Abstract
Mucin-like 1 (MUCL1) was first identified as a breast-specific gene over a decade ago. Based on its highly restricted mRNA expression in breast tissue and continued expression during breast tumorigenesis and progression, MUCL1 is an attractive tumor-associated antigen and a potential therapeutic target. However, very little is known about the cellular location, biological functions and regulation of the MUCL1 protein, which will have a major impact on its druggability. Here we describe our efforts to fully characterize the cellular localization of MUCL1, investigate its regulation by key breast cancer oncogenes such as human epidermal growth factor receptor 2 (HER2) and discover its functional roles in breast cancer. Although some mucins are membrane bound, our data indicate that MUCL1 is secreted by some breast cancer cells, whereas others only express high levels of intracellular MUCL1. MUCL1 expression is highest in HER2-amplified breast tumors and inhibiting HER2 activity in tumor cells resulted in a decreased MUCL1 expression. In-depth investigation demonstrated that phosphoinositide3-kinase/Akt pathway, but not Ras/MEK pathway, controls MUCL1 expression downstream of HER2. Phenotypic assays revealed a strong dependence of HER2-positive cells on MUCL1 for cell proliferation. We further identified the mechanism by which MUCL1 regulates cell growth. Knockdown of MUCL1 induced a G1/S phase arrest concomitant with decreased cyclin D and increased p21 and p27 levels. Finally, we investigated the impact of MUCL1 loss on kinase signaling pathways in breast cancer cells through phospho-kinase array profiling. MUCL1 silencing abrogated phospho-focal adhesion kinase (FAK), Jun NH2-terminal kinase (JNK) and c-Jun signals, but not extracellular signal-regulated kinase or Akt pathway activities, thereby pointing to FAK/JNK pathway as the downstream effector of MUCL1 signaling. We are the first to identify an important role for MUCL1 in the proliferation of breast cancer cells, probably mediated via the FAK/JNK signaling pathway. Taken together, these data suggest a potential utility for therapeutic targeting of this protein in breast cancer.
Introduction
Mucin-like 1 (MUCL1) was first identified as a breast-specific gene by three groups in 2001–2002.1, 2, 3 Its mRNA expression pattern has been further defined in subsequent years, with both the breast and salivary glands expressing high levels, the lung and skin expressing low levels and all other normal tissues absent of MUCL1 transcript. Early studies demonstrated by reverse transcription–PCR analysis that >90% of breast cancer cell lines express MUCL1.1, 3 Further, Colpitts et al.1 confirmed that MUCL1 protein was present in the majority of breast tumors by immunohistochemistry. There is mounting evidence supporting an important role of MUCL1 in the progression or metastasis of breast cancer. Several studies have now shown that MUCL1 expression strongly correlates with higher tumor grade,4 TNM (tumor, node, metastases) staging and lymph node metastasis.3, 5 Most recently, it was shown that high MUCL1 expression is significantly correlated with high recurrence and death rates in triple negative breast cancer patients.6
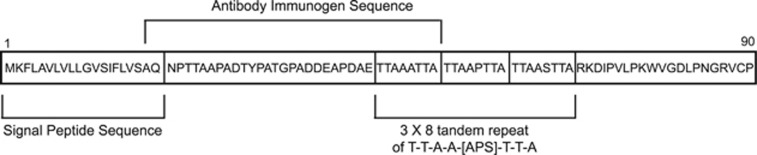
Based on its highly restricted mRNA expression, along with its conservation during breast tumorigenesis and progression, several groups have proposed the utility of the MUCL1 transcript as a biomarker for disease progression and metastasis in breast cancer patients.7, 8, 9, 10 Its limited normal tissue expression also renders MUCL1 an attractive tumor-associated antigen for targeted therapy of breast cancers. Despite our understanding of the expression of MUCL1 in breast cancer, the cellular localization of the MUCL1 protein has remained largely unstudied, which will have a major impact on drug developmentability. Although most mucins are secreted, several members of this protein family such as MUC1 and MUC4 are tethered to the plasma membrane with a hydrophobic membrane-spanning domain. MUCL1 was detected while assessing expression of tumor-derived cDNA fragments on yeast surface by screening with breast cancer patient sera, suggesting that it is membrane bound.11 Protein sequence analysis software yielded an ambiguous prediction that MUCL1 contains an N-terminal peptide signal sequence for targeting to the endoplasmic reticulum/Golgi secretory pathway, which could also double as a weak transmembrane domain (Figure 1). Whether the protein is secreted or tethered to the plasma membrane remains unknown. Early studies reported a secreted form of the protein in engineered NIH293 cells,1 but this was done in an artificial ectopic overexpression system and has not yet been verified in breast cancer cells. In addition to our lack of understanding of MUCL1 localization, a MUCL1 cellular function has not yet been characterized. Here we describe our efforts to fully define the cellular localization of MUCL1 and discover the biological function and signaling network of MUCL1 in breast cancer.
Figure 1.
A schematic of the MUCL1 amino acid sequence is presented. A hydrophobic signal peptide is present at residues 1–20 and a triple serine- and threonine-rich tandem repeat is present at residues 46–69. The antibody used for the current studies was generated against amino acids 19–53.
