR-145, miR-199a, and miR-221 were all significantly downregulated after viral infection, indicating that these miRNAs might be of biological importance and thus actively inhibited upon virus infection.
Figure 5.
Umbilical mesenchymal stem cell (uMSC) exosomal miRNA mixture of let-7f, miR-145, miR-199a, and miR-221 conceived an anti-HCV property. (A): Human dermal fibroblast 7 (Huh7) cells were incubated with exosomes derived from several cell types (uMSC-Exo, human embryonic kidney 293-Exo and HF-Exo) (50 μg/ml) for 24 hours. Expression level of the four miRNAs was tested by real-time quantitative polymerase chain reaction (RT-qPCR). ∗∗, p < .01 compared with mock treated group. (B): Expression level of the four miRNAs was detected in both uninfected and infected Huh7 cells by RT-qPCR. ∗, p < .05; ∗∗, p < .01 compared with mock control. Huh7 cells were transfected with synthesized inhibitors of the four miRNAs (Inh-let-7f, Inh-miR-145, Inh-miR-199a, Inh-miR-221) or a mixture of them (Inh-mix), and infected with HCVcc of JFH-1 in the absence (C) or presence (D, E) of uMSC-Exo (50 μg/ml). At 48 hours after infection, HCV RNA level was detected by RT-qPCR and viral core expression by Western blotting. ∗, p < .05; ∗∗, p < .01 compared with negative control (2 × 105 copies/1 μg RNA for mock-treated group). (F): Ago2-mediated RNA pulldown assay. The amount of HCV RNA in the pulldown products in each group was analyzed using qPCR and compared with the input. IgG groups were used as negative control. Efficiency of the pulldown experiments was shown by Western blotting. ∗∗, p < .01 compared with negative control. Data are shown as mean ± SD. Abbreviations: Exo, exosomes; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCV, hepatitis C virus; HCVcc, cell-derived hepatitis C virus; HEK, human embryonic kidney; HF, human dermal fibroblast; miR and miRNA, microRNA; NC, negative control; uMSC-Exo, umbilical mesenchymal stem cell-derived exosomes.
To further clarify the functions and importance of these four miRNAs, the host cells were transfected with synthesized inhibitors of relevant miRNAs and infected with HCVcc. Inhibitors of the four miRNAs (Inh-let-7f, Inh-miR-145, Inh-miR-199a, Inh-miR-221), especially when combined as a mixture (Inh-mix), significantly promoted HCV infection compared with the negative control group (Inh-NC) (Fig. 5C). When the transfected cells were infected with HCVcc in the presence of uMSC-Exo, the inhibitory effect of exosomes was partially reversed, with the inhibitors showing more eminent effect of promoting HCV infection compared with Inh-NC (Fig. 5D, 5E). The results also indicated the importance of these uMSC-Exo specific miRNAs in anti-HCV infection.
To gain direct evidence of these miRNAs in targeting HCV, we performed an Ago2, a vital member of the RISC-mediated RNA pulldown assay. Infected cells treated with both uMSC-Exo and inhibitors were lysed, then subjected to the anti-Ago2 pulldown assay. The amount of HCV RNA in the pulldown products in each group was analyzed using qPCR and referenced to the input level of HCV RNA. IgG-mediated pulldown products were used as negative control. Results showed that HCV RNA was highly enriched to the Ago2-mediated pulldown products, and inhibition of let-7f, miR-145, or miR-199a significantly reduced the amount of Ago2 binding to the HCV RNA, which indicates that these miRNAs mediate RISC binding to the HCV RNA (Fig. 5F). However, inhibition of miR-221 did not significantly inhibit Ago2 binding to the HCV RNA (Fig. 5F), indicating that it functions through an indirect way to inhibit HCV infection, which is consistent with our previous report [47].
