Abstract
Toxin/antitoxin (TA) systems reduce metabolism under stress; for example, toxin YafQ of the YafQ/DinJ Escherichia coli TA system reduces growth by cleaving transcripts with in-frame 5’-AAA-G/A-3’ sites, and antitoxin DinJ is a global regulator that represses its locus as well as controls levels of the stationary sigma factor RpoS. Here we investigated the influence on cell growth at various temperatures and found that deletion of the antitoxin gene, dinJ, resulted in both reduced metabolism and slower growth at 18°C but not at 37°C. The reduction in growth could be complemented by producing DinJ from a plasmid. Using a transposon screen to reverse the effect of the absence of DinJ, two mutations were found that inactivated the toxin YafQ; hence, the toxin caused the slower growth only at low temperatures rather than DinJ acting as a global regulator. Corroborating this result, a clean deletion of yafQ in the ΔdinJ ΔKmR strain restored both metabolism and growth at 18°C. In addition, production of YafQ was more toxic at 18°C compared to 37°C. Furthermore, by overproducing all the E. coli proteins, the global transcription repressor Mlc was found that counteracts YafQ toxicity only at 18°C. Therefore, YafQ is more effective at reducing metabolism at low temperatures, and Mlc is its putative target.
Introduction
Most Bacteria and Archaea contain toxin/antitoxin (TA) systems [1–3] which reduce cell growth to enable the cells to cope with stress [4]. For example, the MqsR/MqsA TA system enables the cell to withstand oxidative [5] and bile acid stress in the gastrointestinal tract [6]. Usually the genes for TA systems occur in pairs, and many antitoxins regulate the TA locus [7]. In addition, some antitoxins such as MqsA of the MqsR/MqsA TA system are intertwined with the general stress response by regulating other loci including levels of the stationary phase sigma factor RpoS [5, 8, 9], and some toxins exhibit a general regulatory effect via post-transcriptional differential mRNA decay [10, 11]. In addition to the general stress response, TA systems also have roles in biofilm formation [9, 10, 12, 13] and in inhibiting the propagation of phage [14–16].
In Escherichia coli, there are at least 39 TA systems [17–19]. Among them, the YafQ/DinJ TA system has been associated with several physiological roles. YafQ is an endoribonuclease that cleaves mRNA at in-frame 5’-AAA-G/A-3’ sites in conjunction with ribosomes [20]. Specifically, YafQ binds the 70S ribosome at the A site via three surface-exposed patches of basic residues that appear to directly interact with 16SrRNA, and YafQ residues H50, H63, D67, and H87 participate in acid-base catalysis during mRNA hydrolysis [21]. Its antitoxin is DinJ [22], and like the mqsRA locus [23], the dinJ-yafQ locus is not subject to conditional cooperativity [24], a form of regulation in which the binding of the first toxin to the antitoxin represses the TA locus whereas additional toxin molecules induce transcription of the locus [25]. DinJ binding to DNA as a dimer is facilitated by its N-terminal ribbon-helix-helix motif, and its C terminus binds YafQ as a heterotetramer [24, 26]. Also, the SOS regulator LexA binds the dinJ promoter suggesting a link of expression of this operon after DNA damage [24] although experimentally this has not been seen [27, 28].
The physiological roles of the YafQ/DinJ TA system include that it actively participates in the general stress response through the regulation of RpoS by antitoxin DinJ via direct repression of cspE [29]; cold-shock protein CspE enhances translation of RpoS mRNA. YafQ and DinJ are also involved in regulating persistence in E. coli; persister cells evade antibiotics by reducing their metabolism via toxins [30, 31] and are responsible for recurring infections [32]. Inactivation of YafQ reduces the persistence of biofilm cells [33], and dinJ-yafQ are induced in persisters [34]. The interspecies [35, 36] and interkingdom signal indole [37] reduces persistence [38, 39], and toxin YafQ increases persistence by reducing indole by cleaving tryptophanase mRNA [38]. Notably, indole is most active as a signal in E. coli at low temperatures [40]. Critically, aside from erythromycin stress leading to DinJ degradation [29], little is known about the conditions that activate YafQ and make it important for its role in the stress response and persistence.
Since there are few reports of temperature affecting the activity of a TA system [41–44], and since little is understood about what activates toxin YafQ, we explored the effect of temperature on the YafQ/DinJ TA system of E. coli. We found that the deletion of the gene that encodes antitoxin DinJ reduces metabolism and growth only at low temperature and that the mechanism is due to activation of toxin YafQ at low temperature. In addition, it appears the global transcription repressor Mlc may play a role in regulating YafQ activity.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids are listed in Table 1. Lysogeny broth (LB) [45] was used for all the experiments. We used the Keio collection [46] for isogenic mutants and pCA24N [47] for expressing genes in E. coli. One-step inactivation of the dinJ and yafQ genes in BW25113 using polymerase chain reaction (PCR) products [48] was used to create the double deletion strain, BW25113 ΔdinJ ΔyafQ (Table 1). The kanamycin resistance cassette from ΔdinJ, ΔyafQ, and ΔdinJ ΔyafQ was removed by using plasmid pCP20 [49]. Gene deletions were verified by DNA sequencing using primers dinJyafQ-F(CTGGATTTGGAAGGCTCAC) and dinJyafQ-R (CATGGATTGTCGCTGTTGC).
Table 1. E. coli bacterial strains and plasmids used in this study.
| Strains | Genotypea | Source |
|---|---|---|
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | [46] |
| BW25113 ΔyafQ | ΔyafQ Ω KmR | [46] |
| BW25113 ΔyafQ ΔKmR | ΔyafQ ΔKmR | this study |
| BW25113 ΔdinJ | ΔdinJ Ω KmR | [46] |
| BW25113 ΔdinJ ΔKmR | ΔdinJ ΔKmR | this study |
| BW25113 ΔdinJ ΔyafQ ΔKm | ΔdinJ ΔyafQ Ω KmR | this study |
| BW25113 ΔrpoS | ΔrpoS Ω KmR | [46] |
| BW25113 Δmlc | Δmlc Ω KmR | [46] |
| Plasmids | ||
| pCA24N | CmR; lacIq, pCA24N | [47] |
| pCA24N-dinJ | CmR; lacIq, pCA24N PT5-lac::dinJ | [47] |
| pCA24N-yafQ | CmR; lacIq, pCA24N PT5-lac::yafQ | [47] |
| pCA24N-mlc | CmR; lacIq, pCA24N PT5-lac::mlc | [47] |
| pCA24N-yhbU | CmR; lacIq, pCA24N PT5-lac::yhbU | [47] |
| pCA24N-sspA | CmR; lacIq, pCA24N PT5-lac::sspA | [47] |
| pCP20 | ApR, CmR; FLP+, λcI857+, λpRRepts | [49] |
a CmR and KmR are chloramphenicol and kanamycin resistance, respectively.