Results
MUCL1 characterization in breast cancer
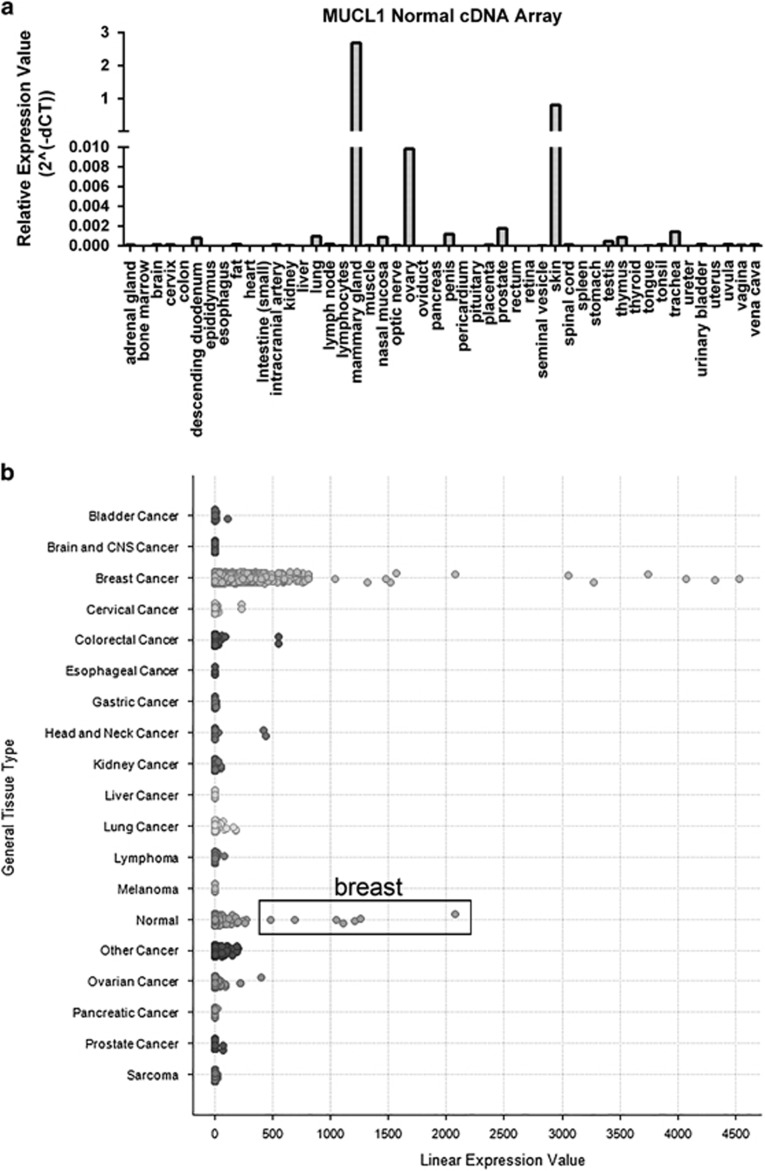
Earlier characterizations of MUCL1 expression examined a limited number of breast cancer and normal tissue samples. To build on these studies, we assessed the levels of MUCL1 expression across 48 normal tissue types using a cDNA array. The highest expression was found in the mammary gland, verifying the previously reported findings (Figure 2a). Significant mRNA expression was also detected in the skin but at a level three times lower than in the mammary gland. All other normal tissues either exhibited undetectable MUCL1 or at least 30 times lower expression compared with the mammary gland. A further survey of 147 normal body tissues was performed using the NextBio Body Atlas application. Results showed the highest expressing samples were the parotid gland, breast tissue, skin, salivary gland, nipple cross-section and mammary gland (Supplementary Figure S1a). We next assessed the expression of MUCL1 RNA in over 1000 cancer cell lines representing 37 cancer types in the Broad-Novartis Cancer Cell Line Encyclopedia. As expected, the highest level of MUCL1 expression was observed in breast cancer cell lines (Supplementary Figure S1b). Correspondingly, when we examined the expression of MUCL1 across a panel of human cancer samples using Oncomine Power Tools, breast cancer displayed the highest expression level of all cancers surveyed (Figure 2b). Further highlighting its restricted expression, breast tissue exhibited the highest MUCL1 gene expression among all the normal tissues included in the Oncomine analysis. Together, these multipronged genomic analyses suggest a restricted expression profile of MUCL1, and that it warrants further exploration as a tumor-associated antigen.
Figure 2.
MUCL1 is highly expressed in normal breast tissue and breast cancer. (a) MUCL1 expression is highest in mammary gland in a cDNA array from Origene. (b) MUCL1 expression examined across a panel of human cancer samples shows breast cancer having the highest expression level. In addition, the normal tissue samples exhibiting the highest expression were all from breast samples and are highlighted in the box. Analyses were done using the Oncomine Power Tools database (powertools.oncomine.com).
MUCL1 protein detection
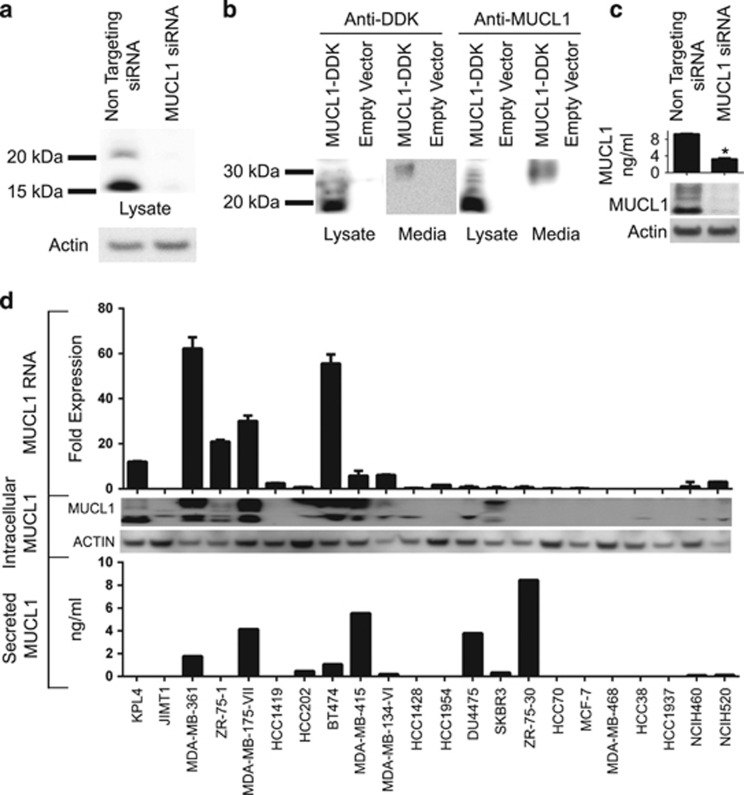
Very little is known about the properties of MUCL1 protein. The MUCL1 gene is predicted to code for a low-molecular-weight glycoprotein containing serine- and threonine-rich tandem repeats that are a characteristic feature of mucins (Figure 1). The commercially available antibodies for MUCL1 are indicated for use in immunohistochemistry, but their utility for this or other applications has not yet been confirmed. Thus, we tested the specificity of a polyclonal antibody targeting the internal region of MUCL1 protein for western blotting (Figure 1). KPL4 cell lysates were used, as it was shown by our above analysis to express a high level of MUCL1 mRNA (Table 1). Cells were transfected with non-targeting (NT) small interfering RNA (siRNA) or MUCL1 siRNA and the cell lysates were probed for MUCL1 expression by western blotting. MUCL1 was detected as two forms with the molecular masses of 17 and 22 kDa in the control cells (Figure 3a). Antibody specificity was validated by abrogation of the MUCL1 expression following siRNA knockdown. Interestingly, both bands are larger than the predicted protein size of 9 kDa, suggesting posttranslational modifications. To further confirm the antibody specificity, we next transfected HEK293 cells with an expression plasmid encoding myc/DDK(flag)-tagged MUCL1. Western blot analysis using an anti-DDK antibody revealed a 19-kDa band in the cell lysates of transfected cells, which was not present in cells transfected with an empty vector control (Figure 3b). The small increase in mass corresponds to the 17-kDa band in KPL4 cells with the addition of the 2.2-kDa myc/DDK tag. The same size bands were detected in these lysates using the anti-MUCL1 antibody, suggesting that it is indeed detecting the MUCL1 protein. In addition, a single band of around 33 kDa was detected in the cell culture media by both the anti-DDK and anti-MUCL1 antibodies. This secreted form corresponds to a previously reported 35 kDa myc-tagged MUCL1 observed in culture supernate of transfected HEK293 cells.1 The specificity of a commercially available enzyme-linked immunosorbent assay for MUCL1 was also validated by MUCL1 silencing (Figure 3c). Together, these data confirm that the commercial antibodies are indeed specific for MUCL1.