Removal of Exosomal Let-7f, miR-145, miR-199a, and miR-221 From the uMSC-Exo Abolished Their Antiviral Activity
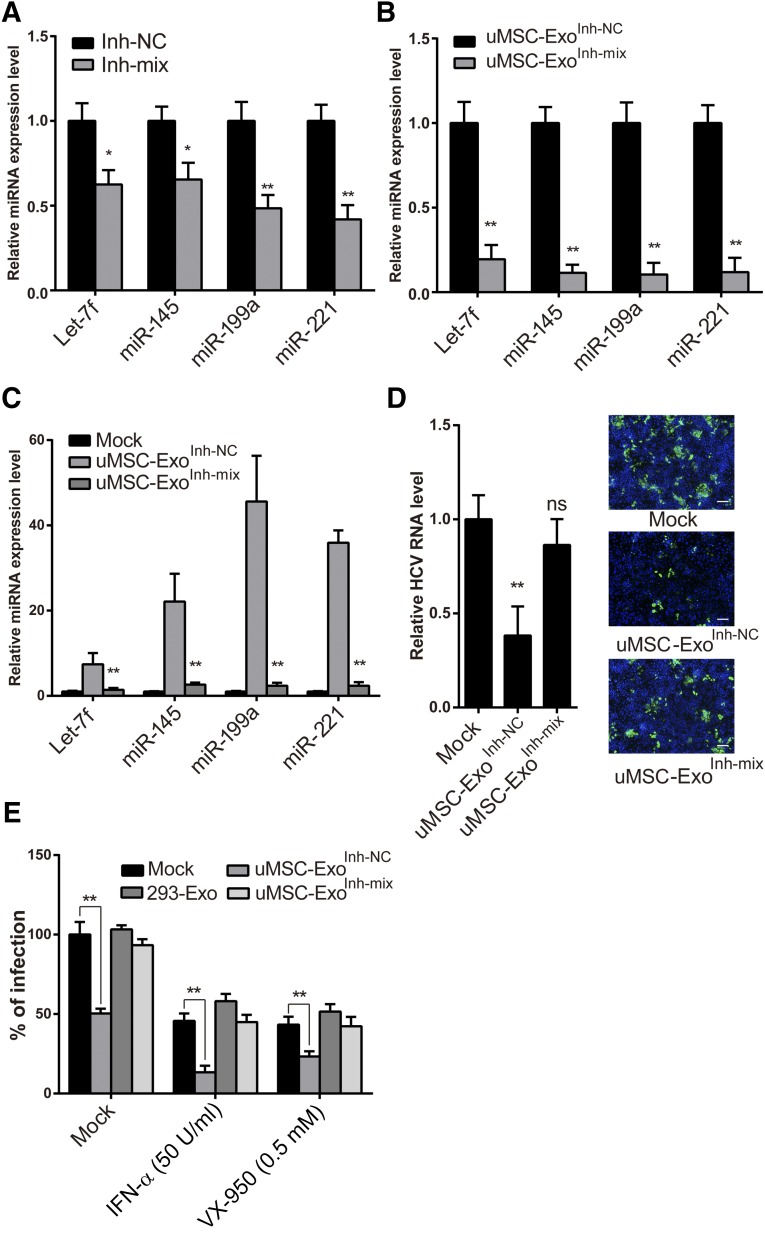
To demonstrate that uMSC-Exo exerted its antiviral effect through these specific miRNAs, we removed these miRNAs from the exosomes. Because the exosomes were derived from uMSCs, we therefore transfected uMSCs with a mixture of the inhibitors targeting these specific miRNAs (uMSC-ExoInh-mix). The expression levels of these miRNAs both in MSCs and secreted exosomes were detected by qPCR. Our result showed that although in uMSCs the expression of these four miRNAs was downregulated to approximately 50% after the inhibitor treatment, the levels in secreted exosomes all significantly decreased to less than 20%. Therefore, the secreted exosomes displayed a reduced potency, probably because of the lack of valid miRNAs in the cells (Fig. 6A, 6B). Huh7 cells were then treated with uMSC-ExoInh-mix and with a scramble inhibitor transfected uMSC-derived exosome control (uMSC-ExoInh-NC). The qPCR result of these four miRNAs suggested that cells expressed a low level of the four miRNAs under uMSC-ExoInh-mix treatment as compared with the mock-treated group (Fig. 6C).
Figure 6.
