Abstract
Obesity and malnutrition are associated with decreased fecundity in women. Impaired reproductive capacity in obese women is often attributed to anovulation. However, obese women with ovulatory cycles also have reduced fertility, but the etiology of their impaired reproduction is only partially understood. Accumulating evidence suggests that obesity directly impairs oocyte and embryo quality as well as endometrial receptivity. In obese women, urinary progesterone metabolite excretion is decreased, but in excess of what can be explained by suppressed gonadotropin secretion, suggesting that apart from its central effect obesity may directly affect progesterone (P4) production. These observations have led to the novel hypothesis that obesity directly affects corpus luteum (CL) function. Similarly, we hypothesize that weight loss may contribute to luteal dysfunction. Here, we propose a non-human primate model, the vervet monkey, to examine the effect of weight gain and loss on menstrual cycle parameters and CL gene expression. In this model, weight gain and loss did not significantly alter menstrual cyclicity, however, both induced alterations in the CL transcriptome. In the weight gain monkey, we observed that impaired mid-luteal P4 secretion was associated with downregulation of steroidogenic pathways in CL. Collectively, these preliminary findings support our hypothesis that weight gain and loss may contribute to CL dysfunction. The vervet model described and preliminary observations provide a basis for a larger study to address this important question. Understanding the mechanisms by which weight gain and loss contribute to reproductive dysfunction can assist in the development of targeted treatments to enhance women’s reproductive capability when it is desired.
Keywords: Corpus luteum, gene expression, microarray, obesity, vervet monkey
Introduction
Female reproductive physiology is affected by extremes in weight such as obesity and malnutrition. It is estimated that over half of reproductive age women are overweight or obese. In addition to other health problems, obesity impairs female fertility. Obese women are less likely to achieve spontaneous pregnancy than normal-weight women and have reduced chances of pregnancy with assisted reproductive technologies. In fact, obesity appears to negatively influence female reproductive function at multiple levels of the hypothalamic-pituitary-ovarian (HPO) axis, and also at the level of embryo development and the endometrium. The impact of obesity on fertility appears to be complex and many mechanistic aspects responsible for compromised fertility in obese women remain elusive.
Weight loss and its resulting negative energy balance are known as a major cause of amenorrhea, anovulation, and infertility in women. Perturbations in HPO axis signaling, including aberrant hypothalamic release of gonadotropin releasing hormone (GnRH) and pituitary luteinized hormone (LH) secretion, appear to be the leading causes for anovulation. Whether weight loss affects any other steps in the reproductive cascade remains to be investigated.
Further understanding of the mechanisms underlying impaired fertility is vital to develop interventions to mitigate obesity and underweight-associated reproductive failure. In this review and hypothesis, we first discuss the current understanding of female reproductive failure as associated with obesity and malnutrition/negative energy balance state. Additionally, we propose a novel hypothesis that metabolic changes linked to weight gain and loss directly target the corpus lutem (CL), thereby affecting its function. We describe in detail the vervets, non-human primates, as a research model to address this question and present the preliminary findings of our concept study upon which the hypothesis drives strength.
Background
Reproductive targets of obesity
HPO axis
Increased weight induces chronic oligo-anovulation that is at least partially due to perturbation of central gonadotropin secretion [Jain et al. 2007]. Anovulatory menstrual cycles are one major reason for infertility in obese women [Grodstein et al. 1994]. However, obese women with regular menstrual cycles also have reduced fecundity and experience a longer time to conception compared to normal weight women [Bolumar et al. 2000; Gesink Law et al. 2007; Polotsky et al. 2010]. The effects of increased body weight on the reproductive axis in ovulatory obese women are not completely understood, but emerging data support the involvement of both pituitary and ovarian dysfunction. In particular, decreased excretion of the urinary progesterone metabolite, pregnanediol 3-glucuronide (PDG), and reduced serum and urinary LH levels were observed in obese, eumenorrheic women compared to normal weight, ovulatory women [Santoro et al. 2004]. In addition, our group has previously studied a cohort of morbidly obese women prior to their scheduled bariatric surgery and demonstrated a greater than 50% reduction in urinary PDG and a 30% reduction in urinary LH in obese women compared to normal weight controls [Jain et al. 2007]. These results indicate that the deficit in PDG exceeds the deficit in LH, implying that, apart from its possible central effects at the hypothalamic and pituitary level, obesity may additionally have a direct impact on the ovary, and in this case, oocyte and corpus luteum function.
Oocyte and embryo quality
Women undergoing assisted reproductive technology (ART) offer a unique opportunity to study the association of obesity with reproductive outcomes. Accordingly, most human data on obesity’s impact on postovulatory events come from women undergoing ART therapies in which exogenous gonadotropins are administered to induce ovarian follicle growth, followed by egg retrieval. Obese women who undergo in vitro fertilization (IVF) using autologous oocytes have reduced clinical intrauterine pregnancy rates [Jungheim et al. 2009; Luke et al. 2011], increased miscarriage rates (Rittenberg et al., 2011a; Rittenberg et al., 2011b), and lower live birth rates [Luke et al. 2011] compared to their normal-weight peers, after controlling for age. Moreover, mature oocytes harvested from obese women have lower fertilization rates than oocytes obtained from age-matched normal-weight women [Shah et al. 2011], and furthermore, their fertilized oocytes have a lower chance of developing to top quality embryos in vitro [Metwally et al. 2007], suggesting compromised oocyte maturation and embryo development. Thus, we can assume the inferior reproductive outcomes after IVF in obese women are due to obesity related metabolic changes directly impairing oocyte quality and/or uterine receptivity.
Follicle development and oogenesis are tightly linked, with an optimal follicular micro-environment being crucial for oocyte developmental competency. Animal and translational studies have investigated the impact of adiposity on the ovarian environment. In mouse studies using the diet-induced obesity model, obesity increased the number of apoptotic ovarian follicles, and had an adverse effect on oocyte and embryo quality [Jungheim et al. 2010]. Female adiposity in the mouse has been shown to alter the intrafollicular environment with elevated levels of fatty acids and triglycerides that are detrimental to oocytes and viability of follicle supporting cells, granulosa and theca cells [Wu et al. 2010; Wu et al. 2012; Yang et al. 2012]. In an interesting translational study, Yang et al. [2012] exposed immature mouse cumulus-oocyte complexes to human lipid-rich follicular fluid and demonstrated compromised in vitro oocyte nuclear maturation, increased oocyte lipid content and induction of endoplasmic reticulum stress markers in these mouse oocytes. Collectively, the data support the notion that the follicular micro-environment determines the health of an oocyte and steroidogenic capacity of the follicle; aberrations from the optimal follicular milieu hamper oocyte developmental competency, ultimately resulting in poor embryo quality.
Endometrium
The impact of obesity on the endometrium and implantation is emerging, but less clear. A systematic review and meta-analysis published by Jungheim et al. [2013] found obese women undergoing donor oocyte recipient cycles to have comparable implantation, pregnancy, and live birth rates to normal-weight women. In contrast, Bellver et al. [2013] reported significantly reduced implantation, clinical pregnancy, and live birth rates with increasing BMI in a large number of women undergoing therapy with donor oocytes. Using previously collected data from the Society for Assisted Reproductive Technologies (SART) database, Luke et al. [2011] demonstrated that obese women using donor oocytes had similar pregnancy rates, but a lower chance of live birth than normal-weight women using donor oocytes. The expression of endometrial genes has been studied in the obese compared to normal weight women to further clarify the role of the endometrium in obesity-associated reproductive impairment [Bellver et al. 2011]. The results suggest that obesity is associated with altered expression of some genes important to endometrial function during the implantation window. Consistent with other studies, these results suggest that obesity not only adversely affects embryo quality but also influences endometrial receptivity, thus highlighting the complex pathophysiology that links obesity to reproductive dysfunction.
Reproductive dysfunction associated with weight loss
Significant weight loss, especially when associated with strenuous physical activity and/or heightened cortisol secretion in women, causes hypothalamic dysfunction, with decreased GnRH pulsatility and impaired gonadotropin secretion, subsequently leading to arrested follicle development, oligo-anovulation, and prolonged amenorrhea [Fourman and Fazeli 2015]. In extreme cases of underweight or weight loss, fertility is therefore highly unlikely and measurement of specific pathways for impaired fertility beyond the HPO axis has not been performed. Infertility therapies for women of borderline weight with functional hypothalamic amenorrhea (FHA) consist of administration of exogenous gonadotropins, both FSH and LH, to induce ovarian follicular growth. While increasing the risk of multiple gestation, this therapy is quite successful for ovulation induction in FHA women and many also achieve conception. Therefore, it is unlikely that the oocytes of the FHA women are severely affected, although this has never been rigorously investigated. While the sex-steroid producing capacity of the CL is impaired after ovulation induction in part due to decreased gonadotropin stimulation in FHA women, the direct impact of weight loss in CL functional capacity remains to be elucidated.