Table 1. The breast and lung cancer cell lines we examined are ranked by relative MUCL1 mRNA expression as determined in NextBio.com.
| Cell line | Cancer type | Fold expression change | ER/PR/HER2 status | Molecular subtype |
|---|---|---|---|---|
| KPL-4 | Breast | 269 | −/−/++ | HER2 |
| JIMT-1 | Breast | 204.1 | −/−/++ | HER2 |
| MDA-MB-361 | Breast | 174.3 | +/+/++ | LUM B |
| ZR-75-1 | Breast | 116.6 | +/+/+ | LUM B |
| MDA-MB-175-VII | Breast | 58.6 | +/−/− | LUM A |
| HCC1419 | Breast | 46.4 | −/−/++ | HER2 |
| HCC202 | Breast | 31.7 | −/−/++ | HER2 |
| BT-474 | Breast | 30.7 | −/+/++ | LUM B |
| MDA-MB-415 | Breast | 29.6 | +/−/− | LUM A |
| MDA-MB-134-VI | Breast | 14.2 | +/−/− | LUM A |
| HCC1428 | Breast | 14.1 | +/+/− | LUM A |
| HCC1954 | Breast | 12.6 | −/−/+ | HER2 |
| DU4475 | Breast | 11.3 | −/−/− | Basal |
| SK-BR-3 | Breast | 9.4 | −/−/++ | HER2 |
| ZR-75-30 | Breast | 9.3 | +/−/++ | LUM B |
| HCC70 | Breast | 6.4 | +/−/− | LUM A |
| MCF7 | Breast | 5.4 | +/+/− | LUM A |
| MDA-MB-468 | Breast | 4.0 | −/−/− | Basal |
| HCC38 | Breast | 3.7 | −/−/− | Basal |
| HCC1937 | Breast | 0.9 | −/−/− | Basal |
| NCI-H460 | Lung | 213.5 | HER2 + | Large cell |
| NCI-H520 | Lung | 150.2 | HER2 − | Squamous cell |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; MUCL1, Mucin-like 1; PR, progesterone receptor.
Fold expression change represents the level of MUCL1 in each cell line divided by the median of the gene across all normal cell lines. The ER, PR and HER2 status as well as the molecular subtype of each cancer cell line are reported.
Figure 3.
MUCL1 RNA and protein expression was examined in a panel of breast and lung cancer cell lines as described in Table 1. (a) KPL4 cells were transiently transfected with either NT siRNA or MUCL1 siRNA for 48 h. Cell lysates were probed using a rabbit polyclonal anti-MUCL1 antibody. β-Actin was used as a loading control. (b) HEK-293 cells were transfected with a DDK-tagged MUCL1 expression vector or empty vector. After 48 h, cell lysates and culture supernatant were immunoblotted using anti-DDK and anti-MUCL1 antibodies. (c) MDA-MB-361 cells were transiently transfected with either NT siRNA or MUCL1 siRNA for 48 h. Cell media was changed and conditioned media was collected 48 h later and assessed by MUCL1 enzyme-linked immunosorbent assay (ELISA). *P<0.01 (n=3). Cell lysates were probed using an anti-MUCL1 antibody. (d) RNA was extracted from cells and assessed for MUCL1 expression by reverse transcription PCR. RNA expression is shown as fold expression in each cell line divided by the median of MUCL1 expression across the panel of cell lines ±s.d. (n=3 technical replicates). For assessing protein levels, cells were grown for 48 h in serum-free media. Culture supernatant was collected and the secreted MUCL1 was measured by ELISA and normalized to the cell number. Intracellular MUCL1 levels were examined in whole-cell lysates by western blotting for comparison. The experiments were repeated twice with similar results.
Endogenous expression of MUCL1 protein
We next assessed the RNA and protein expression of MUCL1 in several cell lines that are reported to have high levels of MUCL1 mRNA by NextBio (Table 1). RNA extracts from a panel of lysates comprising 20 breast cancer and 2 lung cancer cell lines were examined by reverse transcription–PCR and cell lysates were probed by immunoblotting using the MUCL1 antibody described above. As shown in the middle panel of Figure 3d MUCL1 protein expression was observed in many of the cell lines tested. Western blot analysis revealed both a 17-kDa and a 22-kDa band in nine of the cell lines tested and a single 17-kDa band in five cell lines. Both bands represent MUCL1, as they can be efficiently ablated by MUCL1-targeted but not a non-targeted siRNA (Figure 3a). The relative abundance of the two species was not consistent across cell lines. Some cells displayed a greater quantity of the 17-kDa protein, whereas others expressed more of the 22-kDa band. The highest expression observed by western blotting was in KPL-4, MDA-MB-361, MDA-MB-175-VII, BT474 and MDA-MB-415 cells. Interestingly, all of these lines, except MDA-MB-175-VII, harbor high human epidermal growth factor receptor 2 (HER2) expression. A strong concordance was observed between the mRNA expression (top panel Figure 3d) and the protein expression detected by western blotting. However, two of the cell lines with detectable MUCL1 mRNA expression displayed little to no protein by western blotting. In addition, one cell line displayed robust MUCL1 protein expression, although it had much lower gene expression than many of the other cell lines tested. Together, these data indicate that there is an imperfect correlation between RNA and protein expression, potentially due to context-dependent translational or posttranslational controls.
Initial experiments demonstrated a faint MUCL1 reactive band with the mass of ~33 kDa in the media collected from HEK-293 cells transfected with DDK-tagged MUCL1. To determine whether endogenous MUCL1 is secreted by cancer cells, conditioned media were collected from cells grown in serum-free media. MUCL1 was measured by enzyme-linked immunosorbent assay and normalized to the total cell number. As shown in the lower panel of Figure 3d, 11 out of the 22 cell lines tested secreted MUCL1. Surprisingly, MUCL1 was not detected in the culture supernatant of some cells lines such as KPL4 and ZR-75-1, even though they have high MUCL1 expression in their cell lysates. In contrast, the ZR-75-30 cell line had little to no MUCL1 present in the cell lysates but demonstrated high levels of secreted MUCL1.