Cell growth was assayed using the turbidity at 600 nm. Kanamycin (50 μg/mL) was used for the Keio mutants, chloramphenicol (30 μg/mL) was used to maintain the pCA24N-based plasmids [47], and ampicillin (100 μg/mL) was used to maintain pCP20.
Transposon mutant selection for increased growth of the ΔdinJ ΔKmR strain
To identify the protein that caused toxicity at 18°C in the ΔdinJ ΔKmR strain, an EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit (Epicentre) was used to make the mutant library of BW25113 ΔdinJ ΔKmR according to the manufacturer’s instructions. In brief, 1 μL of transposome was electroporated into 50μL of competent cells (1 μL of TypeOne™ Restriction Inhibitor was added into the mixture to increase transduction efficiency). Warm SOC medium (1 mL) was used and the cells were incubated for 37°C for 60 min. The recovered cells (100 μL) were diluted 10-fold and spread on LB plates with kanamycin (50 μg/mL) to enumerate the number of transposon insertion clones. The remaining 900 μL of recovered cells (around 6,000 mutant cells) was inoculated into 20 ml fresh medium and cultured at 18°C for 36 hours. After 36 hr, 100 μL culture was transferred to 5 mL of fresh medium and cultured at 18°C for 36 hours (this was repeated four more times for six total enrichment cultures).
After six rounds of growth to enrich for faster-growing BW25113 ΔdinJ ΔKmR transposon mutants, colonies were formed, and 500 independent colonies were cultured in 0.5 mL of fresh medium in 2 mL microcentrifuge tubes for 24 h at 37°C. One μL of culture was inoculated into 100 μL of fresh medium in 96 well plates (two replicates) and BW25113 ΔdinJ ΔkmR was used for comparison. After culturing at 18°C and 250 rpm for 24h, growth was detected by measuring the turbidity at 600 nm. Colonies that yielded cultures with significant increases in growth (growth ratio greater than 50%) were selected for a second round of screening (in triplicate). The position of the transposon insertion was identified following the method of Ducey and Dyer (http://microgen.ouhsc.edu/forms/Epicentre.pdf) using random amplification of transposon ends (RATE) PCR (primers are listed in Table A in S1 File).
Metabolic activity assay
Overnight cultures were inoculated in LB medium, grown to a turbidity of 1.0 at 600 nm, and resuspended in IF-10 (BioLog, Hayward, CA, USA). Samples were diluted in IF-10 to a turbidity of 0.07 at 600 nm and further diluted 200-fold (turbidity of 0.00035 at 600 nm) into a medium containing IF-10, BioLog Redox Dye D (BioLog) and rich medium (0.1% yeast extract, 0.2% tryptone, and 0.1% NaCl). Cultures were grown at 37°C, 30°C and 18°C in 96-well microtiter plates (100 μL per well) and metabolic activity was monitored by measuring the optical density at 590 nm, indicating both reduction of tetrazolium dye to formazan [50] and sample turbidity. Experiments were performed with at least two independent cultures.
Pooled ASKA selection for increased growth of the ΔdinJ ΔKmR strain
To identify proteins that reduce YafQ toxicity in the absence of DinJ at 18°C, we produced each E. coli protein via the ASKA clone set in BW25113 ΔdinJ ΔKmR and selected for faster growth through successive cultures. One μL (40 ng) of pooled ASKA (GFP-) plasmid DNA [47] was transformed into 50 μL of BW25113 ΔdinJ ΔKmR competent cells by electroporation (1 μL of TypeOne™ Restriction Inhibitor was added into the mixture to increase transduction efficiency). Warm SOC medium (1 mL) was added and the cells were shaken at 37°C for 60 min. The cells were diluted 10-fold, spread on LB Cm30 plates to calculate the number of transposon insertion clones, and 900 μL was inoculated into 20 ml fresh LB Cm30 media with IPTG (200 μM) and cultured at 18°C for 36 hours. 100 μL was then transferred to 5 mL of fresh LB Cm30 IPTG (200 μM) and cultured at 18°C for another 36 hours. This was repeated 5 more times for a total of six successive cultures.
After six rounds of enrichment for faster growth, 100 μL of the cell suspension was diluted and plated on LB Cm30 plates, and 300 colonies were selected. Each colony was cultured in 0.5 ml fresh LB Cm30 media at 37°C for 24 hours, then 1 μL of each culture was inoculated into 100 μL of LB Cm30 media with IPTG 200 μM in 96 well plates. Each colony was assayed as two replicate wells and BW25113 ΔdinJ ΔKmR/pCA24N was used for comparison. After culturing at 18°C and 250 rpm for 24 h, growth was detected by measuring the turbidity at 600 nm. Colonies that yielded cultures with increased growth (increase growth ratio ≥ 50%) were selected for a second round of screening. 40 μL of overnight cultures of BW25113ΔdinJ ΔKmR/pCA24N and the best colonies obtained by the first round of 96-well plate screening were inoculated into 4 mL of fresh LB Cm30 IPTG 200 μM media. After culturing at 18°C and 250 rpm for 24h, growth was detected by measuring the turbidity. Cultures with increased growth (increase growth ratio ≥ 50%) were selected for plasmid extraction. Primers pCA24N-F (5’-GCCCTTTCGTCTTCACCTCG) and pCA24N-R 5’(GAACAAATCCAGATGGAGTTCTGAGGT) were used to identify genes encoded by pCA24N via sequencing. The plasmids of the positive cultures were re-transformed into 50 μL of BW25113 ΔdinJ ΔKmR competent cells to ensure the phenotype was due to the ASKA plasmid. Overnight cultures of BW25113 ΔdinJ ΔKmR /pCA24N and the positive cultures obtained by re-transformation were inoculated into 25 mL of fresh LB Cm30 IPTG (200 μM) media at an initial turbidity of 0.05. After culturing at 18°C and 250 rpm for 24 h, growth was detected by turbidity.