Corpus luteum, a novel target of weight gain and loss in reproduction
The corpus luteum is a transient endocrine gland that forms from cells remaining in the follicle after ovulation. Therefore, luteinized cells of the CL are potential targets of metabolic changes associated with obesity and perhaps also weight loss. The corpus luteum is the primary source of progesterone (P4) during the menstrual cycle and early pregnancy in primates. The mid-cycle LH surge initiates a cascade of events that culminates in ovulation and follicular rupture. After ovulation, the corpus luteum continues to secrete hormones and is therefore essential for establishment and maintenance of early pregnancy. If conception does not occur, timely regression of the CL, called luteolysis, is necessary to allow the initiation of the next ovarian cycle. The process of luteolysis involves a sequence of distinct events that first lead to a cessation of P4 synthesis (functional regression) and then culminate in the degradation and structural remodeling of the CL (structural regression). Dynamic, sequential changes in gene expression have been reported during the lifespan of the rhesus macaque CL [Bogan et al. 2009; Bogan et al. 2008] and these changes reflect the molecular functions important to normal luteal physiology during its function and regression [Bogan et al. 2008].
The regulation of CL function in primates differs from rodents and domestic animals. For example, only in primates does sensitivity to LH decrease as the CL ages [Brannian and Stouffer 1991; Cameron and Stouffer 1982; Eyster et al. 1985], and an increasing number of LH pulses or more potent signals (such as chorionic gonadotropin) are required to maintain the CL at or beyond the end of the normal luteal phase [Duffy et al. 1999]. Additionally, LH is an absolute requirement for luteinization and maintenance of P4 synthesis only in primates [Dubourdieu et al. 1991; Fraser et al. 1987]. Because of the differences between rodent and primate species, non-human primates offer a more applicable and translatable model for the study of human reproduction. Female endocrine control of reproduction exhibits many similarities to the menstrual cycle and reproductive physiology of non-human primates that are different from the estrous cycle in rodents. The vervets (Chlorocebus aethiops sabaeus) are Old World monkeys that belong to the same subfamily as macaques and baboons [Jasinska et al. 2007], and they are phylogenetically closely related to the human. In a fully pedigreed and genotyped colony of over 400 animals (the Vervet Research Colony, VRC), adult vervets have the potential to exhibit spontaneous obesity and an associated metabolic profile similar to humans. Therefore, vervets represent a particularly important animal model to elucidate the effects of weight fluctuations on endocrine target organs in primates, such as corpus luteum.
Building upon previous research, we propose a novel hypothesis that weight changes, either weight gain or loss, are linked to impaired corpus luteum development and function in primates, thereby providing one plausible mechanism by which weight changes impair fertility in women with regular menstrual cycles. As an initial step to test this hypothesis, we designed a small concept study using sexually mature vervet monkeys as a non-human primate model and examined the impact of weight changes, both gain and loss, on menstrual cycle parameters and CL gene expression. By establishing a baseline cycle length prior to dietary intervention, we aimed to target the CL collections to the mid-luteal cycle phase, when the CL is fully functional and has reached its peak progesterone-producing capacity. Additionally, we tested the feasibility of utilizing human high-density microarrays to examine weight change-induced alterations in specific CL transcripts by comparing the transcript profile of the post-intervention CL specimen to the one at baseline. Herein, we present the methods and preliminary results of our concept study formulating the hypothesis that weight changes induce changes in luteal gene expression, thereby potentially impairing CL function.
The concept study
Our concept study was specifically designed to begin to test the hypothesis that weight changes, weight gain and loss, alter menstrual cycle parameters and CL gene expression in primates. For this study, we used two sexually mature female vervets. Following the baseline morphometric evaluation and CL collection, each vervet was assigned to receive either ad libitum or caloric restriction diet for a ten-month period. Morphometric measures including weight, trunk height, and BMI were measured at baseline and at one month intervals until the conclusion of the study. Menstrual cycle length and steroid hormone levels were potential variables to change after weight gain and loss, and therefore they were carefully assessed during the study. To allow comparisons of luteal gene expressions between pre- and post-dietary intervention, the CL tissue was collected in the mid-luteal phase of the cycle: at baseline and after the 10-month intervention induced weight change. Luteal gene expressions were assessed from total RNA using human Affymetrix microarrays, followed by confirmation of the expression of select genes by quantitative real time PCR (qrtPCR) using vervet genomic primers.
Results
Morphometric measures
Two adult vervet monkeys (Chlorocebus aethiops sabaeus) were each assigned to one of the two experimental groups: Ad libitum (monkey 1030) or calorie restriction group (monkey 1031). Table 1 depicts data for body weight and body composition over the course of the study. Over 10 months, monkey 1030 gained 16% of her basal body weight (4.4 kg –> 5.1 kg) with 16% increase in her body mass index (BMI). Under calorie restriction, monkey 1031 lost 17% of her basal body weight (6.9 kg –> 5.7 kg) and her BMI decreased by 17% over the course of 10 months.
Table 1.
Body weight and composition of vervet monkeys at baseline and post-dietary intervention at 10 months.
| Ad Libitum | Caloric Restriction | ||
|---|---|---|---|
| Body weight (kg) | |||
| Baseline | 4.4 | 6.9 | |
| 10 months | 5.1 | 5.7 | |
| % change | 15.9 | −17.3 | |
|
| |||
| Body mass index (kg/m2) | |||
| Baseline | 52.7 | 77.6 | |
| 10 months | 61.1 | 64.2 | |
| % change | 15.9 | −17.3 | |
Effects of the intervention on menstrual cyclicity and serum hormone levels
Menses were recorded for 10 menstrual cycles prior to the first CL tissue collection and for up to 14 cycles following the start of ad libitum or calorie restriction intervention. Menstrual cycle length remained essentially unchanged after weight gain (M1030) when comparing the average cycle length of the cycles 1–10 to the cycles 11–24 (Figure 1). In contrast, weight loss (M1031) was associated with cycle length fluctuations with the average cycle length being 24–28 days, but with occasional long cycles lasting 34–47 days and a single short, 22 day cycle. As expected, CL collection resulted in the disruption of P4 secretion and subsequent early menstrual bleeding that started 2–5 days after CL collection.
Figure 1.
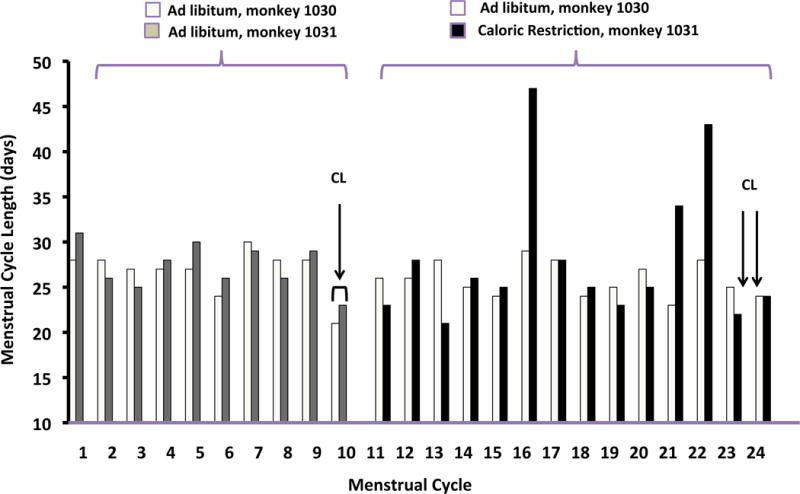
Menstrual cycle length in days for baseline cycles (cycle #1–10) and cycles during dietary intervention (cycles #11–24). Arrows indicate the timing of CL collection for each monkey during the baseline and post-dietary intervention (post-int.) cycles.
Mid-luteal serum P4 decreased with weight gain from 8.6 ng/mL on CD#20 of the baseline cycle to 2.4 ng/mL on CD#19 after weight gain (Table 2). There was no change in mid-luteal P4 level after the weight loss. There was no difference of mid-luteal serum estradiol (E2) or follicle-stimulating hormone (FSH) levels between the baseline cycle and the cycles studied after weight gain or loss.
Table 2.
Serum hormone levels drawn during the mid-luteal phase four cycles after the baseline CL biopsy and on the day of post-intervention CL collection.
| Monkey 1030 | Monkey 1030 | Monkey 1031 | Monkey 1031 | |
|---|---|---|---|---|
| Cycle number | #14 | #23 | #14 | #24 |
| Cycle length (days) | 25 | 24 | 26 | 22 |
| Cycle day | 20 | 19 | 22 | 21 |
| Progesterone (ng/mL) | 8.6 | 2.4 | 4.3 | 3.7 |
| Estradiol (pg/mL) | 9.71 | 11.24 | 25.1 | 19.8 |
| FSH (ng/mL) | 0.6 | 0.52 | 0.65 | 0.8 |
CL: corpus luteum; FSH: follicle-stimulating hormone.