Cellular localization of MUCL1
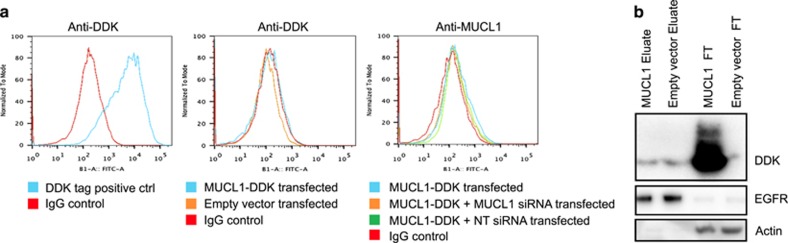
The exact cellular localization of MUCL1 remains largely unknown. As mentioned earlier, MUCL1 contains an N-terminal signal peptide that could also double as a weak membrane-spanning domain. To determine whether MUCL1 protein is expressed on the cell surface, we carried out live-cell staining and flow cytometry (FCM) analysis. HEK-293 cells were transfected with plasmid encoding tagged (DDK) MUCL1. Forty-eight hours later, cells were collected, stained using an anti-DDK antibody and analyzed by FCM. No shift in fluorescence was detected compared with the IgG control (Figure 4a). This is in contrast to the significant shift observed in the positive control cells expressing a known membrane-bound DDK-tagged protein. Three additional anti-MUCL1 antibodies raised against two different epitopes were tested and also resulted in no shift in fluorescent signal (data not shown). Based on the very high levels of expression reported for KPL4 cells, we subsequently tested for the presence of endogenous MUCL1 or overexpressed MUCL1-DDK on the surface of this cell line. Again, no signal was detected by FCM using either anti-DDK or anti-MUCL1 antibodies (Supplementary Figure S2).
Figure 4.
(a) HEK-293 cells were transfected with a DDK-tagged MUCL1 expression vector or empty vector. After 48 h, live cells were stained with anti-DDK or anti-MUCL1 antibodies followed by staining with a fluorescently labeled secondary antibody and analyzed by FCM. Cells expressing a known DDK-tagged membrane protein (positive control) demonstrated a significant shift in fluorescence. No shift in fluorescence was detected in HEK-293 cells transfected with DDK-tagged MUCL1 by either antibody. (b) HEK-293 cells were transfected with a DDK-tagged MUCL1 expression vector or empty vector. After 48 h, DDK-tagged MUCL1 was probed for using the ThermoScientific Cell Surface Protein Isolation Kit. Cells were treated with a biotinylation reagent to label surface proteins and then collected, lysed and labeled proteins were purified using neutravidin agarose resin. The eluate contains the isolated, labeled cell surface proteins and the flow-through (FT) contains unlabeled, intracellular proteins. The experiments were repeated twice with similar results.
To verify the FCM results, we characterized the ability of MUCL1 to be labeled by a cell-impermeable biotinylation probe. HEK-293 cells were transfected with MUCL1-DDK or empty vector and grown for 48 h. Cells were then incubated with a cell-impermeable biotinylation reagent (Sulfo-NHS-SS-Biotin) to label exposed primary amines of proteins on the extracellular surface of cells. MUCL1 contains three lysines potentially amenable to this labeling if exposed to the extracellular space. Cells were collected, lysed and the labeled surface proteins were affinity purified with avidin beads. Anti-DDK western blottings were carried out on the resulting two fractions of cellular extract: the eluate containing isolated, labeled cell surface proteins and the flow-through containing any unlabeled, hence intracellular proteins. As shown in Figure 4b, an intense band was detected in the flow-through (unlabeled) fraction of MUCL1-DDK-expressing cells, which was not present in the flow-through of cells transfected with empty vector. A faint band of a similar molecular weight was seen in the eluate fraction of MUCL1-overexpressing cells, but this was deemed nonspecific, as it was also observed in empty vector-transfected cells. The validity of the assay was confirmed by demonstrating that epidermal growth factor receptor, a well-established membrane protein, is primarily captured in the labeled eluate fraction, whereas actin, a cytoplasmic protein is only present in the flow-through/unlabeled fraction. Together with the FCM results, we conclude that MUCL1 is not exposed on the cell surface in this cell context.
HER2 signaling regulates MUCL1 expression
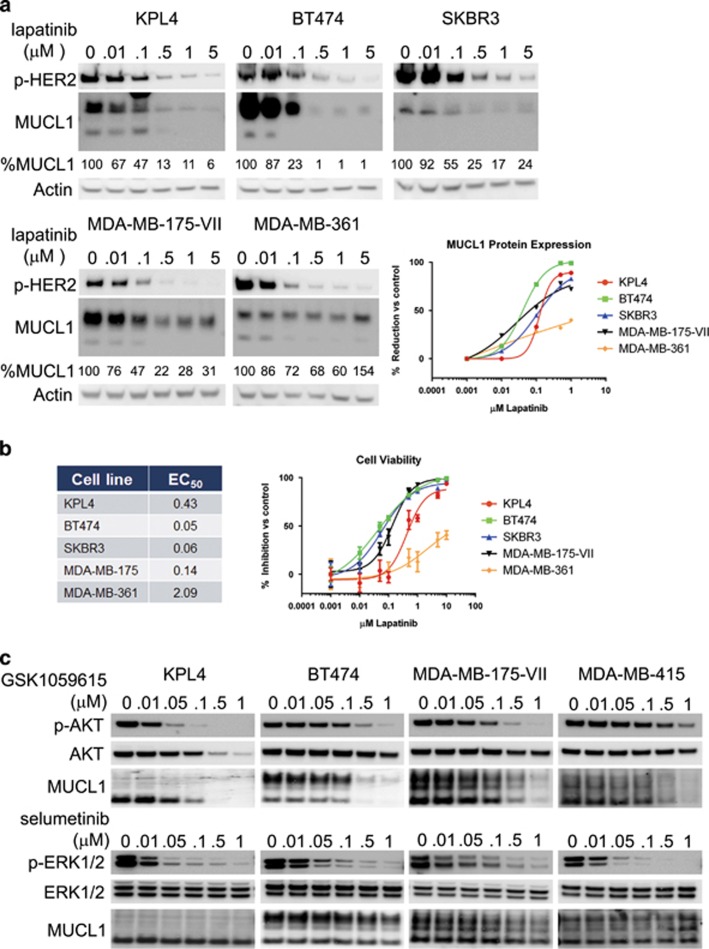
Further examination across breast cancer subtypes in Oncomine revealed that MUCL1 RNA expression was highest in HER2-overexpressing patient tumor samples. Data from four independent microarray studies demonstrated a consistent increase in MUCL1 expression in HER2-amplified tumors ranging between 3.5- and 5.5-fold higher than in HER2 non-amplified tumors (Table 2 and Supplementary Figure S3).12, 13, 14, 15 To examine whether HER2 signaling may regulate MUCL1 expression in breast cancer cells, we treated cells with increasing concentrations of lapatinib, a specific tyrosine kinase inhibitor of HER2, from 0.01 to 5 μM for 72 h. As expected, lapatinib treatment decreased the phosphorylation of HER2 in a dose-dependent manner (Figure 5a). In addition, inhibition of HER2 by lapatinib resulted in the dose-dependent decrease in MUCL1 expression in four of five cell lines tested (Figure 5a). We next tested for the anti-proliferative effect of lapatinib on these cell lines and calculated the half maximal effective concentration of growth inhibition (Figure 5b). The cell lines showing the greatest growth inhibition with lapatinib also showed the greatest reduction in MUCL1 protein, relative to controls (Figures 5a and b). Interestingly, despite a decrease in HER2 phosphorylation following lapatinib treatment in the MDA-MB-361 cell line, these cells were both significantly more resistant to the anti-proliferative effect and displayed significantly less reduction in MUCL1 levels compared with the other four cell lines. Contrary to the clear downregulation of MUCL1 protein following lapatinib treatment, no clear pattern of decrease in the MUCL1 RNA levels were observed in response to treatment (Supplementary Figure S4), suggesting regulation occurs at the posttranscriptional level. As HER2 stimulates downstream signaling primarily through the phosphoinositide3-kinase–Akt–mTOR and Ras–Raf–MEK–ERK pathways, we sought to clarify which of these pathways controls MUCL1 expression. Treatment of either HER2-overexpressing or HER2 normal cells with GSK1059615, a small molecule inhibitor of phosphoinositide3-kinase and mTOR, drastically reduced the expression of MUCL1, while inhibiting pAkt signal. In contrast, treatment with selumetinib, a small molecule inhibitor of MEK1/2, had no effect on MUCL1 levels despite efficient abrogation of phosphorylated extracellular signal-regulated kinase (ERK) activity (Figure 5c). These data indicate that in a majority of HER2-amplified breast cancer cell lines tested in this study, MUCL1 expression is dependent on HER2 signaling via phosphoinositide3-kinase/Akt, and that MUCL1 may contribute to HER2-mediated tumor cell growth.