Elimination of the four miRNAs from uMSC-Exo abolished their original anti-HCV activity and synergistic effect to combine with IFN or VX-950. (A, B): Umbilical mesenchymal stem cells (uMSCs) were transfected with a mixture of the inhibitors (Inh-mix) targeting the four miRNAs. Expression levels of these miRNAs were detected in both MSCs (A) and secreted exosomes (B) by quantitative polymerase chain reaction (qPCR). ∗, p < .05; ∗∗, p < .01 compared with NC (Inh-NC). (C): Human dermal fibroblast 7 (Huh7) cells were treated with uMSC-ExoInh-NC or uMSC-ExoInh-mix for 24 hours before expression of the four miRNAs was detected by real-time qPCR. ∗∗, p < .01 compared with mock control. (D): Huh7 cells were infected with cell-derived HCV (HCVcc) in the presence of uMSC-ExoInh-mix or uMSC-ExoInh-NC. At 48 hours after virus inoculation, infection was determined by detecting HCV RNA level or viral protein expression. ∗∗, p < .01 compared with mock control (1.5 × 105 copies/1 μg RNA for mock treated group). The scale bar represents 200 μm. (E): Huh7 cells were infected with HCVcc of JFH-1 under the treatment of uMSC-Exo, 293-Exo, or uMSC-ExoInh-mix (20 μg/ml). At 12 hours after infection, IFN-α (50 U/ml) or VX-950 (0.5 μM) was added to culture medium together with the exosomes. HCV infectivity was detected 48 hours later by immunofluorescence. ∗∗, p < .01 compared with mock control (505 colonies per well for mock-treated group). Data are shown as mean ± SD. Abbreviations: HCV, hepatitis C virus; IFN, interferon; miR and miRNA, microRNA; NC, negative control; ns, not significant; uMSC-Exo, umbilical mesenchymal stem cell-derived exosomes; uMSC-ExoInh-mix, inhibitor mixture transfected umbilical mesenchymal stem cell-derived exosome; uMSC-ExoInh-NC, scrambled inhibitor transfected umbilical mesenchymal stem cell-derived exosome.
After identifying that modified uMSC-Exo indeed lost the ability to increase the expression level of these miRNAs in targeted cells, we again examined their antiviral effect using the HCVcc infection model. The result showed that the inhibitory effect on HCV infection of uMSC-Exo was significantly reversed upon depleting its exosomal miRNAs, as shown in the uMSC-ExoInh-mix group compared with the mock-treated group (Fig. 6D). Together, these data suggested that the antiviral activity of uMSC-Exo against HCV was mainly due to the exosomal let-7f, miR-145, miR-199a, and miR-221.
uMSC-Exo Synergistically Inhibited HCV Infection When Combined With IFN-α or Telaprevir
uMSC-Exo were produced and secreted from MSCs under normal culture conditions. Thus, the constituents of exosomes were also derived from MSCs under normal culture conditions. The cell viability assay confirmed that uMSC-Exo had low cytotoxic effect on the host cells, even at the dose of 1,000 μg/ml (supplemental online Fig. 2C), which indicates that normal culture-derived uMSC-Exo are significantly less toxic in vitro.
IFN-α is a broad-spectrum antiviral and indispensable constituent of the current standard HCV treatment. VX-950, also known as telaprevir, is an HCV NS3-4A protease inhibitor, which is clinically used in treating patients with HCV genotype 1. We therefore explored the possibility of combining uMSC-Exo with either of the two U.S. Food and Drug Administration (FDA)-approved drugs. Huh7 cells were infected with HCVcc in the presence of uMSC-Exo combined with IFN-α or VX-950. As shown in Figure 6F, uMSC-Exo blocked HCV infection more potently when added together with IFN or VX-950, and the synergy effect was almost abolished when the four miRNAs were inhibited in the exosomes (Fig. 6E).
Discussion
In this study, we demonstrated that uMSC-Exo might be a potent antiviral agent against HCV. The antiviral activity was mediated by the miRNAs wrapped in uMSC-Exo. These exosomal miRNAs suppressed viral infection mainly by targeting viral replication stage and showed a synergistic effect when combined with clinically approved anti-HCV drugs such as IFN or telaprevir. uMSC-Exo, which potently inhibited HCV infection, were collected and purified from the normal human stem cells, and possess the unique advantage to be normal culture-secreted substances with low cytotoxicity.