RNA isolation and quantitative, real-time, reverse transcription polymerase chain reaction (qRT-PCR)
To investigate the changes in yafQ mRNA levels at 18°C and 37°C, overnight cultures of the wild type strain BW25113 and of BW25113 ΔdinJ ΔKmR were inoculated into fresh LB at an initial turbidity of 0.05. Cells were collected after 2 hours culture at 37°C or 16 hours at 18°C at a turbidity of 0.7~0.8 for the wild type strain. To avoid RNA degradation, cells were harvested rapidly, cooled using ethanol/dry ice, centrifuged, then the cell pellets flash frozen in ethanol/dry ice. Total RNA was isolated from cells as described previously [51] with a RNeasy Mini Kit (Qiagen, Valencia, CA, USA) using a bead beater (Biospec, Bartlesville, OK, USA) and RNAlater buffer (Applied Biosystems, Foster City, CA, USA) to stabilize the RNA. 100 ng of total RNA was used for qRT-PCR using the Power SYBR Green RNA-to-CT1-Step Kit and the StepOne Real-Time PCR System (Applied Biosystems). Primers were designed using Primer-BLAST and are listed in Table B in S1 File. The housekeeping gene rrsG was used to normalize the gene expression data. The annealed temperature was 60°C for all the genes in this study.
Western blot analysis
To investigate the influence of dinJ deletion on RpoS at different temperatures, Western blots were performed as described previously [38, 52]. Overnight culture of wild type strain BW25113 and BW25113 ΔdinJ ΔKmR were inoculated into fresh LB with an initial turbidity of 0.05. Cells were collected after 2 hours of culturing at 37°C or 16 hours at 18°C at a turbidity of 0.7~0.8 for the wild type strain. Cell pellets were processed with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma-Aldrich) and sonicated twice on ice for 15 s. The cell lysate was centrifuged at 13,000 g for 10 min, and the protein concentration of each supernatant was quantified by using the Pierce Protein Assay kit. The same amount of protein (40 μg) was loaded into each well of a 12% SDS-PAGE gel, then transferred to a PVDF membrane, which was then blocked with 4% BSA in TBST (10 mM Tris pH 7.5, 100 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. The Western blots were probed with a 1:2000 dilution of anti-RpoS monoclonal antibody (Neoclone) followed by a 1:20 000 dilution of horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (Millipore).
Results
Inactivation of antitoxin DinJ reduces metabolism at 18°C
The PortEco phenotypic database [53] provides scores for growth phenotypes from screening a library of single gene mutants in E. coli against a panel of 324 different chemical treatments covering 114 unique stresses. Hence, we searched the PortEco database for significant growth changes related to temperature that involved TA systems. The database indicated that a strain lacking the gene that encodes antitoxin DinJ had reduced growth at lower temperatures (score -4.2 at 20°C or at 18°C) [53]. Therefore we explored the impact of the dinJ mutation on metabolism at low temperatures. To ensure there were no artifacts, we utilized only strains in which the kanamycin resistance selection marker was deleted so that the only difference was the deletion of the chromosomal copy of the gene. Upon deletion of the kanamycin resistance marker, the promoter regions for the genes were not changed; for example, sequencing of ΔdinJ ΔKmR showed the original promoter and operator region were not altered. In addition, the ribosome binding site for yafQ is preserved in ΔdinJ ΔKmR and located in the residual portion of the dinJ gene (Fig A in S1 File).
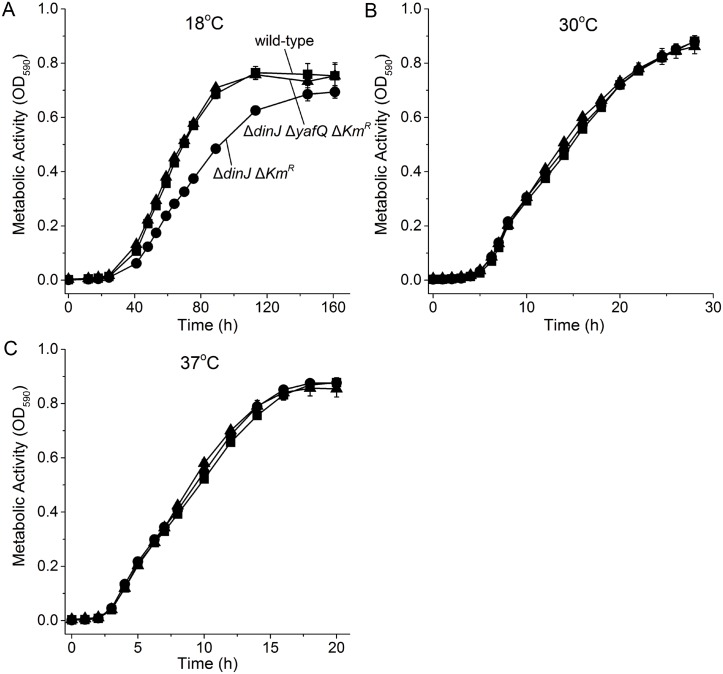
We first investigated the role the YafQ/DinJ TA system at low temperatures by performing the metabolic activity assay which utilizes tetrazolium dye; tetrazolium dye is reduced to the purple compound formazan due to NADH produced during cellular respiration [50]. We found that deletion of both the toxin and antitoxin had no phenotype whereas deletion of only antitoxin dinJ reduced metabolism significantly at 18°C but not at 30°C or 37°C (Fig 1). Therefore, inactivation of antitoxin DinJ reduces cell metabolism only at lower temperatures.
Fig 1. ΔdinJ reduces metabolism only at 18°C.
Comparison of metabolic activity via the Biolog assay for BW25113 wild-type (■), ΔdinJ ΔKmR (●) and ΔdinJ ΔyafQ ΔKmR (▲) at 18°C (A), 30°C (B), and 37°C (C). Data are the average of two independent cultures and one standard deviation is shown.