Table 2. MUCL1 gene expression was examined across four independent data sets where HER2-negative breast cancer patient samples were compared with HER2-positive samples 12, 13, 14, 15 .
| Study | Sample type | Fold change MUCL1 expression | P-value | n (HER2+) | n (HER2−) |
|---|---|---|---|---|---|
| Curtis breast | Invasive ductal breast carcinoma | 3.461 | 7.02E-23 | 1163 | 388 |
| Kao breast | Breast carcinoma | 3.953 | 1.28E-08 | 252 | 75 |
| Gluck breast | Invasive breast carcinoma | 4.544 | 1.03E-05 | 119 | 33 |
| Lu breast | Ductal breast carcinoma | 5.476 | 1.52E-05 | 69 | 26 |
Analyses were done using the Oncomine database (www.oncomine.org).
Figure 5.
(a) Breast cancer cell lines were treated with the indicated doses of lapatinib for 72 h. Phospho-HER2 and MUCL1 levels were assessed by western blotting. β-Actin was used as a loading control. The percentage of MUCL1 relative to the vehicle control (0 μM) is shown. (b) The dose–response effect of growth inhibition following a 6-day lapatinib treatment is shown and the half maximal effective concentration (EC50) values for each cell line were calculated. The mean ±s.d. is plotted (n=5). (c) Breast cancer cells were treated with the indicated doses of the phosphoinositide3-kinase (PI3K) inhibitor GSK1059615 or the MEK1/2 inhibitor selumetinib for 48 h. Phospho-Akt, phospho-ERK1/2 and MUCL1 levels were assessed by western blotting. β-Actin was used as a loading control. The experiments were repeated twice with similar results.
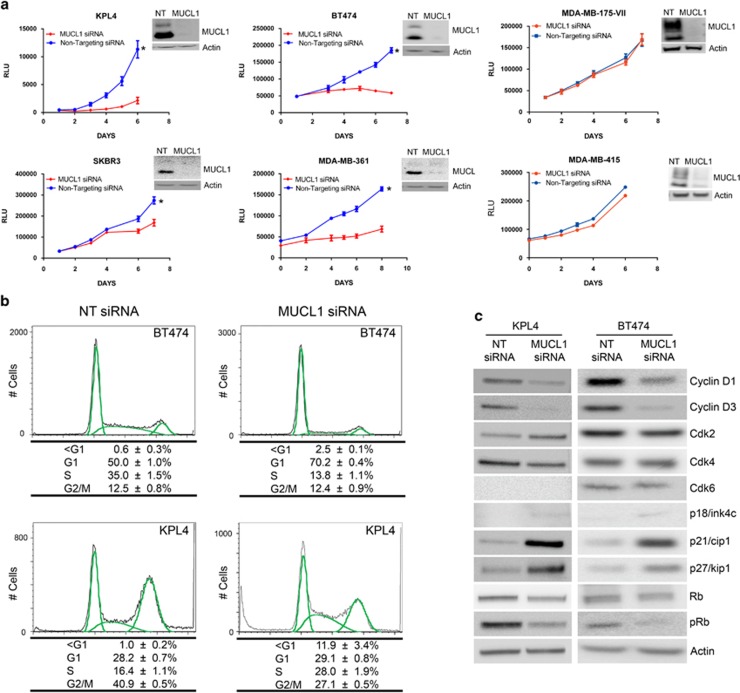
MUCL1 regulates cell proliferation
Despite being cloned more than a decade ago, no cellular functions have been ascribed to MUCL1. To investigate its physiological role, we knocked down MUCL1 using siRNA in several breast cancer cell lines. Proliferation of the transfected cells was tracked over time using cell viability-based CellTiter Glo Assay. Four of the six cell lines tested showed a significant reduction in proliferation when MUCL1 was knocked down (Figure 6a). BT474 and KPL-4 cells displayed the greatest reduction in growth, with 84% and 82% fewer cells, respectively, by day 6. MDA-MB-361 and SKBR3 were less inhibited with 70% and 47% fewer cells, respectively. Interestingly, no effect was seen following knockdown in two HER2 normal (also known in literature as ‘HER2 low') cell lines, MDA-MB-175-VII and MDA-MB-415. Specificity of the siRNA was demonstrated by using both a pooled set of siRNA and two independent siRNAs (Supplementary Figure S5).
Figure 6.
(a) Breast cancer cell lines were transfected with MUCL1 siRNA or NT control siRNA. Cell proliferation was assessed by Cell Titer Glo Assay each day for 1 week. Mean ±s.d. is shown (n=10). *P<0.001 for difference of growth rate. MUCL1 knockdown was confirmed by western blotting. (b) BT474 and KPL4 cells were transfected with MUCL1 or NT siRNA for 96 h, methanol-fixed and stained with propidium iodide for cell cycle analysis. The percent of cells in each cell cycle phase is shown as the mean ±s.d. (n=3). (c) Western blots of cell cycle regulators 72 h post transfection shows significant decreases in cyclins D1 and D3, as well as increases in the cyclin-dependent kinase (Cdk) inhibitors p21cip1 and p27kip1. The experiments were repeated twice with similar results.