Antiviral agents against HCV have improved the treatment efficacy of HCV infection to an unprecedented level in recent years. However, many crucial issues still need to be solved such as antiviral effect in refractory patients, safety problems and concomitant side effects of the antiviral therapy, all of which also cause low patient adherence. Novel anti-HCV agents, such as small molecule compounds, cell-derived peptides, miRNAs, and long noncoding RNAs, have optimized the current antiviral research on HCV [38, 47–49]. However, it has never been reported that cellular secreted vesicles are of antiviral potency. Cell-based therapy that uses normal cells for therapeutic reasons has been widely studied in recent years, becoming a promising alternative for regenerative medicine. Traditionally, stem cell-based therapy uses massive amounts of cells to achieve replacement or regeneration, either by direct differentiation into specific tissues or indirect secretion of proteins, cytokines, or RNAs [17]. MSCs are reported to produce various cytokines, chemokines, and growth factors, some of which contribute to MSCs’ immunomodulatory effects, including downregulation of IFN-γ and antibody production [5]. It is also reported that MSC-derived IFN-γ could compensate for immune-suppressive functions of MSCs as well as facilitate cytotoxic T lymphocytes responses during viral infection [50]. However, Exofree from MSCs as well as the digestion assay of uMSC-Exo with proteinase K largely diminished the possibility of soluble factors in the supernatant of uMSC or protein components in uMSC-Exo to be the major elements with anti-HCV potency. Until now, MSC treatments have caused slight or no adverse reactions in clinical trials [51, 52].
Recent findings on the regulatory effect of MSC-based therapy suggest that the paracrine signaling of these cells exerts far more profound effects on the recipient than MSCs in modulating immune response [11]. Exosomes have become recognized as an important mode of paracrine cell signal [17]. Exosomes are secreted from various types of cells, with the cell membrane representing the outside shell. Therefore, the lipid bilayer originated from the cell was able to protect exosomes from degrading the biological substances during their transport and to fuse or be smoothly absorbed into the recipient cells [53]. These advantages have made exosomes optimal transmitters of biological molecules from donor to recipient cells. Exosomes derived from MSCs have already been applied in research on various diseases [22, 54, 55]. Although it was reported that exosomes were involved in HCV transmission [25], here we discuss stem cell-derived exosomes (uMSC-Exo) carrying a pack of miRNAs with anti-HCV potency. Incubation of uMSC-Exo during an early phase of the life cycle of HCV infection did not cause the anti-HCV effect, whereas the addition of exosomes in a late postentry stage of viral infection, even with a long period of incubation, led to significant suppression of HCV infection. It demonstrated that uMSC-Exo affected the postentry step of viral infection in a fairly stable and robust manner, probably because of the protectant property of exosomes to maintain the stability of the inside miRNAs. Moreover, the liver is an organ abundant in blood flow. Hence, the exosomes in circulation can easily be assimilated into the hepatocytes, providing an ideal transport tool to delivery specific elements to the liver with high efficiency and low cytotoxicity in the body. We evaluated the delivery ability of exosomes to mouse liver with indicated concentrations of uMSC-Exo and found that intravenous injections of uMSC-Exo ranging from 50 μg to 500 μg were sufficient to enter the livers of ICR mice in a dose-dependent manner without obvious cytotoxicity (data not shown).
In our study, we explored additional function of MSC-Exo in viral infection via miRNA transferring to the target cells. The antiviral activity of uMSC-Exo was also tested on dengue virus and enterovirus 71 (EV71). However, we did not observe potent antiviral effect on these viruses (data not shown), ruling out the possibility of uMSC-Exo being broad-spectrum antivirals. The roles of miRNAs in the establishment and maintenance of HCV infection have been well illustrated [56]. These small RNAs can target specific cellular factors or directly bind to viral genomes to block productive HCV replication. miRNA dysregulation represents a novel approach to treat diseases by overexpression or replacement of the miRNAs. Among the mixture of the four miRNAs with anti-HCV potency, the let-7 family was able to suppress HCV infection by targeting insulin-like growth factor 2 mRNA-binding protein 1, an essential host factor required for HCV replication [44]. miR-199a is closely related to HCV replication [45, 46], whereas miR-145 was downregulated in HCV-associated hepatocellular carcinoma [57]. miR-221 was reported by our laboratory to accentuate the anti-HCV effect of IFN by targeting the inhibitory factors of IFN pathway [47]. Together, the combination of the four miRNAs showed a potent inhibitory effect on HCV replication. A recent study revealed the role of miR-27a in suppressing HCV [58]. However, because the amount and fold changes of miR-27a were not among the top in sequencing data, it may not be critical for the antiviral ability of uMSC-Exo.