Inactivation of antitoxin DinJ reduces growth at 18°C
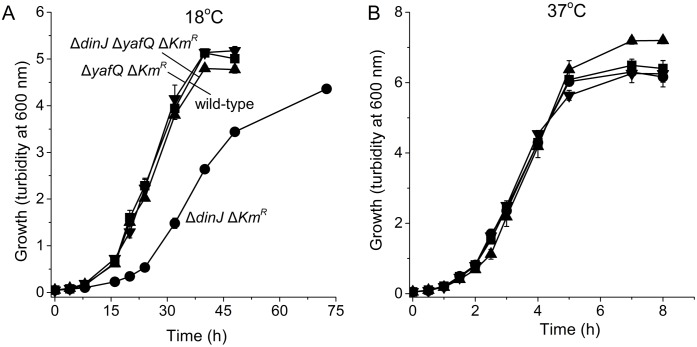
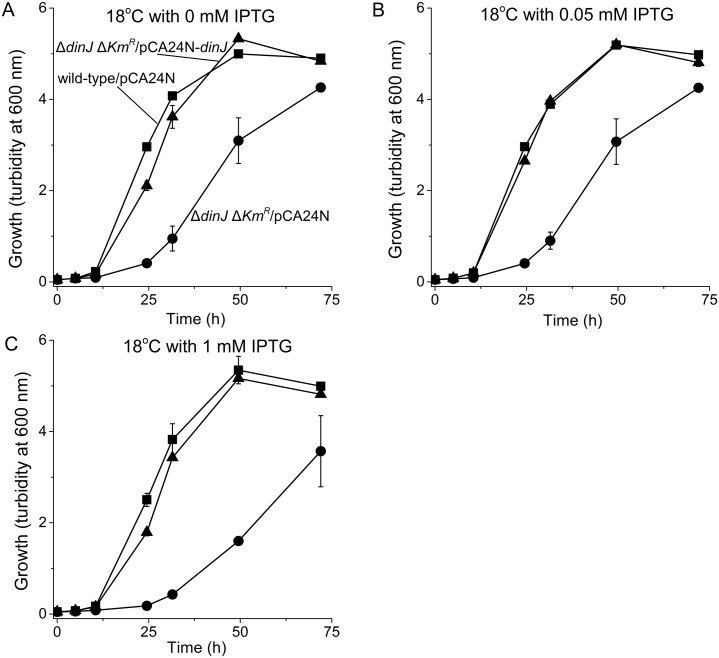
Since the dinJ deletion reduced metabolism only at low temperatures, we checked growth for this strain at 37°C and 18°C. We found that like metabolism, inactivation of DinJ reduced growth only at 18°C (Fig 2). This phenotype could be complemented by producing DinJ from plasmid pCA24N-dinJ (Fig 3). Note that low levels of DinJ sufficed to reduce the toxicity since similar results were seen via the leaky PT5-lac promoter at 0 mM IPTG as well as at 0.05 and 1 mM IPTG (Fig 3); some leakiness has been reported for this promoter [47]. As a negative control, the empty plasmid pCA24N did not affect growth of both the wild-type and the dinJ strain. Therefore, inactivation of antitoxin DinJ reduces cell growth only at lower temperatures.
Fig 2. ΔdinJ reduces growth only at 18°C.
Comparison of growth in LB medium for BW25113 wild-type (■), ΔdinJ ΔKmR (●), ΔdinJ ΔyafQ ΔKmR (▲), and ΔyafQ ΔKmR (▼) at 18°C (A) and 37°C (B). Data are the average of three independent cultures and one standard deviation is shown.
Fig 3. Complementation of the ΔdinJ lower growth phenotype at 18°C.
Comparison of growth in LB medium for BW25113/pCA24N (■), BW25113 ΔdinJ ΔKmR/pCA24N (●) and BW25113 ΔdinJ ΔKmR/pCA24N-dinJ (▲)at 18°C with IPTG induction of 0 mM IPTG (A), 0.05 mM IPTG (B), and 1 mM IPTG (C). Data are averaged from three independent cultures and one standard deviation is shown.
Toxin YafQ reduces metabolism and growth at 18°C
The cause of the reduced metabolism and growth only at low temperatures may be due to DinJ acting as a repressor of loci other than its own by binding at a palindrome (e.g., cspE which affects RpoS levels) [29] or due to activation of toxin YafQ. Hence, we randomly inactivated genes via transposon mutagenesis in the slow-growing ΔdinJ ΔKmR strain and selected for mutations that increased growth at 18°C; the transposon mutants were cultured together in LB at 18°C for six successive serial dilution cultures, and the DNA of the fastest growing strains was sequenced to determine where the mutations lie. Approximately 6,000 transposon mutants were formed. Because the wild-type strain grows at least as twice as the dinJ deletion strain at 18°C (Fig 2A), after six rounds of enrichment (i.e., six rounds of growth for 36 hr), the faster growing strains with a transposon insertion would be enriched by 33 million fold. We plated the cells after five rounds of enrichment, screened 500 of the transposon mutants in 96 well plates for faster growth, and found 10 mutants that showed a significant increase in growth compared with BW25113 ΔdinJ ΔKmR. After sequencing out from the transposon insertion, along with mutations in rapA, hupA, gcvT, gcvP, and orfE (Table 2), we found that growth was restored by mutations in the gene which encodes toxin YafQ (mutant found twice). For the two yafQ mutants, one had the Tn5 insertion upstream of the start codon (TR2-17), and for the second, the Tn5 insertion (TR2-40) removed the last 22 amino acids of YafQ (from aa 71 to 92). Based on the mutagenesis study by Armalyte J et al. [54], Arg83, His87, and Phe91 are active-site residues of YafQ since the Arg83Ala, His87Ala, and Phe91Ala substitutions abolished mRNA cleavage activity in vivo.
Table 2. Gene inactivations that restore growth at 18°C.
| Mutant | Gene | Function of gene product | Insertion site |
|---|---|---|---|
| TR2-17 | yafQ | YafQ is a sequence-specific mRNA endoribonuclease | -3/279 (RBS) of yafQ |
| TR2-40 | yafQ | 212/279 | |
| TR3-43 | rapA | RNA polymerase-associated, ATP-dependent RNA translocase; RNA polymerase PTC complex remodeling/recycling factor | 268/2907 |
| TR3-17 | rapA | 803/2907 | |
| TR3-6 | rapA | 1868/2907 | |
| TR4-45 | rapA | 260/2907 | |
| TR4-49 | rapA | 1372/2907 | |
| TR6-1 | hupA | Histone-like protein HU-alpha | 113/273 |
| TR6-19 | hupA | 1/273 | |
| TR3-37 | gcvT | Aminomethyl transferase | 446/1095 |
| TR3-29 | gcvP | Glycine decarboxylase | 347/2874 |
| TR4-53 | orfE | Pseudogene reconstruction, RNase PH | 352/717 |
Tn5-based random mutagenesis was performed with BW25113 ΔdinJ ΔKmR, and mutations that increase growth were identified by DNA sequencing. Insertion site numbers indicate the position of insertion of Tn5; for example, 216/279 means Tn5 is inserted at the 216nt where position one is start codon.