We next examined whether the decrease in cell proliferation following MUCL1 knockdown was attributable to cell cycle arrest. KPL4 and BT474 cells were transfected with either NT or MUCL1 siRNA and subjected to cell cycle analysis 4 days later. We observed a significant decrease in S phase cells from 35 to 14% and reciprocal increases in G1 phase from 50 to 70% in BT474 cells, suggesting a G1 arrest (Figure 6b). In KPL4 cells, there was a decrease in G2 phase from 41 to 27% and an increase in S phase from 16 to 28%, suggesting an S-phase/replicative arrest. Interestingly, there was also an increase in the sub-G1 fraction from 0 to 12% in KPL4 cells, suggesting a concomitant apoptotic induction along with the S-phase arrest. To examine which cell cycle proteins control the response following MUCL1 knockdown, a panel of cell cycle regulators were examined 48 h after transfection with siRNA. In both BT474 and KPL4 cells, reduced phosphorylated retinoblastoma protein (a marker of G1/S transition) confirmed the cell cycle arrests we observed by cell cycle analysis (Figure 6c). Although there were no observed changes in G1 phase cyclin-dependent kinase levels, there were significant decreases in the cyclin-dependent kinase activators cyclins D1 and D3. In addition, a robust increase was seen in the levels of the cyclin-dependent kinase inhibitors p21cip1 and p27kip1, and a slight increase in p18ink4c. Together, these changes are consistent with a reduction in proliferation due to a cell cycle arrest.
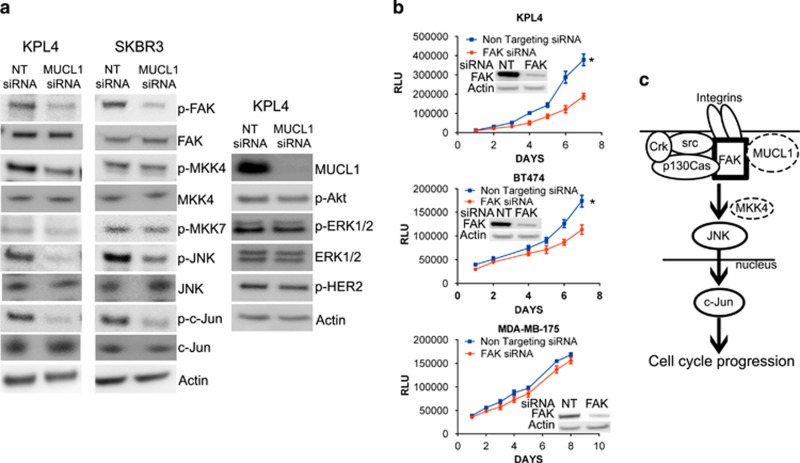
We next investigated which signaling pathways might be disrupted by MUCL1 silencing leading to the observed cellular effects. We first analyzed the phosphorylation profiles of 49 receptor tyrosine kinases in lysates from NT or MUCL1 siRNA-transfected KPL4 and BT474 cells using a phospho-protein array, and no significant changes were detected (data not shown). We next examined the phosphorylation status of intracellular protein kinases by carrying out an intracellular phospho-protein array. Several proteins displayed a significant reduction in their phosphorylation following MUCL1 knockdown. These included focal adhesion kinase (FAK), Jun NH2-terminal kinase (JNK) and c-Jun, and were verified by western blotting (Figure 7a). In addition, KPL4 cells exhibited decreased phospho-MKK4, one of the major kinases that binds to and phosphorylates JNK. No changes were observed in the phosphorylation of ERK or Akt, two major oncogenic pathways that are involved in cell proliferation and survival. In addition, we failed to detect changes in HER2 activation upon MUCL1 knockdown, consistent with our findings that HER2 acts upstream but not downstream of MUCL1 to regulate its expression. Finally, we examined whether the inhibition of FAK mimics the effects of silencing MUCL1 in these cells. Indeed, when HER2-overexpressing cell lines KPL4 and BT474 were transfected with FAK siRNA, cell proliferation is inhibited, while the HER2 normal expressing cell line MDA-MB-175-VII was not affected by FAK knockdown (Figure 7b).
Figure 7.
(a) KPL4 and SKBR3 cells were transiently transfected with MUCL1 siRNA or NT control siRNA and lysed for western blotting 48 h later. Immunoblot analysis showed a decrease in the phosphorylation of FAK, JNK and c-Jun in MUCL1 cells as compared with control cells, and phosphorylated MKK4 was decreased in KPL4 cells only. No changes were detected in activation of MKK7, Akt, ERK1/2 or HER2. β-Actin was used as a loading control. (b) Breast cancer cells were transfected with FAK siRNA or NT control siRNA. Cell proliferation was assessed by Cell Titer Glo Assay each day for 1 week. Mean ±s.d. is shown (n=10). *P<0.001 for difference of growth rate. FAK knockdown was confirmed by western blotting. (c) Proposed model depicting MUCL1-mediated FAK activation and signaling to downstream JNK. A potential interaction between MUCL1 with FAK is likely to be intracellular. Following integrin engagement, JNK activation requires association of FAK with a Src kinase and p130Cas, and the recruitment of Crk. The activation of JNK may be through MKK4 or other mediators depending on the cell type. On stimulation by JNK, c-Jun translocates to the nucleus and mediates G1/S phase transition leading to cell proliferation. The experiments were repeated twice with similar results.
Discussion
In this study, we demonstrate the first effort to define the localization and signaling pathways of human MUCL1 in breast cancer. We detected the MUCL1 protein in cell lysates from breast cancer cell lines, ranging between 17 and 22 kDa. Interestingly, the MUCL1 polypeptide sequence predicts a theoretical molecular mass of only 9 kDa. The higher molecular weights detected are probably the result of O-glycosylation on Thr-rich sequences of the protein. Additional posttranslational modifications, including potential dimerization might also contribute to the increased molecular weights. Our findings additionally describe MUCL1 as a secreted protein in a subset of cancer models and not displayed on the cell membrane. Surprisingly, not all cell lines with high levels of cellular MUCL1 secrete the protein. In fact, no secreted protein was detected in two of the highest expressing cell lines, KPL4 and ZR-75-1. However, our assays are limited by the specificity of the antibody used, which may not detect all forms of the protein with posttranslational modifications such as glycosylation. In addition, several alternatively spliced isoforms have been reported for the MUCL1 transcript, including at least one variant that lacks the predicted peptide signal sequence. It is possible that the breast cancer cells vary in which isoforms are predominantly expressed, thus dictating whether the protein is secreted from the cells or retained intracellularly. One interesting topic for future research is to elucidate the function of the secreted MUCL1 in the subset of breast cancers with such a population. Our current siRNA platform depletes both intracellular and secreted populations, thus making it difficult to discriminate any potentially distinct functionality between the two forms.