Identification of differentially expressed luteal genes after weight changes
To establish gene expression profiles of the mid-luteal CL samples, microarray analyses were performed on the CL collected from each monkey at baseline and post-dietary intervention. To identify the most informative set of differentially expressed genes, we ranked each gene based on its fold change in expression between the baseline and after weight change specimens. Because we included only one monkey per group in this concept study, no statistical significance could be calculated. Gene expression values were compared within each monkey. After performing pair-wise comparisons, 764 individual gene transcripts were differentially expressed by 3-fold or greater difference between the CL specimens at baseline and after weight gain (M1030). Of these 764 transcripts, 327 were upregulated and 437 downregulated. There were only 64 genes showing 3-fold or greater difference between the CL samples at baseline and after weight loss. Therefore, for the weight loss monkey, we utilized a cutoff level of 2-fold or greater difference in transcript abundance. In this fashion, we identified 317 unique transcripts that were differentially expressed between the CL at baseline and after weight loss (M1031), with 150 showing increased and 167 decreased transcript abundance. The transcriptome profiles for the CL at baseline and after the weight changes are illustrated for each monkey in the heatmap (Figure 2). The gene expression profiles suggest that weight gain induces more changes in the CL transcriptome, altering a greater number of CL genes expressions than did weight loss.
Figure 2.
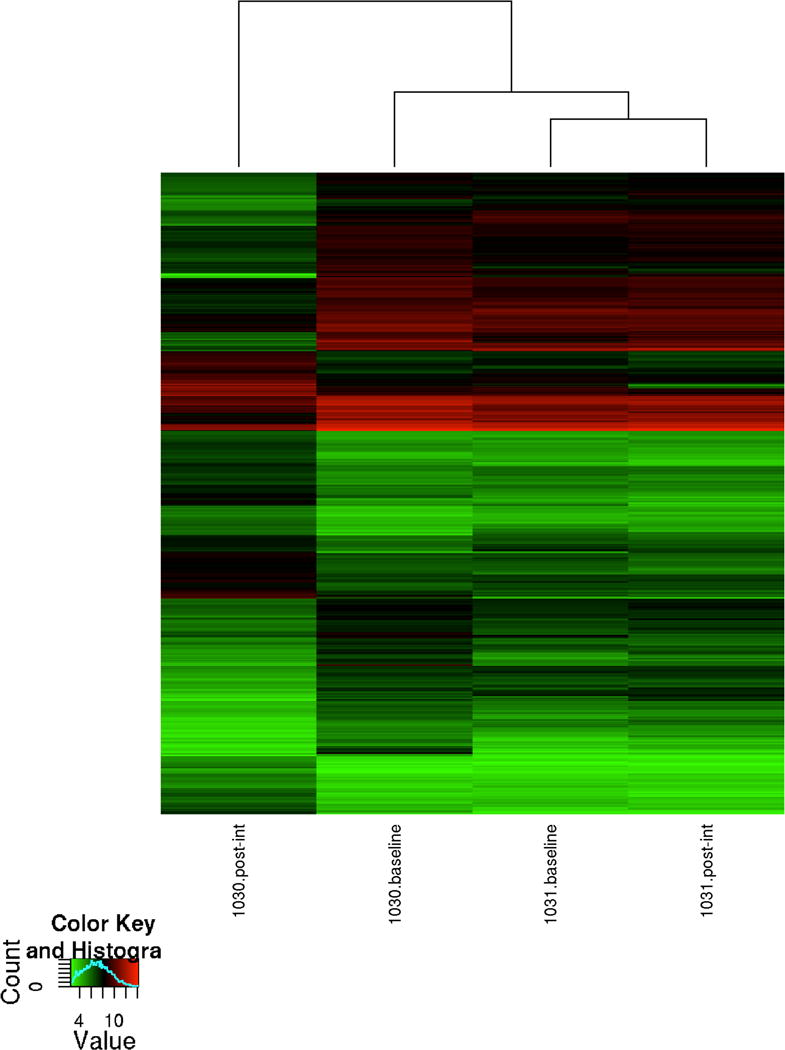
Dendrogram and unsupervised hierarchical clustering of the array data. Data from differentially expressed mRNA transcripts showing a 3-fold or greater change in expression for the monkey 1030 and a 2-fold or greater change in expression for the monkey 1031. The top dendrogram shows the relationship between the samples based on gene expression patterns. Hierarchical clustering indicates that the CL transcriptome after weight gain shows the greatest number of expression changes compared to the other CL transcriptomes. The expression intensity of each gene varies from red (high) to green (low).
Weight gain associated changes in gene expression affect LH induced P4 secretion and angiogenesis
We analyzed the transcriptome of the CL after weight gain compared to the CL transcriptome at baseline using the DAVID bioinformatics program. The expression of several genes known to be involved in LH-induced P4 biosynthesis was decreased. Key components of the steroid hormone biosynthetic pathway, including LH receptor (LHCGR), cholesterol side-chain cleavage enzyme (CYP11A1), and 3 beta-hydroxysteroid dehydrogenase type II (HSD3B2) were decreased in transcript abundance (Table 3). We did not detect the expression of vervet aromatase by the human Affymetrix array. Additionally, we observed reduced expression of several genes involved in LH-regulated cholesterol transport in luteinized granulosa cells (GCs), and these genes include STAR, LDL receptor (LDLR), and scavenger receptor B1 (SCARB1) (Table 3). The LDL receptor and LDL complex mediate the entry of low-density lipoproteins into cells via receptor-mediated endocytosis; whereas high-density lipoproteins interact with membrane protein, SCARB1 to allow the uptake of cholesterol esters from HDL complexes into cells [Connelly and Williams 2004]. In contrast, the expression of luteal genes encoding for proteins of cholesterol efflux, including members of ATP-binding cassette subfamilies, ABCA1 and ABCG1 [Bogan and Hennebold 2010], were unaffected by weight gain. Of the cholesterol acceptor molecules, apolipoprotein A1 (APOA1) transcript abundance was reduced in the CL after weight gain. Furthermore, in the post-weight gain CL we observed a decrease in the expression of genes involved in de novo cholesterol biosynthesis, including DHCR24 and the rate-limiting enzyme in cholesterol biosynthesis, HMGCR. Collectively, these findings indicate suppression of hormone biosynthesis in the CL after weight gain.
Table 3.
Differentially expressed gene transcripts regulate corpus luteum hormone biosynthesis and lutenized granulosa cell survival after weight gain.
| Gene symbol | Fold change by the array data |
|---|---|
| LHCGR | −8.4 |
| LDLR | −10.1 |
| SCARB1 | −10.3 |
| STAR | −11.3 |
| CYP11A1 | −4.8 |
| HSD3B2 | −29 |
| AREG | −585 |
| EREG | −74 |
Similar findings were obtained when differentially expressed genes were uploaded to the Ingenuity Pathway Analysis (IPA) database to explore enriched biological functions (Figure 3A). The molecular and cellular functions that were suppressed in the CL after weight gain included cellular movement, cellular growth and proliferation, and lipid metabolism. Particularly, the molecular signals affecting decreased cholesterol influx as a sign of reduced cholesterol intake to cells and reduced expression of enzymes directing P4 production were in accordance with the diminished steroid biosynthesis of the CL after weight gain.
Figure 3.
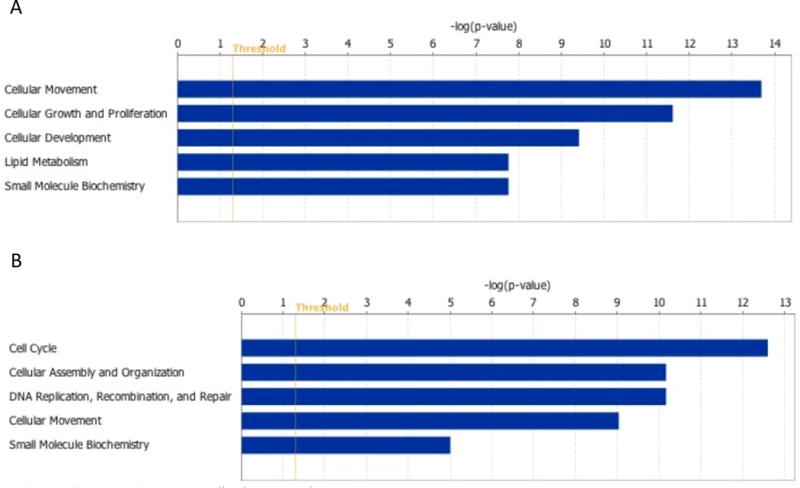
Molecular and cellular functions enriched in corpus luteums (CLs) after dietary intervention compared to CL at baseline. (A) Molecular and cellular functions enriched among the downregulated CL transcripts of the weight gain monkey. Transcript regulating lipid transport and metabolism are downregulated in CL after weight gain. Suppression of small molecular biochemistry involved functions related to steroid hormone biosynthesis. Also, growth and proliferation of endothelial cells is suppressed in CL after weight gain. (B) Molecular and cellular functions enriched among the upregulated CL transcripts of the weight loss monkey. Function related cell cycle regulation and mitosis are upregulated in the CL after weight loss. Threshold indicates right-tailed Fisher exact test p=0.05.