Corroborating these results, deletion of yafQ in the ΔdinJ ΔKmR background restored both metabolism (Fig 1A) and growth (Fig 2A) at 18°C. Also, deletion of the toxin gene yafQ (along with the kanamycin marker used to make it) restored the growth seen by the wild-type strain at low temperatures (Fig 2A). These results suggest the reduction in growth seen in the dinJ mutant is due to activity of toxin YafQ, rather than repression of other loci by DinJ. Therefore, YafQ reduces growth at low temperatures upon deletion of dinJ.
YafQ is more toxic at lower temperatures
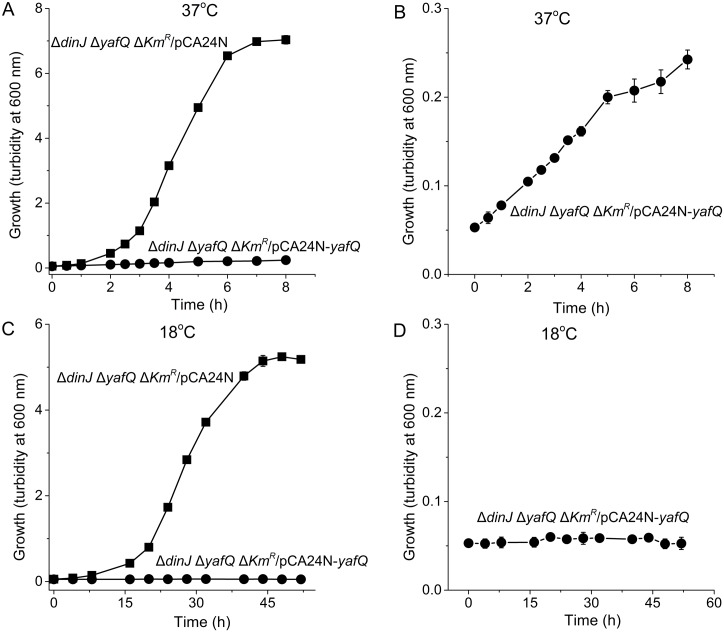
To address why YafQ reduces growth at low temperatures upon deletion of dinJ at 18°C, YafQ mRNA and toxicity were assayed in BW25113 wild-type and ΔdinJ ΔKmR at 18°C vs. 37°C. Based on our qRT-PCR result, YafQ mRNA levels are unchanged in BW25113 wild-type and ΔdinJ ΔKmR at both 18°C or 37°C (Table B in S1 File). We also explored YafQ toxicity at 37°C and 18°C and found that YafQ is more toxic at lower temperatures (Fig 4). Hence, the increase in YafQ toxicity upon dinJ deletion at 18°C is not due to changes in YafQ mRNA levels; however, it is unclear whether different amounts of YafQ protein contribute to this difference.
Fig 4. YafQ is more toxic at 18°C.
Comparison of growth in LB medium for BW25113 ΔdinJ ΔyafQ ΔKmR/pCA24N (■) and ΔdinJ ΔyafQ ΔKmR/pCA24N-yafQ (●) at 37°C (A) and 18°C (C). Panels (B) and (D) expand data for the ΔdinJ ΔyafQ ΔKmR/pCA24N-yafQ curves for (A) and (C), respectively. Data are averaged from three independent cultures, and one standard deviation is shown.
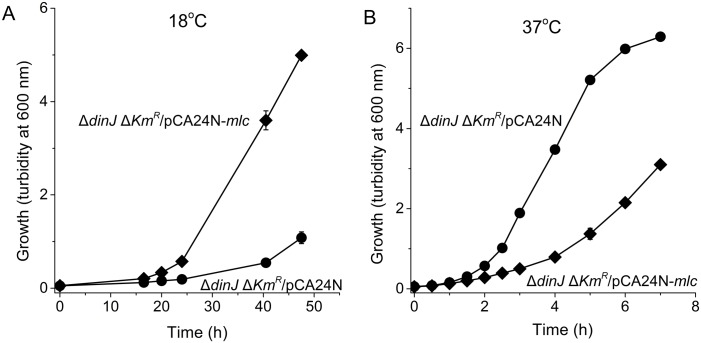
Mlc suppresses YafQ toxicity
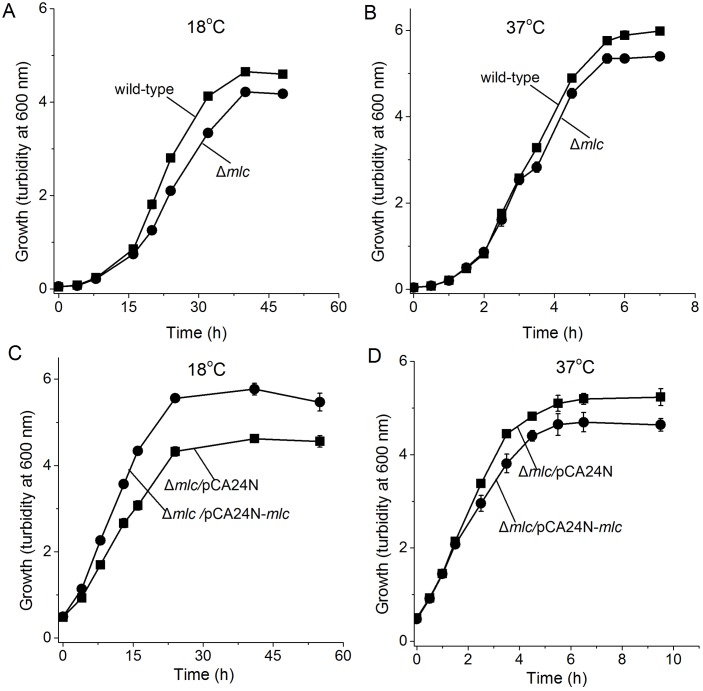
Since YafQ was not toxic at 37°C in the absence of DinJ, we investigated whether a protein beyond DinJ masks YafQ toxicity. Hence, we selected for faster growth at 18°C (where YafQ was activated and slowed growth) after electroporation of a pooled ASKA library into ΔdinJ ΔKmR. After enrichment from six rounds of growth, four plasmids of the ASKA clone set were found that restored BW25113ΔdinJ ΔKmR growth; sequencing revealed that the pCA24N plasmids encoded YhbU, SspA, and Mlc (Table 3). To avoid the influence of spontaneous mutations during the enrichment, the plasmids carrying yhbU, sspA, and mlc were re-electroporated into ΔdinJ ΔKmR, and Mlc was found to increase the growth of ΔdinJ ΔKmR the most. Critically, Mlc increased growth in the absence of dinJ at 18°C while it decreased growth at 37°C (Fig 5); hence Mlc is toxic at 37°C. Corroborating these results, deletion of mlc in the wild-type strain decreased growth more at 18°C (Fig 6A and 6B), and this phenotype could be complemented by producing Mlc from plasmid pCA24N-mlc only at 18°C (Fig 6C and 6D). These results demonstrate that Mlc reduces YafQ toxicity in the absence of antitoxin DinJ but only at low temperatures.