This report serves as the first demonstration of a functional role of MUCL1 in breast cancer. Significantly, the expression of MUCL1 protein is necessary for the proliferation of HER2-overexpressing breast cancer cell lines tested here. Knockdown of MUCL1 in these breast cancer cells resulted in a cell cycle arrest coinciding with decreased expression of cell cycle promoting cyclins D1 and D3, and increased expression of cell cycle inhibitors p21cip1 and p27kip1. Interestingly, our studies have revealed that MUCL1 knockdown results in decreased FAK phosphorylation as well as JNK and c-Jun activation in cells. We propose a model in which MUCL1 interacts with FAK governing its phosphorylation and activation, to allow subsequent downstream activation of JNK and its substrate c-Jun, ultimately resulting in cell cycle progression by inducing G1 phase cyclins and reducing cyclin-dependent kinase inhibitors (Figure 7c). Although FAK is primarily known for its roles in cell adhesion and migration, it has been shown to function as a mediator of cell cycle regulation through FAK/Src complex formation, leading to transcription of cyclin D1 (Zhao et al.16) and decreased p21 expression.17 Although studies have demonstrated that FAK can regulate cell cycle progression through either ERK or JNK activation,17, 18 here we show that MUCL1 acts via the FAK–JNK pathway. It is reported that following integrin engagement, JNK activation requires association of FAK with an Src kinase and p130Cas, the phosphorylation of p130Cas and the subsequent recruitment of Crk.18 Significantly, integrin-mediated activation of FAK and downstream JNK signaling was shown to be required for cell cycle progression from the G1 to S phase in adherent cells.18 Further, the phosphorylation of c-Jun by JNK is known to be required for activation of AP-1, which can directly regulate transcription of cyclin D1, cyclin D3, p21cip1 and p27kip1.
The connection of FAK signaling to cell proliferation is not unexpected, as human breast cancers frequently overexpress FAK19 and studies in mouse models of breast cancers have shown evidence for a direct role for FAK and associated effectors such as Src and p130Cas in tumor initiation and development.20, 21 For example, ablation of FAK in the MMTV-PyVmT model of mammary tumorigenesis leads to impaired maintenance and progression of tumor cells as a result of decreased proliferation.22 Deletion of Src in the same tumor model has similar effects including defects in proliferation and cell cycle.23 Although the mechanism by which MUCL1 regulates the activation of FAK remains unknown, it is interesting that knockdown of MUCL1 resulted in reduced proliferation and reduced FAK signaling in cells that do not secrete the MUCL1 protein. This suggests that MUCL1 is not interacting with extracellular components to induce the phosphorylation of FAK. In addition, we did not observe any changes in a phospho-receptor tyrosine kinase array following MUCL1 knockdown.
The studies presented here show that high MUCL1 expression is associated with HER2 amplification in breast cancer cells. HER2 overexpression is clearly associated with tumor metastasis and worse clinical outcome.24, 25 Our findings established a novel role for HER2 signaling in regulating the expression of MUCL1 through the phosphoinositide3-kinase/Akt signaling pathway and describe MUCL1 as a potential mediator of HER2's tumor growth effects. Although MUCL1 expression was controlled by HER2 signaling in both HER2-overexpressing and HER2 normal cell lines, the requirement for MUCL1 for full cell proliferation was only seen in HER2-overexpressing cells. Similarly, FAK activation was only required for proliferation of HER2-overexpressing cells. The effect of FAK silencing in these cell lines was not as dramatic as MUCL1 silencing, suggesting that additional pathways downstream of MUCL1 are responsible for mediating its role in cell proliferation. All together, these results indicate that HER2 regulates MUCL1 expression and in turn, MUCL1 promotes breast cancer growth and survival of HER2-overexpressing cells in part due to sustaining FAK activity and cell cycle promotion.
Materials and methods
Cell lines and reagents
Cells were obtained from American Type Culture Collection (Manassas, VA, USA). Cell culture media and supplements are listed in Supplementary Figure S6. Cells were grown at 37 °C and 5% CO2 in a humidified incubator. Cell line authentication was conducted by short tandem repeat-based DNA fingerprinting and multiplex PCR, and cells were tested for mycoplasma.
Enzyme-linked immunosorbent assay kits were from US Biological (Salem, MA, USA). MUCL1 TruORF cDNA clone in pLenti-C-Myc-DDK, pCMV6-Entry vector and DDK antibody were from Origene (Rockville, MD, USA). MUCL1 and NT siRNAs were from GE Dharmacon (Lafayette, CO, USA). All antibodies are listed in Supplementary Figure S6. Human Phospho-Kinase Antibody Array was from R&D Systems (Minneapolis, MN, USA).
Transfection with MUCL1 ORF and siRNA
HEK-293 cells were seeded at 50% confluence and allowed to attach overnight. MUCL1 open reading frame (ORF) vector was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer's protocol. Medium was refreshed the day of transfection. siRNA transfection of HEK-293 was performed using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. Cells were reverse transfected with siRNA oligos targeting MUCL1. Briefly, transfection mixes were prepared and siRNA–lipid complexes incubated in six-well culture dishes for 20 min. Cells were then added to wells with transfection mixes. Medium was replaced 24 h post transfection and lysates were collected 24 h later.
Cell surface protein isolation and gel electrophoresis
The Pierce Cell Surface Protein Isolation Kit (ThermoScientific, Waltham, MA, USA) was used following the manufacturer's instructions. Cells were reverse transfected with MUCL1-DDK or empty vector plasmids as described above and each treatment group was cultured in four 75 cm2 flasks. After 72 h, cells were incubated with Sulfo-NHS-SS-Biotin for 30 min at 4 °C after which the reaction was quenched. Cells were washed, scraped and lysed using the provided lysis buffer with protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA). To capture biotinylated (surface) proteins, lysates were incubated with Neutravidin Agarose gel columns for 2 h. The unbound (unbiotinylated) proteins, representing the intracellular fraction, were separated from the captured surface proteins by centrifugation. Captured surface proteins were eluted from the biotin-Neutravidin Agarose by incubation with dithiothreitol and were collected by centrifugation. Cellular equivalents were run on SDS–polyacrylamide gel electrophoresis and immunoblotted.
FCM and cell cycle analysis
Surface expression of DDK-MUCL1 on AD293 cells was tested by FCM. Cells were trypsinized and washed in phosphate-buffered saline (PBS) containing 1% bovine serum albumin. Live cells were incubated with mouse-anti-DDK (Origene) in PBS containing 1% bovine serum albumin for 1 h at 4 °C, rinsed and incubated with rabbit-anti-mouse-488 antibody (Molecular Probes, Life Technologies, Carlsbad, CA, USA) for 30 min at room temperature in the dark. Finally, cells were rinsed and diluted in PBS containing 1% bovine serum albumin and analyzed using a MACSQuant VYB Flow Cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany).