The uMSC-Exo study has been an intriguing area of research in recent years. It has been found that the specific exosomal miRNAs from uMSCs contribute to the enhancement of the original effect of the miRNAs inside the cells [59]. In addition, their specific miRNA expression profile shows that the suppressing nucleic acids against HCV are included in the uMSC secreted exosomes, which enables them to be desirable and inexhaustible recycling sources for cell-based therapy to produce robust anti-HCV agents. In addition, the synergistic effect of the exosomes with the FDA-approved anti-HCV drugs confers an additional potential power in the HCV therapy, which could be valuable in future clinical application. The culture environment is also critical for cell type-specific exosomes isolation, for serum exosomes could contaminate the purity of isolated products. However, the use of exosomes-free media or chemical-defined media could solve this problem [60, 61], which enhances the clinical potential of exosomes
Conclusion
We have shown that exosomes secreted from uMSC have potent antiviral activity against HCV, especially by targeting viral replication. The high-throughput miRNA sequencing of uMSC-Exo and subsequent functional assay suggest that the antiviral process is mainly mediated by a series of miRNAs (let-7f, miR-145, miR-199a, and miR-221) transported specifically through exosomes. This work provides new insights and prospects to uMSC-Exo-based therapy to combat viral infection by delivering normal culture-produced substances. Therefore, the present study illustrates a promising method for the development of anti-HCV therapy.
Supplementary Material
Acknowledgments
We thank C.M. Rice, Ph.D., J. Zhong, Ph.D., F.L. Cosset, Ph.D., and T. Wakita, Ph.D., for research materials. This study was supported by research grants from the National Natural Science Foundation of China (No. 81273557, 81572096, and 81372017), Shanghai Key Clinical Center Construction Project (No. 000002), and Joint Study Project of Important Diseases (2013ZYJB050) of the Shanghai Municipal Commission of Health and Family Planning. We thank Shanghai NovelBio Bio-Pharm Technology Co. Ltd for help with high-throughput-sequencing experiments.
Author Contributions
X.Q.: conception and design, collection of data, data analysis and interpretation, manuscript writing; C.X.: provision of study material, data analysis and interpretation, collection of data, manuscript writing; S.F.: experiments, collection of data, manuscript writing; P.Z.: experiments, collection of data; Y.W. and H.L.: provision of study material; W.Y.: data analysis and interpretation, manuscript writing; Z.Q.: conception and design, data analysis and interpretation, financial support, administrative support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Ippolito G, Capobianchi MR, Lanini S, et al. Is hepatitis C virus eradication around the corner only 25 years after its discovery? Int J Antimicrob Agents. 2015;45:111–112. doi: 10.1016/j.ijantimicag.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Cox AL. MEDICINE. Global control of hepatitis C virus. Science. 2015;349:790–791. doi: 10.1126/science.aad1302. [DOI] [PubMed] [Google Scholar]
- 3.Kohli A, Shaffer A, Sherman A, et al. Treatment of hepatitis C: A systematic review. JAMA. 2014;312:631–640. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- 4.Petta S, Craxì A. Current and future HCV therapy: Do we still need other anti-HCV drugs? Liver Int. 2015;35(suppl 1):4–10. doi: 10.1111/liv.12714. [DOI] [PubMed] [Google Scholar]
- 5.Kode JA, Mukherjee S, Joglekar MV, et al. Mesenchymal stem cells: Immunobiology and role in immunomodulation and tissue regeneration. Cytotherapy. 2009;11:377–391. doi: 10.1080/14653240903080367. [DOI] [PubMed] [Google Scholar]
- 6.Baglio SR, Pegtel DM, Baldini N. Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol. 2012;3:359. doi: 10.3389/fphys.2012.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’souza N, Rossignoli F, Golinelli G, et al. Mesenchymal stem/stromal cells as a delivery platform in cell and gene therapies. BMC Med. 2015;13:186. doi: 10.1186/s12916-015-0426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaher W, Harkness L, Jafari A, et al. An update of human mesenchymal stem cell biology and their clinical uses. Arch Toxicol. 2014;88:1069–1082. doi: 10.1007/s00204-014-1232-8. [DOI] [PubMed] [Google Scholar]