Table 3. Proteins whose production from pCA24N increased growth at low temperatures in the ΔdinJ ΔKmR strain as identified by screening the complete ASKA overexpression library.
| Colony | Growth increase (%) | Protein | Function |
|---|---|---|---|
| 1-3-57 | 32 | YhbU | U32 peptidase family protein. yhbUV divergent operon promoter region binds FNR and yhbUV operon expression is activated by FNR |
| 2-3-25 | 19 | SspA | Stringent starvation protein A, phage P1 late gene activator; RNAP-associated acid-resistance protein; inactive glutathione S-transferase homolog |
| 2-6-45 | 65 | Mlc (DgsA) | Global transcriptional repressor; regulates pts operon expression at P0; required for anaerobic growth on glucosamine, binds nagC promoters; regulates manX and malT; makes large colonies; autorepressor |
| 2-6-64 | 62 |
Fig 5. Mlc suppresses YafQ toxicity.
Comparison of growth in LB medium for BW25113 ΔdinJ ΔKmR/pCA24N (●) and BW25113 ΔdinJ ΔKmR/pCA24N-mlc (♦) with 200 μM IPTG induction at 18°C (A) and 37°C (B). Data are averaged from three independent cultures, and one standard deviation is shown.
Fig 6. Δmlc reduces growth at 18°C and 37°C.
Comparison of growth in LB medium for BW25113 wild-type (■) and BW25113 Δmlc (●) at 18°C (A) and 37°C (B). Comparison of growth in LB medium for BW25113 Δmlc/pCA24N (■) and BW25113 Δmlc/pCA24N-mlc (●) with 0.5 mM IPTG induction at 18°C (C) and 37°C (D). Data are averaged from three independent cultures and one standard deviation is shown.
RpoS increases at 18°C when dinJ is deleted
Since DinJ reduces stationary phase sigma factor (RpoS) levels to affect the general stress response [29], we investigated if RpoS levels were altered by growth in the BW25113 ΔdinJ ΔKmR mutant relative to the wild-type strain at 37°C and 18°C using a Western blot. As expected at 37°C [29], we found that DinJ inactivation results in roughly two fold more RpoS, and similar results were found at 18°C (Fig B in S1 File). Therefore, the quantity of RpoS is not the major reason for elevated toxicity of YafQ at 18°C.
Discussion
In this study, we show clearly that inactivation of antitoxin DinJ results in slower growth and metabolism only at low temperatures. This reduction in growth is not from the interaction of DinJ and the stress response sigma factor RpoS [29] but instead from activation of toxin YafQ since both metabolism (Fig 1) and cell growth (Fig 2) were decreased only at this temperature and inactivation of YafQ (identified by a transposon search) restored growth at low temperatures (Fig 2). Therefore, the YafQ/DinJ TA system seems to be utilized primarily at low temperatures in a manner similar to the activation at low temperatures of toxin GraT of the type II GraT/GraA TA system of Pseudomonas putida [42]; toxin GraT inhibits ribosome assembly at low temperatures by interacting with the chaperone DnaK [55]. The other TA systems that are affected by temperature changes are the type I BsrG/SR4 TA system of Bacillus subtilis [41] in which toxin BsrG mRNA is degraded upon heat shock, and the single polypeptide toxin and antitoxin EzeT which only has toxicity at low temperatures due to changes in protein folding [43].
Since YafQ was not toxic at 37°C (in the absence of antitoxin DinJ, Fig 2B) but YafQ has activity both at 18°C and 37°C when it is overproduced from its non-native promoter (Fig 4), these results suggest that there is another protein (beyond DinJ) that inactivates YafQ at 37°C or that the primary target of YafQ is produced only at 18°C. Our attempt to find this suppressor/target, by overproducing all the E. coli proteins in a dinJ mutant, determined that the global transcription repressor Mlc suppresses YafQ toxicity effectively at 18°C (Fig 5); hence, the cell may use more than the toxin YafQ to regulate antitoxin activity or, more likely, overproducing one of the main targets of YafQ at 18°C, Mlc, prevents YafQ from slowing growth at low temperatures. In support of this hypothesis, Mlc has nine in-frame YafQ cleavage sites and no DinJ binding motifs so it is very likely the Mlc mRNA is degraded by YafQ. Also, Mlc is a ROK family transcriptional regulator that binds DNA via a helix-turn-helix motif and regulates glucose transport via its regulation of the phosphotranserase system [56]. In agreement with our hypothesis that Mlc may play a role regulating the activity of YafQ, mlc is induced during cold shock [57]. Therefore, the mechanism of slow growth only at low temperatures upon deleting antitoxin gene dinJ is activation of toxin YafQ (rather than DinJ interactions with other genes), and the ensuing toxicity due to activation of YafQ is probably cleavage of the cold-shock-related regulator Mlc.