The Ingenuity category of cellular development as well as cellular growth and proliferation involved functions related to endothelial cell proliferation, tubulogenesis and branching, and the expression of genes, such as VEGFA, VEGFC, VEGFR1, and TIMP metallopeptidase inhibitor 1 (TIMP1), were all observed to be reduced (Figure 3). These results suggest suppression of angiogenesis in the mid-luteal CL after weight gain.
Next, we compared our data on the differentially expressed CL genes after weight gain to genes that are not involved in steroid hormone biosynthesis or cholesterol transport/metabolism but whose expression has been detailed across luteal phases in non-human primates. In our study, two such genes, amphiregulin (AREG) and epiregulin (EREG), showed the greatest level of reduction in CL transcript abundance after weight gain. Other LH downstream effector genes with greater than 3-fold reduction in expression in the CL after weight gain were ADAMTS1 and TIMP3. The expression of ADAMTS1 and TIMP3 has been previously detailed in the macaque CL across the luteal phase [Young et al. 2004]. ADAMTS1 expression at the mRNA and protein level has been demonstrated to predominate during CL formation and TIMP3 expression during CL regression in macaque [Young et al. 2004]. In the present study, we observed reduced expression of both ADAMTS1 and TIMP3 in the CL after weight gain by the array data. Similarly, the expression of matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMPs) exhibit dynamic changes during primate luteal regression [Young et al. 2002]. Matrix metallopeptidase 9 (MMP9) expression is low in the early luteal stage but increases in the very late CL; whereas TIMP1 is expressed at high levels in the CL until the very late stage [Young et al. 2002]. We found 4.6-fold reduction of mid-luteal TIMP1 and increased MMP9 expression by 3.7-fold in the CL after weight gain, indicating expression patterns more similar to the late luteal phase than mid-luteal phase CL. In contrast, TIMP2 and MMP2 showed no difference in transcript abundance in the CL after the weight gain.
We found that few genes involved in the formation of reactive oxygen species (ROS) were altered by weight gain. The mRNA levels of cytochrome b-245, beta polypeptide (CYBB or NOX2), and NADH oxidase (NCF2 or NOXA2) encoding for NADPH oxidase proteins in neutrophils showed elevated expression after weight gain. The luteal expression of IgG Fc receptors IIb (FCGR2B) and IIc (FCGR2C) was increased and ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) and RAB27A was reduced in transcript abundance after the weight gain, and a similar pattern of expression has been demonstrated in the regressing macaque CL [Bishop et al. 2011].
Very few changes were noted in the expression of genes associated with prostaglandin biosynthesis, metabolism, and signaling. The expression of these genes dynamically changes during the transition from early to late and very late luteal phase in the macaque CL [Bogan et al. 2009; Bogan et al. 2008]. We observed reduction in the expression of hydroxyprostaglandin dehydrogenase (HPGD) and prostaglandin-endoperoxidase synthase 1 (PTGS1) in the CL after weight gain. Bogan et al. [2009] observed a significant decrease in HPGD mRNA but not protein level from functionally regressed to functionally late regressed macaque CL. In contrast, PTGS1 showed no expression changes from early to very late phase macaque CL [Bogan et al. 2008].
Weight loss associated changes in gene expression
The functions related to the changes in luteal gene expression after weight loss differed from those found after the weight gain. Weight loss induced the cell cycle and DNA replication and suppressed the expression of genes promoting cell cycle arrest, thus resulting in a net effect towards cell proliferation (Figure 3B). In addition, expression of molecules associated with cell differentiation and cell death were suppressed in the CL of the weight loss monkey (data not shown). Although endogenous cholesterol synthesis of the CL has been reported to be critical for maintenance of steroidogenesis under severely lipid-deprived states, we did not observe any changes in the mRNA levels of the genes participating in cholesterol synthesis in the CL of the weight loss monkey. This finding likely indicates that the diet composition and the magnitude of weight loss did not result in severe lipid depletion in the weight loss monkey.
Confirmation of microarray data
A total of 7 genes were selected for the quantification of the transcript abundance by qrtPCR to validate the array results (Figure 4). We focused on differentially expressed genes with potential biological relevance to CL function and for which vervet genomic sequence information was available. Although not statistically significant due to the sample size of one per group, qrtPCR confirmed the direction of expression data of all genes except for the gene encoding for nuclear receptor subfamily 5, group A, member 2 (NR5A2) (Figure 4A). The expression of NR5A2 showed a 3.7-fold reduction in expression after the weight gain by the array; however, up to 2-fold increase in transcript abundance was found by qrtPCR with two different PCR primer pairs. This discrepancy in the array and qrtPCR results may simply reflect sequence differences between the human and vervet NR5A2 gene because we used human arrays but validated the data using qrtPCR primers based on vervet sequences.
Figure 4.
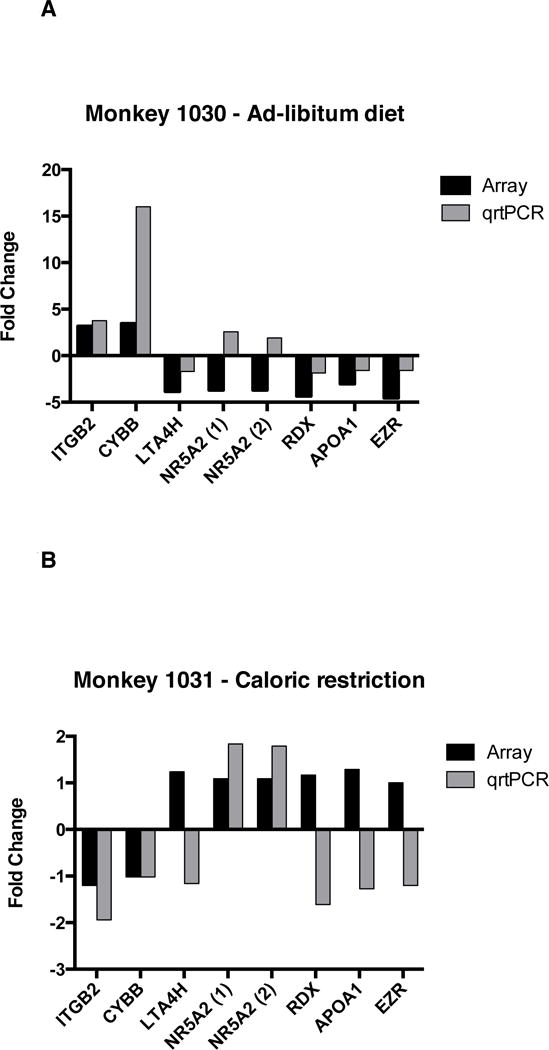
Quantitative real-time PCR confirms array data for select transcripts. Real-time primers were designed using vervet genomic sequences. All samples were normalized to the housekeeping genes, GAPDH and β-actin. Data shown indicates the relative expression of the post-intervention CL sample with respect to the pre-intervention sample for each transcript from microarray (gray bar) and qrtPCR (black bar). (A) Data for the weight gain vervet monkey and (B) for the weight loss vervet monkey. ITGB2: integrin B2; CYBB: cytochrome b-245, beta polypeptide; APOA1: apolipoprotein A; LTA4H: leukotriene A4 hydrolase; RDX: radixin; EZR: ezrin.
In addition to the data confirmation, we also studied the transcript abundance of the same 7 genes in the CL tissue after weight loss even though they were not differentially expressed by the array data (Figure 4B). Interestingly, the integrin beta 2 (ITGB2) transcript abundance by qrtPCR decreased by 2-fold after weight loss, thus showing an expression pattern opposite to that in the CL after weight gain. The expression of NR5A2 was increased almost by 2-fold after weight loss by qrtPCR, although the array data showed no change in expression between the baseline and post-intervention CL samples.