- 9.Tan X, Gong YZ, Wu P, et al. Mesenchymal stem cell-derived microparticles: A promising therapeutic strategy. Int J Mol Sci. 2014;15:14348–14363. doi: 10.3390/ijms150814348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caplan AI. MSCs: The sentinel and safe-guards of injury. J Cell Physiol. 2016;231:1413–1416. doi: 10.1002/jcp.25255. [DOI] [PubMed] [Google Scholar]
- 11.Liang X, Ding Y, Zhang Y, et al. Paracrine mechanisms of mesenchymal stem cell-based therapy: Current status and perspectives. Cell Transplant. 2014;23:1045–1059. doi: 10.3727/096368913X667709. [DOI] [PubMed] [Google Scholar]
- 12.Konala VB, Mamidi MK, Bhonde R, et al. The current landscape of the mesenchymal stromal cell secretome: A new paradigm for cell-free regeneration. Cytotherapy. 2016;18:13–24. doi: 10.1016/j.jcyt.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Liu J, Manuchehrabadi N, et al. Umbilical cord and bone marrow mesenchymal stem cell seeding on macroporous calcium phosphate for bone regeneration in rat cranial defects. Biomaterials. 2013;34:9917–9925. doi: 10.1016/j.biomaterials.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Pham P, Truong NC, Le PT, et al. Isolation and proliferation of umbilical cord tissue derived mesenchymal stem cells for clinical applications. Cell Tissue Bank. 2015 doi: 10.1007/s10561-015-9541-6. Dec 17 [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Jung Y, Bauer G, Nolta JA. Concise review: Induced pluripotent stem cell-derived mesenchymal stem cells: Progress toward safe clinical products. Stem Cells. 2012;30:42–47. doi: 10.1002/stem.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biancone L, Bruno S, Deregibus MC, et al. Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol Dial Transplant. 2012;27:3037–3042. doi: 10.1093/ndt/gfs168. [DOI] [PubMed] [Google Scholar]
- 17.Yu B, Zhang X, Li X. Exosomes derived from mesenchymal stem cells. Int J Mol Sci. 2014;15:4142–4157. doi: 10.3390/ijms15034142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bobrie A, Colombo M, Raposo G, et al. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 19.O’Loughlin AJ, Woffindale CA, Wood MJ. Exosomes and the emerging field of exosome-based gene therapy. Curr Gene Ther. 2012;12:262–274. doi: 10.2174/156652312802083594. [DOI] [PubMed] [Google Scholar]
- 20.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Mathivanan S, Fahner CJ, Reid GE, et al. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Yin Y, Lai RC, et al. Mesenchymal stem cells secrete immunologically active exosomes. Stem Cells Dev. 2014;23:1233–1244. doi: 10.1089/scd.2013.0479. [DOI] [PubMed] [Google Scholar]
- 23.Lai FW, Lichty BD, Bowdish DM. Microvesicles: Ubiquitous contributors to infection and immunity. J Leukoc Biol. 2015;97:237–245. doi: 10.1189/jlb.3RU0513-292RR. [DOI] [PubMed] [Google Scholar]
- 24.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosset FL, Dreux M. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol. 2014;60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA. 2014;111:E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang G, Liu G, Kitamura K, et al. TGF-β suppression of HBV RNA through AID-dependent recruitment of an RNA exosome complex. PLoS Pathog. 2015;11:e1004780. doi: 10.1371/journal.ppat.1004780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madison MN, Jones PH, Okeoma CM. Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology. 2015;482:189–201. doi: 10.1016/j.virol.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan CY, Lai RC, Wong W, et al. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res Ther. 2014;5:76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, He Z, Yuan J, et al. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell Microbiol. 2015;17:105–118. doi: 10.1111/cmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Liu K, Liu Y, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 32.Yeo RW, Lai RC, Zhang B, et al. Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander M, Hu R, Runtsch MC, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Y, Luan J, Li H, et al. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590:185–192. doi: 10.1002/1873-3468.12024. [DOI] [PubMed] [Google Scholar]