For cell cycle analysis, cells transfected with siRNA for 96 h were trypsinized, collected and washed with PBS. Pellets were suspended with 70% methanol at −20 °C overnight, washed with PBS, then incubated with 0.1% Triton X-100, 0.2 mg/ml RNase A (Sigma-Aldrich) and 20 μg/ml propidium iodide (Sigma-Aldrich) for 30 min at room temperature. FCM (LSR-II system, BD Biosciences, San Jose, CA, USA) was used for detection. Data were analyzed with FlowJo (FlowJo, LLC, Ashland, OR, USA).
In vitro cell proliferation assays
Cells were reverse transfected with MUCL1siRNA, FAK siRNA or NT siRNA as described above. Viability of cells was measured in quadruplicate, using CellTiter-Glo kits (Promega, Madison, WI, USA) according to the manufacturer's instructions. Relative cell viability was determined by dividing the luminescence values for MUCL1 siRNA cells by the average luminescence for NT siRNA cells.
cDNA arrays
An array with 48 normal tissues (Origene) was probed for MUCL1 and GAPDH mRNA levels using the Fluidigm system (San Francisco, CA, USA). cDNA was pre-amplified with gene-specific probes before quantitative PCR and data were normalized to GAPDH. Samples were made relative using 2−ΔCt method.
Statistical analysis
For analysis of growth curves, the data were log transformed to obtain a linear growth curve and tested for significance between growth rates of target and NT siRNA. The P-value is provided under the linear mixed model considering the correlation structure between repeated measurements for same subject (linear mixed model with repeated measures).
Acknowledgments
Funding for research was provided by MedImmune. We thank John Meekin and Rosa Carrasco for the RNA library, and Terry O'Day and Xin Cao for statistical analysis.
SC, EB, DT, RH, RH and ZX are employees of MedImmune. Funding for all research was provided by MedImmune.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Colpitts TL, Billing P, Granados E, Hayden M, Hodges S, Roberts L et al. Identification and immunohistochemical characterization of a mucin-like glycoprotein expressed in early stage breast carcinoma. Tumour Biol 2002; 23: 263–278. [DOI] [PubMed] [Google Scholar]
- Houghton RL, Dillon DC, Molesh DA, Zehentner BK, Xu J, Jiang J et al. Transcriptional complementarity in breast cancer: application to detection of circulating tumor cells. Mol Diagn 2001; 6: 79–91. [DOI] [PubMed] [Google Scholar]
- Miksicek RJ, Myal Y, Watson PH, Walker C, Murphy LC, Leygue E. Identification of a novel breast- and salivary gland-specific, mucin-like gene strongly expressed in normal and tumor human mammary epithelium. Cancer Res 2002; 62: 2736–2740. [PubMed] [Google Scholar]
- Skliris GP, Hube F, Gheorghiu I, Mutawe MM, Penner C, Watson PH et al. Expression of small breast epithelial mucin (SBEM) protein in tissue microarrays (TMAs) of primary invasive breast cancers. Histopathology 2008; 52: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZZ, Xie XD, Qu SX, Zheng ZD, Wang YK. Small breast epithelial mucin (SBEM) has the potential to be a marker for predicting hematogenous micrometastasis and response to neoadjuvant chemotherapy in breast cancer. Clin Exp Metastasis 2010; 27: 251–259. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu Z, Qu S, Zheng Z, Liu Y, Xie X et al. Small breast epithelial mucin tumor tissue expression is associated with increased risk of recurrence and death in triple-negative breast cancer patients. Diagn Pathol 2013; 8: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayerbes MV, Diaz-Prado S, Ayude D, Campelo RG, Iglesias P, Haz M et al. In silico and in vitro analysis of small breast epithelial mucin as a marker for bone marrow micrometastasis in breast cancer. Adv Exp Med Biol 2008; 617: 331–339. [DOI] [PubMed] [Google Scholar]
- Valladares-Ayerbes M, Iglesias-Diaz P, Diaz-Prado S, Ayude D, Medina V, Haz M et al. Diagnostic accuracy of small breast epithelial mucin mRNA as a marker for bone marrow micrometastasis in breast cancer: a pilot study. J Cancer Res Clin Oncol 2009; 135: 1185–1195. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Verduijn P, Bosma AJ, Rutgers EJ, Peterse HL, van't Veer LJ. Detection of metastases in sentinel lymph nodes of breast cancer patients by multiple mRNA markers. Br J Cancer 2004; 90: 1531–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Cao J, Yang NW, Liu JL, Zhang CM, Chen JS et al. [Expression of small breast epithelial mucin mRNA in peripheral blood of breast cancer patients and its clinical significance]. Ai Zheng 2005; 24: 842–845. [PubMed] [Google Scholar]
- Wadle A, Mischo A, Imig J, Wullner B, Hensel D, Watzig K et al. Serological identification of breast cancer-related antigens from a Saccharomyces cerevisiae surface display library. Int J Cancer 2005; 117: 104–113. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer 2011; 11: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck S, Ross JS, Royce M, McKenna EF Jr., Perou CM, Avisar E et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/- trastuzumab. Breast Cancer Res Treat 2012; 132: 781–791. [DOI] [PubMed] [Google Scholar]
- Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat 2008; 108: 191–201. [DOI] [PubMed] [Google Scholar]
- Zhao J, Pestell R, Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol Biol Cell 2001; 12: 4066–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol 1998; 143: 1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktay M, Wary KK, Dans M, Birge RB, Giancotti FG. Integrin-mediated activation of focal adhesion kinase is required for signaling to Jun NH2-terminal kinase and progression through the G1 phase of the cell cycle. J Cell Biol 1999; 145: 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cance WG, Harris JE, Iacocca MV, Roche E, Yang X, Chang J et al. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: correlation with preinvasive and invasive phenotypes. Clin Cancer Res 2000; 6: 2417–2423. [PubMed] [Google Scholar]
- Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett 2010; 289: 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Ozturk S, Thiagalingam S. Integrin signaling in mammary epithelial cells and breast cancer. ISRN Oncol 2012; 2012: 493283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci USA 2007; 104: 20302–20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Smith HW, Sanguin-Gendreau V, McDonough RV, Muller WJ. Mammary epithelial-specific disruption of c-Src impairs cell cycle progression and tumorigenesis. Proc Natl Acad Sci USA 2012; 109: 2808–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari RK, Borgen PI, Wong GY, Cordon-Cardo C, Osborne MP. HER-2/neu amplification and overexpression in primary human breast cancer is associated with early metastasis. Anticancer Res 1992; 12: 419–425. [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science (New York, NY) 1987; 235: 177–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.