Hypothesis
The direct impact of weight changes on primate CL function remains unknown; however, disruption of normal CL function induced by weight gain and loss is a plausible mechanism by which reproductive function is compromised in women and thus warrants investigation. So far, research studying the influences of weight gain or loss on reproductive function has mainly focused on the effects of weight changes at the level of hypothalamus/pituitary and oocytes. Obesity has been shown to impair reproductive outcomes after IVF and is suggested to have a direct impact on the ovary and endometrium. Herein, we hypothesize that metabolic changes associated with weight changes can impair CL development and function. Our small concept study was intended to specifically design a non-human primate model to examine the impact of weight gain and loss on menstrual cycle parameters and the gene expression profile of the CL of cycling vervet monkeys. In this model, we used sexually mature female vervets that were exposed to dietary intervention with either an ad libitum diet or calorie restriction that caused weight gain or loss over time, respectively. The preliminary data suggests that weight gain and loss induce alterations in the CL transcriptome of the studied vervets and that these transcript changes may be associated with impaired CL function supporting our hypothesis. As described, weight gain affected pathways involved in steroid hormone biosynthesis, which was concomitant with reduced mid-luteal P4 secretion observed in the study vervet monkey. In contrast, the gene expression changes of CL after weight loss may point to the perturbation in the CL development. As ovarian intra-follicular environment has such a critical influence on follicular development and its supporting cell viability, we can speculate that weight changes and related metabolic changes may alter follicular environment and thereby not only compromise oocyte health but also result in dysfunctional CL cells.
Because the sequencing effort of the vervet genome [Warren et al. 2015] was not completed at the time of the study, we were unable to use vervet gene expression arrays. In this concept study, we successfully employed human high-density arrays to establish the vervet CL gene expression profiles and indeed obtained vervet CL transcriptome profiles that represent molecular markers of several luteal functions, including P4 biosynthesis and CL regression. The preliminary results indicate that the correct tissue material was biopsied from the vervet ovary by the technique and timing described. Quantification of mRNA levels by qrtPCR with PCR primers of the vervet sequences validated our array data for all except one gene transcript. Collectively, these observations confirmed important aspects of our concept study: 1) successful collection of vervet CL, 2) feasibility of profiling vervet tissue using human arrays, and 3) improved accuracy of data validation by using primers for qrtPCR based on available vervet genomic sequences.
The preliminary data of our small study indicates that while weight gain impaired mid-luteal P4 production, the menstrual cycle length remained unchanged in the weight gain monkey. In contrast, weight loss resulted in cycle length variability and this monkey experienced both longer and shorter menstrual cycles during the calorie restriction compared to the baseline cycles. Previously, Lujan et al. [2005] have demonstrated that chronic caloric restriction inhibits tonic gonadotropin secretion, attenuates the LH and FSH surge, and results in anovulatory cycles in rhesus monkeys. They also reported a significant increase in cortisol secretion in the calorically restricted rhesus monkeys, reflecting heightened activation of the HPA axis [Lujan et al. 2005]. Notably, these endocrine findings are very similar to human models of chronic energy restriction, such as anorexia nervosa and exercise-induced hypothalamic amenorrhea. Although we did not measure cortisol or gonadotropin secretion in this study, the cycle length variability observed in the weight loss monkey was most likely due to mild impairment in GnRH and gonadotropin secretion, which in turn led to occasional delayed follicular development and prolonged cycle length. However, most cycles of the weight loss vervet monkey remained ovulatory, as evidenced by the overall normal cycle length and appropriate mid-luteal P4 level. The level of caloric restriction we employed led to significant weight loss but probably only modest negative energy balance and therefore this monkey experienced cycle irregularity but did not become amenorrheic.
The comparisons of the expression data between the weight gain and loss vervet pointed to a difference with weight gain inducing changes in a greater number of CL transcripts than did weight loss. Of note is that the CL gene expression changes are based on observations of only one vervet in weight gain and loss group, and therefore no statistically significant results were obtained. After weight gain, transcript abundance of genes encoding for proteins essential for progesterone hormone biosynthesis, cholesterol transport into cells, and luteinized GC survival were all decreased in the mid-luteal phase CL. Thus, in our weight gain monkey model, the mid-luteal transcriptome of this vervet CL suggests a dysfunctional CL with impaired progesterone hormone biosynthesis. Consistent with these results, the mid-luteal phase P4 secretion was also decreased in this vervet. The expression profile of the vervet CL after weight gain showed many expression changes similar to the rhesus macaque CL of the late luteal phase [Bogan et al. 2008]. These findings suggest that after weight gain, the vervet mid-luteal CL may undergo functional but not structural regression because P4 secretion is impaired, yet the cycle length remains unchanged. Our observation raises the hypothesis that weight gain may induce premature functional regression of the mid-luteal CL, when its P4 secretion is expected to be at its peak to support potential conception and the consequent functional changes may result in impaired reproductive capacity.
As the factors initiating the process of luteolysis in primates remain unknown, they cannot be directly tested. There appears to be a decrease in responsiveness of the primate CL to LH as it ages [Brannian and Stouffer 1991; Cameron and Stouffer 1982; Eyster et al. 1985]. Luteinizing hormone/chorionic gonadotropin receptor (LHCGR) expression at the mRNA level is highest in the mid to late luteal phase in rhesus macaque [Bogan et al. 2008], suggesting that a decrease in LH responsiveness is not due to a reduced number of receptors in primates. Therefore, other factors may be involved and it has been suggested that they may be intraovarian of origin [Bogan and Hennebold 2010; Bogan et al. 2009]. We speculate that weight gain induces or suppresses molecular factors that negatively impact CL gene expression and disturb CL cellular homeostasis, thereby contributing to CL dysfunction (Figure 5). Studies are needed to determine at which stage the gene expression changes start to occur in the CL after weight gain. Collection of vervet CLs at different stages of luteal lifespan will be important to characterize if there are transcript alterations that occur early during the lifespan of the CL upon weight gain. Another possibility is that the weight gain impairs early follicular development, in particular granulosa cell function. Altered GC competence may translate to poor quality CL that is prone to undergo early luteolysis or development of a CL with normal lifespan but impaired P4 hormone secretion. Indeed, we observed decreased expression of angiogenic factors that can potentially represent poorly developed CL. Interestingly, studies in mice exposed to a high-fat diet have shown that obesity impairs ovulation, oocyte quality, and in vitro fertilization by causing lipotoxicity in cumulus-oozyte complexes (COC) and GCs through induction of endoplasmic reticulum (ER) stress response and subsequent GC apoptosis [Wu et al. 2010]. Similarly, compared to normal weight women, luteinized GCs from obese women showed increased expression of ER stress markers [Wu et al. 2010]. Such ER stress response pathways have been shown to be inducible by the single fatty acid, palmitic acid, during in vitro maturation of mouse COC, and interestingly, this effect was completely reversed by the ER stress inhibitor [Wu et al. 2012]. Collectively, these data suggest that in obesity, factors such as free fatty acids, activate ER stress response and lipotoxicity in ovarian cells with deleterious effects on oocyte developmental potential [Wu et al. 2010; Wu et al. 2012]. In addition to affecting COC, these fatty acids could potentially perturb homeostasis in luteinized GCs of the CL, contributing to its dysfunction after weight gain.
Figure 5.
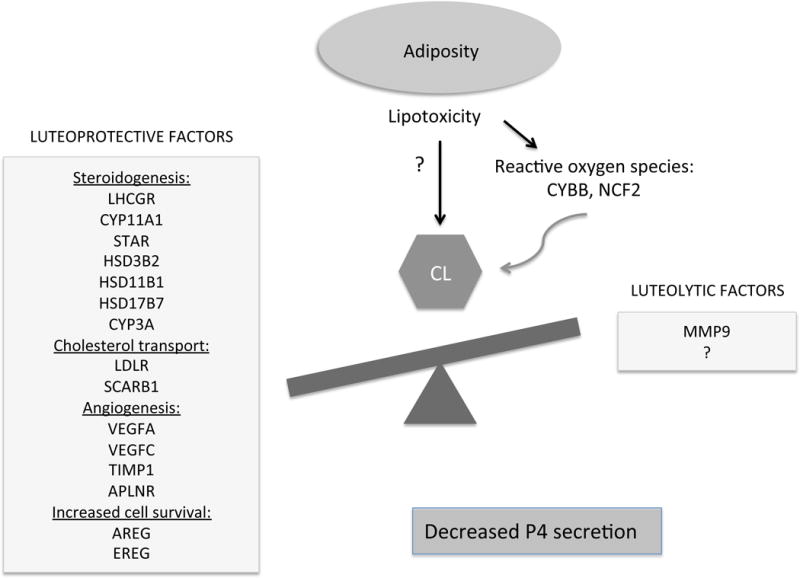
Schematic of the proposed impact of increased adiposity on CL function. After weight gain the mid-luteal vervet CL showed decreased expression of the genes involved in P4 steroidogenesis, cholesterol transport, angiogenesis, and luteinized GC survival. In this model, we hypothesize that adiposity and associated lipotoxic changes impair normal cellular homeostasis of the CL, resulting in its premature functional regression and decreased P4 secretion. The exact mechanisms by which lipotoxicity leads to the CL dysfunction remains to be investigated. Reactive oxygen species may play a role in this process.