In addition to the yafQ deletion, growth was restored to the dinJ mutant at 18°C by mutations in rapA, hupA, gcvT, gcvP, and orfE (Table 2). RapA, which shows the highest frequency of Tn insertion, 5 out of 12, stimulates RNA polymerase recycling in transcription [58]. Hence, Tn insertion in rapA perhaps lowered RNA polymerase recycling and thereby reduced yafQ transcription, resulting in a decrease of YafQ and increased growth.
HU is a major component of the bacterial nucleoid, composed of two non-identical but highly homologous subunits, HUα and HUβ [59, 60]. HUα is encoded by hupA (which shows the second highest frequency of Tn insertion, 2 out of 12). HUα is abundant during growth, while the heterodimeric αβ form predominates at the end of the exponential growth phase and remains as in the stationary phase[61]. HU is required for efficient expression of RpoS [62], and we have shown DinJ decreases RpoS levels [29] (Fig B in S1 File). Hence, Tn insertion in hupA could perhaps decrease rpoS translation and offset the increase in RpoS due to dinJ deletion, resulting in reduced of RpoS and therefore restoring the growth as the wild type strain.
RNase PH, encoded by orfE, could also play a role in structured RNA degradation [63]. YafQ is an endoribonuclease that associates with the ribosome through the 50S subunit and blocks translation elongation through mRNA cleavage at 5'-AAA-G/A-3' sequences. When dinJ is deleted, YafQ would be more toxic with higher endoribonuclease activity at 18°C, and the slow growth of the ΔdinJ ΔKmR mutant is caused by the degradation of RNA in cell. Hence, an insertion of Tn in orfE perhaps slows the degradation structured RNA, which would restore growth of the ΔdinJ ΔKmR mutant.
Proteins encoded by gcvT and gcvP belong to the glycine cleavage system, which is widely distributed in animals, plants, and bacteria [64] and catalyzes the oxidative cleavage of glycine to carbon dioxide, ammonia, and 5,10-methylenetetrahydrofolate as well as provides a secondary pathway for one-carbon biosynthesis [65–67]. It is not clear why growth of the ΔdinJ ΔKmR mutant was restored when the glycine cleavage system is blocked by Tn mutation in gcvT and gcvP.
Supporting Information
Fig A, Sequence analysis of BW25113 ΔdinJ ΔKmR. Fig B, Rpos levels detected by an anti-RpoS antibody. Table A, Oligonucleotides used for random amplification of transposon ends (RATE) PCR and qRT-PCR. Table B, Summary of qRT-PCR results.
(DOC)
Acknowledgments
This work was supported by the ARO (W911NF-14-1-0279) and National Natural Science Foundation of China (31401600). Y. Z. was supported by the China Scholarship Council for study at the Pennsylvania State University (201403250037), and T.K.W. is the Biotechnology Endowed Chair at Pennsylvania State University. We are grateful for the Keio and ASKA strains provided by the National Institute of Genetics of Japan.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the ARO (W911NF-14-1-0279) and National Natural Science Foundation of China (31401600). YZ was supported by the China Scholarship Council for study at the Pennsylvania State University, and TKW is the Biotechnology Endowed Chair at Pennsylvania State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38(11):3743–59. 10.1093/nar/gkq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, et al. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012;40(13):6158–73. 10.1093/nar/gks231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–83. 10.1128/AEM.05068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7:359–66. 10.1038/nchembio.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan BW, Lord DM, Peti W, Page R, Benedik MJ, Wood TK. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ Microbiol. 2015;17:3168–81. 10.1111/1462-2920.12749 [DOI] [PubMed] [Google Scholar]
- 7.Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340(2):73–85. 10.1111/1574-6968.12074 [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Wang X, Zhang X-S, Grigoriu S, Page R, Peti W, et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ Microbiol. 2010;12(5):1105–21. 10.1111/j.1462-2920.2009.02147.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soo VWC, Wood TK. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci Rep. 2013;3:3186 10.1038/srep03186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol. 2006;188:305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009;5(3):e1000390 10.1371/journal.pgen.1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Wang X, Ma Q, Zhang X-S, Wood TK. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–67. 10.1128/JB.01465-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Environ Microbiol. 2004;64:515–24. [DOI] [PubMed] [Google Scholar]
- 14.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178(7):2044–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GPC. The phage abortive infection system, ToxIN, functions as a protein—RNA toxin—antitoxin pair. Proc Natl Acad Sci U S A. 2009;106(3):894–9. 10.1073/pnas.0808832106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics. 2004;272(2):227–34. [DOI] [PubMed] [Google Scholar]
- 17.Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2011;79(1):109–18. 10.1111/j.1365-2958.2010.07433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Lord DM, Cheng H-Y, Osbourne DO, Hong SH, Sanchez-Torres V, et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8:855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Quiroga C, Chen Q, McAnulty MJ, Benedik MJ, Wood TK, et al. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014;42(10):6448–62. 10.1093/nar/gku279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, et al. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol. 2009;71(5):1071–87. 10.1111/j.1365-2958.2008.06572.x . [DOI] [PubMed] [Google Scholar]
- 21.Maehigashi T, Ruangprasert A, Miles SJ, Dunham CM. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res. 2015; 10.1093/nar/gkv791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motiejūnaitė R, Armalytė J, Deputienė V, Sužiedėlienė E. Escherichia coli dinJ-yafQ operon shows characteristic features of bacterial toxin—antitoxin modules. BIOLOGIJA. 2005;4:9–14. [Google Scholar]
- 23.Brown BL, Lord DM, Grigoriu S, Peti W, Page R. The E. coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J Biol Chem. 2013;288:1286–94. 10.1074/jbc.M112.421008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruangprasert A, Maehigashi T, Miles SJ, Giridharan N, Liu JX, Dunham CM. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J Biol Chem. 2014;289(30):20559–69. 10.1074/jbc.M114.573006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loris R, Garcia-Pino A. Disorder- and dynamics-based regulatory mechanisms in toxin—antitoxin modules. Chem Rev. 2014;114(13):6933–47. 10.1021/cr400656f [DOI] [PubMed] [Google Scholar]