- 36.Xin H, Li Y, Buller B, et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–1564. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiao C, Xu W, Zhu W, et al. Human mesenchymal stem cells isolated from the umbilical cord. Cell Biol Int. 2008;32:8–15. doi: 10.1016/j.cellbi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Si Y, Liu S, Liu X, et al. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Guan J, Niu X, et al. Exosomes released from human induced pluripotent stem cells-derived MSCs facilitate cutaneous wound healing by promoting collagen synthesis and angiogenesis. J Transl Med. 2015;13:49. doi: 10.1186/s12967-015-0417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ti D, Hao H, Tong C, et al. LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. J Transl Med. 2015;13:308. doi: 10.1186/s12967-015-0642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong Y, Zhu Y, Xia X, et al. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J Virol. 2011;85:2793–2802. doi: 10.1128/JVI.01818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 44.Cheng M, Si Y, Niu Y, et al. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707–9718. doi: 10.1128/JVI.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami Y, Aly HH, Tajima A, et al. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453–460. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Gao H, Duan S, et al. Inhibition of microRNA-199a-5p reduces the replication of HCV via regulating the pro-survival pathway. Virus Res. 2015;208:7–12. doi: 10.1016/j.virusres.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Xu G, Yang F, Ding CL, et al. MiR-221 accentuates IFN׳s anti-HCV effect by downregulating SOCS1 and SOCS3. Virology. 2014;462-463:343–350. doi: 10.1016/j.virol.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Calland N, Albecka A, Belouzard S, et al. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55:720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- 49.Qian X, Xu C, Zhao P, et al. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology. 2016;492:155–165. doi: 10.1016/j.virol.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 50.Kang HS, Habib M, Chan J, et al. A paradoxical role for IFN-gamma in the immune properties of mesenchymal stem cells during viral challenge. Exp Hematol. 2005;33:796–803. doi: 10.1016/j.exphem.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Sharma RR, Pollock K, Hubel A, et al. Mesenchymal stem or stromal cells: A review of clinical applications and manufacturing practices. Transfusion. 2014;54:1418–1437. doi: 10.1111/trf.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munir H, McGettrick HM. Mesenchymal stem cell therapy for autoimmune disease: Risks and rewards. Stem Cells Dev. 2015;24:2091–2100. doi: 10.1089/scd.2015.0008. [DOI] [PubMed] [Google Scholar]
- 53.Kang K, Ma R, Cai W, et al. Exosomes secreted from CXCR4 overexpressing mesenchymal stem cells promote cardioprotection via Akt signaling pathway following myocardial infarction. Stem Cells Int. 2015;2015:659890. doi: 10.1155/2015/659890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res (Amst) 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Reis LA, Borges FT, Simões MJ, et al. Bone marrow-derived mesenchymal stem cells repaired but did not prevent gentamicin-induced acute kidney injury through paracrine effects in rats. PLoS One. 2012;7:e44092. doi: 10.1371/journal.pone.0044092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Jiang JD, Peng ZG. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol. 2016;22:1487–1496. doi: 10.3748/wjg.v22.i4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 58.Choi JE, Hur W, Kim JH, et al. MicroRNA-27a modulates HCV infection in differentiated hepatocyte-like cells from adipose tissue-derived mesenchymal stem cells. PLoS One. 2014;9:e91958. doi: 10.1371/journal.pone.0091958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee HK, Finniss S, Cazacu S, et al. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014;23:2851–2861. doi: 10.1089/scd.2014.0146. [DOI] [PubMed] [Google Scholar]
- 60.Hoshino D, Kirkbride KC, Costello K, et al. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Reports. 2013;5:1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimbo K, Miyaki S, Ishitobi H, et al. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem Biophys Res Commun. 2014;445:381–387. doi: 10.1016/j.bbrc.2014.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.