Local P4 action is required for luteal development and its role as a potent promoter of CL survival has been demonstrated. Progesterone receptor (PGR) antagonists induce increased caspase production, DNA fragmentation, and apoptotic cell death in luteinized granulosa cells [Svensson et al. 2001]. Recent studies indicate that P4 dependent granulosa cell survival is mediated by the induction of EGF family members, including EGF-like ligands, AREG and EREG. In rhesus and human cultured GCs, LH stimulation induced mRNA expression for AREG and EREG [Motola et al. 2008; Puttabyatappa et al. 2013], thus exhibiting a temporal expression pattern coinciding with P4 and PGR. Further, P4 was shown to promote the expression of AREG and EREG in macaque GCs [Puttabyatappa et al. 2013]. Additionally, these EGF-like ligands can partially prevent RU-486-induced cell death of cultured human luteinized GCs, implicating their importance as mediators of P4’s pro-survival actions [Puttabyatappa et al. 2013]. Based on the currently available primate data, we hypothesize that the decreased mRNA level of AREG and EREG observed in our vervet CL after weight gain is likely due to reduced luteal P4 synthesis rather than a result of negative transcriptional regulation in response to weight gain. However, further studies are required to investigate this possibility.
We also observed elevation in the luteal expression of CYBB and NCF2 in the weight gain monkey. CYBB and NCF2 are genes that encode for NADPH oxidase proteins and participate in the production of reactive oxygen species (ROS) in neutrophils. Interestingly, the expression of these oxidases has been reported to increase during GnRH antagonist (Antide) induced luteal regression in the macaque CL [Bishop et al. 2009]. While ROS are formed periodically within the ovary as by-products of normal steroid synthesis during the follicular and luteal phase of the rat CL [Behrman et al. 2001], they have also been implicated to influence luteal P4 production [Carlson et al. 1995; Gatzuli et al. 1991]. Concurrently, a parallel increase in ovarian antioxidant defenses is necessary to preserve the CL activity in the luteal phase [Rizzo et al. 2012]. Studies in rats have provided substantial evidence that ROS play an important role in the regression of the CL [Behrman and Aten 1991; Gatzuli et al. 1991; Musicki et al. 1994; Sawada and Carlson 1989] and regulate the lifespan of the CL. Besides the physiological functions exerted by ROS, in high concentration they can have potential negative effects. In cells, lipotoxicity occurs through the accumulation of ROS, which induces ER stress, unfolded protein response and cell death [Borradaile et al. 2006]. If weight gain can induce excess ROS production by neutrophils in vervet CL, as indicated by the increased mRNA levels of CYBB and NCF2, the elevated ROS presence may cause oxidative stress and thereby potential damage to the CL cells. Alternatively, accumulation of ROS may provide a potential mechanism by which weight gain can induce premature luteolysis.
Herein, we have examined changes on the gene expression profiles of whole CL homogenates in response to changes in adiposity. Due to limitations of our study size and the array, we have unlikely identified all expressed genes and this should be addressed in a future study. Moreover, the corpus luteum comprises several cellular components, including large luteal cells, small luteal cells (stromal cells), and vessels, each with distinct gene expression patterns reflecting cell-type specific functions. Accordingly, in future studies, it will be important to focus on isolated cell populations to elucidate the cell-specific CL functions in primates. Also, the timing of vervet CL collections can be improved by monitoring daily serum E2 levels to detect the late follicular phase rise and subsequent drop as a marker of the luteal transition [Kundu et al. 2013].
The main limitation of this concept study is the small sample size, and therefore the altered gene expressions after the weight changes could be a result of random variation and cannot be generalized. Nevertheless, our findings are intriguing and warrant investigation in larger groups of non-human primates. This study also allowed us to establish protocols in which caloric restriction and ad libitum diet led to reasonable weight loss and gain in vervets, respectively. Another limitation of this study is the weight difference of the two study vervets at baseline that may represent a confounding factor (difference in the amount of adipose tissue), although this effect may have been partially reduced as each vervet served as its own control. Admittedly, selection of animals with similar baseline weight and body composition is critical when planning future studies.
In conclusion, we established study protocols in which caloric restriction and ad libitum diet led to reasonable weight loss and gain in non-human primates, vervet monkeys. Intriguingly, we found that weight fluctuations induce transcriptomic alterations in the mid-luteal CL collected from vervet monkeys. While weight gain did not alter menstrual cycle length in this model, the preliminary data supports impairment in mid-luteal P4 secretion and reduced expression of several genes involved in P4 hormone biosynthesis and cholesterol metabolism of the CL. These preliminary results strengthen our hypothesis that CL is a potential target organ of adiposity and weight loss in primates. To confirm these intriguing preliminary findings and ultimately test the novel hypothesis, future studies need to be carried out in a larger group of vervet monkeys. The described vervet model has proven feasible to test this hypothesis. If CL indeed is found to be a direct target of the metabolic changes induced by weight gain and loss and specific markers responsible for CL dysfunction can be defined, these data can be utilized for the development of targeted therapies for women desiring conception or even for the development of novel methods of contraception. Furthermore, we propose that the vervet model of weight gain and loss is suitable for studies investigating effects of weight gain and loss on metabolic changes and other reproductive target organs, such as endometrium.
Materials and methods
Ethics
The Wake Forest University Primate Center (WFRC) Friedberg Campus is an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)- accredited facility and all housing is AAALAC- and FDA- approved. AAALAC provided approval for this study protocol (Assurance number A3391-01). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the WFRC Animal Care and Use Committee. All policies and procedures were conducted in accordance with state and federal laws, the National Research Council (NRC) publication Guide for the Care and Use of Laboratory Animals, and regulations and guidelines established by the Wake Forest University Animal Care and Use Committee. No adverse events occurred. After the study, all animals were examined and released back into the colony.
Animal protocols and study design
Two adult female vervet monkeys 16 years of age from the Vervet Research Colony (VRC) at the Wake Forest University Primate Center (WFRC, Winston-Salem, NC) were used for this study. The monkeys were housed at Wake Forest School of Medicine Primate Center/Center for Comparative Medicine and Research/Friedberg Campus (Winston, Salem). The animals were pair-housed indoors in a climate controlled, temperature and humidity monitored room and building, They were exposed to a 12 h light (0600–1800 h) and 12 h dark (1800–0600 h) schedule. Both monkeys were within normal weight range for this species and were sexually mature at study entry [Kavanagh et al. 2007]. Baseline characteristics of the monkeys are depicted in Table 4. Prior to the intervention, the monkeys were transitioned from a commercial diet (Purina Monkey Chow) to a semi-purified, nutritionally complete, low cholesterol, low fat, and high fiber diet (similar to monkey chow but without isoflavones, Supplemental Table 1) prepared at the Wake Forest Diet Laboratory.
Table 4.
Baseline characteristics of vervet monkeys.
| Monkey ID | Age (years) | Weight (kg) | Dietary Intervention |
|---|---|---|---|
| 1030 | 16 | 4.4 | Ad libitum |
| 1031 | 16 | 6.9 | Caloric restriction |
Each monkey was assigned to one of the two experimental groups: Ad libitum (monkey 1030) or calorie restriction group (monkey 1031). Each vervet served as its own control to measure the effect of weight change within each animal. The ad libitum fed monkey received 120 calories per kg body weight plus 10% for waste. Over a 10 month period, calories were reduced ~8% per month for the calorically restricted monkey until 25% calorie restriction from the baseline feeding was obtained. Low calorie food enrichment (crystal light ice treats, carrots, celery, popcorn, etc.) was provided to both monkeys three times per week. The monkeys had unrestricted access to water and exercise.
Morphometric measures
At baseline and monthly throughout the study, monkeys were weighed (body weight, kg), trunk length was measured, and BMI was calculated as body weight divided by trunk length (m2) as suggested for this species [Jen et al. 1985].
Menstrual cyclicity
Menses were determined using daily vaginal swabbing as previously published for vervets [Kundu et al. 2013] and macaques [Adams et al. 1985]. Monkeys were trained to present for vaginal swabbing using positive reinforcement in the form of food rewards. Once trained, a cotton swab was inserted into the vagina to the level of cervix, withdrawn, and assessed for the presence of blood that was recorded on a scale of 1 to 3 with 1 indicating scant, 2 moderate, and 3 heavy bleeding. The first day of bleeding was counted as day 1 of the menstrual cycle. Cycle length and menses were determined as number of days for each cycle; both monkeys were monitored for 24 cycles.