- 26.Liang Y, Gao Z, Wang F, Zhang Y, Dong Y, Liu Q. Structural and functional characterization of Escherichia coli toxin-antitoxin complex DinJ-YafQ. J Biol Chem. 2014;289(30):21191–202. 10.1074/jbc.M114.559773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armalytė J, Jurėnaitė M, Beinoravičiūtė G, Teišerskas J, Sužiedėlienė E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J Bacteriol. 2012;194(6):1523–32. 10.1128/jb.06104-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, et al. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35(6):1560–72. 10.1046/j.1365-2958.2000.01826.x [DOI] [PubMed] [Google Scholar]
- 29.Hu Y, Benedik MJ, Wood TK. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ Microbiol. 2012;14(3):669–79. 10.1111/j.1462-2920.2011.02618.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–21. 10.1128/aem.02636-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother. 2013;57:1468–73. 10.1128/AAC.02135-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–72. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- 33.Harrison JJ, Wade WD, Akierman S, Vacchi-Suzzi C, Stremick CA, Turner RJ, et al. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob Agents Chemother. 2009;53(6):2253–8. 10.1128/AAC.00043-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186(24):8172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Attila C, Cirillo SL, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol. 2009;2(1):75–90. 10.1111/j.1751-7915.2008.00061.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A. 2010;107(1):228–33. 10.1073/pnas.0906112107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Kwan BW, Osbourne DO, Benedik MJ, Wood TK. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol. 2015;17(4):1275–85. 10.1111/1462-2920.12567 [DOI] [PubMed] [Google Scholar]
- 39.Kwan BW, Osbourne DO, Hu Y, Benedik MJ, Wood TK. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng. 2015;112:588–600. 10.1002/bit.25456 [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Zhang X-S, Hegde M, Bentley WE, Jayaraman A, Wood TK. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2(10):1007–23. 10.1038/ismej.2008.54 [DOI] [PubMed] [Google Scholar]
- 41.Jahn N, Preis H, Wiedemann C, Brantl S. BsrG/SR4 from Bacillus subtilis–the first temperature-dependent type I toxin—antitoxin system. Mol Microbiol. 2012;83(3):579–98. 10.1111/j.1365-2958.2011.07952.x [DOI] [PubMed] [Google Scholar]
- 42.Tamman H, Ainelo A, Ainsaar K, Hõrak R. A moderate toxin, GraT, modulates growth rate and stress tolerance of Pseudomonas putida. J Bacteriol. 2014;196(1):157–69. 10.1128/jb.00851-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rocker A, Meinhart A. A cis-acting antitoxin domain within the chromosomal toxin—antitoxin module EzeT of Escherichia coli quenches toxin activity. Mol Microbiol. 2015;97(3):589–604. 10.1111/mmi.13051 [DOI] [PubMed] [Google Scholar]
- 44.Janssen BD, Garza-Sánchez F, Hayes CS. YoeB toxin is activated during thermal stress. MicrobiologyOpen. 2015;4(4):682–97. 10.1002/mbo3.272. PMC4554461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12(5):291–9. [DOI] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. [DOI] [PubMed] [Google Scholar]
- 50.Berridge MV, Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev. 2005;11:127–52. [DOI] [PubMed] [Google Scholar]
- 51.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64(4):515–24. Epub 2004/01/17. 10.1007/s00253-003-1517-y . [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–83. Epub 2011/06/21. 10.1128/AEM.05068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144(1):143–56. 10.1016/j.cell.2010.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armalyte J, Jurenaite M, Beinoraviciute G, Teiserskas J, Suziedeliene E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J Bacteriol. 2012;194(6):1523–32. Epub 2012/01/17. 10.1128/jb.06104-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ainelo A, Tamman H, Leppik M, Remme J, Hõrak R. The toxin GraT inhibits ribosome biogenesis. Mol Microbiol. 2016:n/a–n/a. 10.1111/mmi.13344 [DOI] [PubMed] [Google Scholar]
- 56.Bréchemier-Baey D, Domínguez-Ramírez L, Plumbridge J. The linker sequence, joining the DNA-binding domain of the homologous transcription factors, Mlc and NagC, to the rest of the protein, determines the specificity of their DNA target recognition in Escherichia coli. Mol Microbiol. 2012;85(5):1007–19. 10.1111/j.1365-2958.2012.08158.x [DOI] [PubMed] [Google Scholar]
- 57.Phadtare S, Inouye M. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol. 2004;186(20):7007–14. 10.1128/jb.186.20.7007-7014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sukhodolets MV, Cabrera JE, Zhi H, Jin DJ. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15(24):3330–41. Epub 2001/12/26. 10.1101/gad.936701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rouviere-Yaniv J, Kjeldgaard NO. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979;106(2):297–300. Epub 1979/10/15. . [DOI] [PubMed] [Google Scholar]
- 60.Rouviere-Yaniv J, Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975;72(9):3428–32. Epub 1975/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claret L, Rouviere-Yaniv J. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J Mol Biol. 1997;273(1):93–104. Epub 1997/11/21. 10.1006/jmbi.1997.1310 . [DOI] [PubMed] [Google Scholar]
- 62.Balandina A, Claret L, Hengge-Aronis R, Rouviere-Yaniv J. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol Microbiol. 2001;39(4):1069–79. Epub 2001/03/17. . [DOI] [PubMed] [Google Scholar]
- 63.Jain C. Novel role for RNase PH in the degradation of structured RNA. J Bacteriol. 2012;194(15):3883–90. Epub 2012/05/23. 10.1128/jb.06554-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Japan Acad, Ser B Phys Biol Sci. 2008;84(7):246–63. Epub 2008/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kikuchi G. The glycine cleavage system: Composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1(2):169–87. 10.1007/bf01659328 [DOI] [PubMed] [Google Scholar]
- 66.Meedel TH, Pizer LI. Regulation of One-Carbon Biosynthesis and Utilization in Escherichia coli. J Bacteriol. 1974;118(3):905–10. PMC246838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sagers RD, Gunsalus IC. Intermediary Metabolism of Diplococcus Glycinophilus I.: Glycine Cleavage and One-Carbon Interconversions. J Bacteriol. 1961;81(4):541–9. PMC279049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig A, Sequence analysis of BW25113 ΔdinJ ΔKmR. Fig B, Rpos levels detected by an anti-RpoS antibody. Table A, Oligonucleotides used for random amplification of transposon ends (RATE) PCR and qRT-PCR. Table B, Summary of qRT-PCR results.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.