Serum hormone measures
Estradiol, P4, and FSH levels were measured during two menstrual cycles: four cycles after the baseline CL collection and on the day of the post-intervention CL collection. E2 was measured from serum using a modification of a commercially available radioimmunoassay (RIA) from Diagnostic Products Corporation [Pazol et al. 2004]. Intra-assay and inter-assay coefficient of variations (CV) were <4% and 10%, respectively. P4 levels were measured by using tritiated steroids in a RIA procedure [Adams et al. 1985]. Intra-assay and inter-assay CVs were 2.7–8.8% and 3.9–8.7%, respectively. Serum was sent to the Biomarkers Core Laboratory (Yerkes National Primate Research Center, Atlanta, GA, USA) for FSH measurements using RIA and macaque primary antibodies. The intra-assay CV was 15.43% at 7.67 ng/mL and the inter-assay CV was 11.98% at 7.13 ng/mL.
Corpus luteum collection
Corpus luteum tissue was collected via laparotomy. Prior to the surgical procedure, the monkeys were pre-medicated with atropine (0.03 mg/kg), sedated with ketamine HCl (15 mg/kg), followed by anesthetization with 3–5% isoflurane gas, and then intubated. Using aseptic surgical technique, a midline vertical abdominal incision was made to allow visualization of both ovaries. The ovary containing CL was stabilized and a small linear incision was made in the ovarian tunica overlying the CL. The CL was then dissected bluntly off from the ovarian tissue. The ovarian tunica was re-approximated with interrupted 5–0 absorbable sutures. The abdominal incision was closed meticulously in three layers (fascia, subcutaneous tissue, and skin) and the monkeys were allowed to recover from the surgery under close observation. The corpus luteum tissue was immediately rinsed in cold sterile normal saline and then placed directly into a sterile vial containing RNAlater (Life Technologies, Grand Island, NY, USA). The vials were rotated for 3–5 times to ensure coverage of the entire tissue and placed in +4°C for 24 h prior to storage in −80°C as per the manufacturer’s instructions.
Timing of corpus luteum collection
CL tissue was collected twice from each monkey over the course of the study. The first CL collection was timed during the mid-luteal phase of the baseline cycle based on the cycle length of the previous several cycles for each monkey. The first CL collection was performed on cycle day 20 for the monkey 1030 and on cycle day 22 for the monkey 1031. The second CL collection was targeted again during the mid-cycle phase after dietary intervention and it was performed on cycle day 19 for the monkey 1030 and on cycle day 21 for the monkey 1031.
RNA extraction
Frozen CL tissue was first minced into small 2 mm pieces in a sterile dish on ice and then placed in TRIzol® Reagent (Life Technologies) followed by a brief homogenization on ice using a tissue homogenizer. Chloroform was added in 1:5 ratio and the samples were centrifuged 12,000× g for 15 min at +4°C to allow the separation of layers as per the manufacturer’s instructions. The aqueous layer containing RNA was collected and 1.25 volumes of ethanol was added to the suspension followed by gentle mixing as recommended for total RNA isolation. Total RNA was further purified using the filters and Wash I and II solutions of the MirVana miRNA isolation kit as per the manufacture’s instructions (Ambion/Life Technologies). Total RNA was recovered in nuclease-free water. The RNA concentration was measured using NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and the integrity and quality of RNA was assessed with Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Gene expression arrays
Human Affymetrix Human Genome U133 Plus 2.0 arrays were used for gene expression profiling to test their feasibility in vervet studies. Labeled cRNA probes were generated from total RNA using MessageAmp Premier RNA Amplification Kit (Ambion/Life Technologies). Briefly, 300 ng of total RNA was reverse transcribed with a T7 oligo(dT) primer bearing a T7 promoter sequence followed by in vitro transcription of the generated DNA with T7 RNA polymerase to produce anti-sense RNA copies of each mRNA. The quality of the cRNA was assessed with Agilent Bioanalyzer and nano-sizing assay (Agilent Technologies). Biotinylated cRNA probes were hybridized onto Affymetrix Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA). Hybridization, staining, and washing of the arrays were performed in the Affymetrix Fluidics station 400. Detection and quantification of target hybridization were performed using an Affymetrix GeneChip scanner 3000. Data were assessed for array performance before analysis. The probe-level intensities on each array were scaled to 500 target intensity using Microarray suite software (Affymetrix). Statistical analyses were performed using the open-source R statistical software (version 2.13) and R statistical package from Bioconductor Consortium (www.bioconductor.org) [Jain et al. 2003]. We used a custom made R library “hgu133plus2cdf_vervet.rda” (http://vervet.bmap.ucla.edu:8080/vervet-1.0/AnalysisResources/index.html) to filter the raw data and retained only transcripts with the highest homology to the vervet genomic sequences. The data were then processed using the robust multiarray average method (RMA) for background correction, normalization and log2 transformation [Irizarry et al. 2003]. Further filtering included selection of only probe sets with significant ’Detection Above Background’ in both samples. Because of the low sample size, we performed local-pooled-error test using ’LPE’ bioconductor library [Jain et al. 2003]. This method uses the pooling errors within genes and between replicate arrays for genes in which expression values are similar to estimate the significant difference. We combined the method with False Discovery Rate (FDR) correction for multiple testing and FDR was imposed at 0.05.
Quantitative real-time PCR (qrtPCR)
Complementary DNA (cDNA) was generated from two micrograms of total RNA using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) according to the manufacture’s protocol. Quantitative real-time PCR reactions were prepared in triplicate using SYBR® Green PCR master mix (Applied Biosystems/Life Technologies) according to the manufacturer’s instructions and the reactions were run on iCycler iQ™ real-time PCR Detection system (Bio-Rad). For designing qrtPCR primers, we first extracted the human target sequences covered by a probe set of genes of interest (Netaffyx query) and these human sequences were aligned to the first generation vervet genomic data assembly. The PCR primers were designed from the vervet sequences to be intron spanning and to amplify ~80–200 bp fragments (Supplemental Table 2). The expression changes for each transcript were normalized to the vervet GAPDH and ß-actin, both selected as endogenous controls, and relative expression changes were calculated using the 2 −ΔΔCT method [Livak and Schmittgen 2001].
Bioinformatics
The Database for Annotation, Visualization, and Integrated Discovery [Huang da et al. 2009a; b] (DAVID; http://abcc.ncifcrf.gov) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems) were used to identify enriched cellular and molecular functions among differentially expressed gene transcripts. In IPA, statistical significance was calculated using the right-tailed Fisher exact test and p-value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We acknowledge Margaret May Long, Dewayne Cairnes, and Lumie Benard for their technical contribution to the study.
Grant support: NIH U54 HD058155 (S.K, A.J.P. and N.S), NIH RR019963/OD010965 (S.E.A.), the Veterans Administration (S.E.A), NIH/NCRR R01RR016300 (A.J), NIH/NCRR P40RR19963 (A.J), and NIH/NINDS P30NS062691 (A.J).
Abbreviations
- CL
corpus luteum
- P4
progesterone
- E2
estradiol
- PDG
pregnanediol 3-glucoronide
- LH
luteinizing hormone
- FSH
follicle-stimulating hormone
- GnRH
gonadotrpoin releasing hormone
- BMI
body mass index
- qrtPCR
quantitative real-time PCR
- PGR
progesterone receptor
- ART
assisted reproductive technology
- IVF
in vitro fertilization
- HPO
hypothalamic-pituitary-ovarianMMPs: matrix metalloproteinases
Gene symbols
- LHGCR
LH receptor
- CYP11A1
cholesterol side-chain cleavage enzyme
- HSD3B2
3 beta-hydroxysteroid dehydrogenase type II
- STAR
steroidogenic acute regulatory protein
- LDLR
LDL receptor
- SCARB1
scavenger receptor B1
- ABCA1
ATP-binding cassette sub-family A member 1
- ABCG1
ATP-binding cassette sub-family G member 1
- APOA1
apolipoprotein A
- DHCR24
24 dehydrocholesterol reductase
- HMGCR
3-hydroxy-3-methylglytaryl-CoA reductase
- VEGFA
vascular endothelial growth factor A
- VEGFC
vascular endothelial growth factor C
- VEGFR1
vascular endothelial growth factor receptor 1
- TIMP1
TIMP metallopeptidase inhibitor 1
- AREG
amphiregulin
- EREG
epiregulin
- CEBPBA
CCAAT/enhancer binding protein alpha
- CREB3L1
cAMP responsive element binding protein 3-like 1
- ADAMTS1
ADAM metallopeptidase with thrombospodin type 1 motif; 1
- MMP9
matrix metallopeptidase 9
- CYBB or NOX2
cytochrome b-245; beta polypeptide
- NCF2 or NOXA2
NADH oxidase
- FCGR2B
of IgG Fc receptor IIb
- FCGR2C
of IgG Fc receptor IIc
- ENPP1
ectonucleotide pyrophosphatase/phosphodiesterase 1
- RAB27A
RAB27A member RAS oncofamily
- HPGD
hydroxyprostaglandin dehydrogenase
- PTGS1
prostaglandin-endoperoxidase synthase 1
- ITGB2
integrin B2
- LTA4H
leukotriene A4 hydrolase
- RDX
radixin
- EZR
ezrin
Footnotes
Declaration of interest
The authors report no declarations of interest.
Presented in part at the Society of Gynecological Investigation 60th Annual Meeting, March 20–23, 2013, Orlando, Florida (Abstract F205).
Notes on contributors
Conceived and designed the experiments: SK, AJP, APB, NS, SEA; Performed the experiments: SK, JC, APB, AJ; Analyzed the data: SK, AJP, APB, TP, NS, SEA, JC; Contributed reagents/materials/analysis tools: AJ, TP; Wrote the manuscript: SK, AJP, APB, NS, SEA; All authors approved revisions and the final paper.
References
- Adams MR, Kaplan JR, Koritnik DR. Psychosocial influences on ovarian endocrine and ovulatory function in Macaca fascicularis. Physiol Behav. 1985;35:935–940. doi: 10.1016/0031-9384(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Behrman HR, Aten RF. Evidence that hydrogen peroxide blocks hormone-sensitive cholesterol transport into mitochondria of rat luteal cells. Endocrinology. 1991;128:2958–2966. doi: 10.1210/endo-128-6-2958. [DOI] [PubMed] [Google Scholar]
- Behrman HR, Kodaman PH, Preston SL, Gao S. Oxidative stress and the ovary. J Soc Gynecol Investig. 2001;8:S40–42. doi: 10.1016/s1071-5576(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Bellver J, Martinez-Conejero JA, Labarta E, Alama P, Melo MA, Remohi J, et al. Endometrial gene expression in the window of implantation is altered in obese women especially in association with polycystic ovary syndrome. Fertil Steril. 2011;95:2335–2341. 2341 e2331–2338. doi: 10.1016/j.fertnstert.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Bellver J, Pellicer A, Garcia-Velasco JA, Ballesteros A, Remohi J, Meseguer M. Obesity reduces uterine receptivity: clinical experience from 9,587 first cycles of ovum donation with normal weight donors. Fertil Steril. 2013;100:1050–1058. doi: 10.1016/j.fertnstert.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Bishop CV, Bogan RL, Hennebold JD, Stouffer RL. Analysis of microarray data from the macaque corpus luteum; the search for common themes in primate luteal regression. Mol Hum Reprod. 2011;17:143–151. doi: 10.1093/molehr/gaq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CV, Hennebold JD, Stouffer RL. The effects of luteinizing hormone ablation/replacement versus steroid ablation/replacement on gene expression in the primate corpus luteum. Mol Hum Reprod. 2009;15:181–193. doi: 10.1093/molehr/gap005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Hennebold JD. The reverse cholesterol transport system as a potential mediator of luteolysis in the primate corpus luteum. Reproduction. 2010;139:163–176. doi: 10.1530/REP-09-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Hennebold JD. Dynamic changes in gene expression that occur during the period of spontaneous functional regression in the rhesus macaque corpus luteum. Endocrinology. 2009;150:1521–1529. doi: 10.1210/en.2008-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Systematic determination of differential gene expression in the primate corpus luteum during the luteal phase of the menstrual cycle. Mol Endocrinol. 2008;22:1260–1273. doi: 10.1210/me.2007-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolumar F, Olsen J, Rebagliato M, Saez-Lloret I, Bisanti L. Body mass index and delayed conception: a European Multicenter Study on Infertility and Subfecundity. Am J Epidemiol. 2000;151:1072–1079. doi: 10.1093/oxfordjournals.aje.a010150. [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, Schaffer JE. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res. 2006;47:2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- Brannian JD, Stouffer RL. Progesterone production by monkey luteal cell subpopulations at different stages of the menstrual cycle: changes in agonist responsiveness. Biol Reprod. 1991;44:141–149. doi: 10.1095/biolreprod44.1.141. [DOI] [PubMed] [Google Scholar]
- Cameron JL, Stouffer RL. Gonadotropin receptors of the primate corpus luteum. II. Changes in available luteinizing hormone- and chorionic gonadotropin-binding sites in macaque luteal membranes during the nonfertile menstrual cycle. Endocrinology. 1982;110:2068–2073. doi: 10.1210/endo-110-6-2068. [DOI] [PubMed] [Google Scholar]
- Carlson JC, Sawada M, Boone DL, Stauffer JM. Stimulation of progesterone secretion in dispersed cells of rat corpora lutea by antioxidants. Steroids. 1995;60:272–276. doi: 10.1016/0039-128x(94)00053-f. [DOI] [PubMed] [Google Scholar]
- Connelly MA, Williams DL. Scavenger receptor BI: a scavenger receptor with a mission to transport high density lipoprotein lipids. Curr Opin Lipidol. 2004;15:287–295. doi: 10.1097/00041433-200406000-00008. [DOI] [PubMed] [Google Scholar]
- Dubourdieu S, Charbonnel B, Massai MR, Marraoui J, Spitz I, Bouchard P. Suppression of corpus luteum function by the gonadotropin-releasing hormone antagonist Nal-Glu: effect of the dose and timing of human chorionic gonadotropin administration. Fertil Steril. 1991;56:440–445. doi: 10.1016/s0015-0282(16)54537-3. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Stewart DR, Stouffer RL. Titrating luteinizing hormone replacement to sustain the structure and function of the corpus luteum after gonadotropin-releasing hormone antagonist treatment in rhesus monkeys. J Clin Endocrinol Metab. 1999;84:342–349. doi: 10.1210/jcem.84.1.5362. [DOI] [PubMed] [Google Scholar]
- Eyster KM, Ottobre JS, Stouffer RL. Adenylate cyclase in the corpus luteum of the rhesus monkey. III. Changes in basal and gonadotropin-sensitive activities during the luteal phase of the menstrual cycle. Endocrinology. 1985;117:1571–1577. doi: 10.1210/endo-117-4-1571. [DOI] [PubMed] [Google Scholar]
- Fourman LT, Fazeli PK. Neuroendocrine causes of amenorrhea–an update. J Clin Endocrinol Metab. 2015;100:812–824. doi: 10.1210/jc.2014-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HM, Nestor JJ, Jr, Vickery BH. Suppression of luteal function by a luteinizing hormone-releasing hormone antagonist during the early luteal phase in the stumptailed macaque monkey and the effects of subsequent administration of human chorionic gonadotropin. Endocrinology. 1987;121:612–618. doi: 10.1210/endo-121-2-612. [DOI] [PubMed] [Google Scholar]
- Gatzuli E, Aten RF, Behrman HR. Inhibition of gonadotropin action and progesterone synthesis by xanthine oxidase in rat luteal cells. Endocrinology. 1991;128:2253–2258. doi: 10.1210/endo-128-5-2253. [DOI] [PubMed] [Google Scholar]
- Gesink Law DC, Maclehose RF, Longnecker MP. Obesity and time to pregnancy. Hum Reprod. 2007;22:414–420. doi: 10.1093/humrep/del400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodstein F, Goldman MB, Cramer DW. Body mass index and ovulatory infertility. Epidemiology. 1994;5:247–250. doi: 10.1097/00001648-199403000-00016. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Polotsky AJ, Rochester D, Berga SL, Loucks T, Zeitlian G, et al. Pulsatile luteinizing hormone amplitude and progesterone metabolite excretion are reduced in obese women. J Clin Endocrinol Metab. 2007;92:2468–2473. doi: 10.1210/jc.2006-2274. [DOI] [PubMed] [Google Scholar]
- Jain N, Thatte J, Braciale T, Ley K, O’Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19:1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Levinson M, Slaten E, Lee O, Sobel E, et al. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus) Mamm Genome. 2007;18:347–360. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- Jen KL, Hansen BC, Metzger BL. Adiposity, anthropometric measures, and plasma insulin levels of rhesus monkeys. Int J Obes. 1985;9:213–224. [PubMed] [Google Scholar]
- Jungheim ES, Lanzendorf SE, Odem RR, Moley KH, Chang AS, Ratts VS. Morbid obesity is associated with lower clinical pregnancy rates after in vitro fertilization in women with polycystic ovary syndrome. Fertil Steril. 2009;92:256–261. doi: 10.1016/j.fertnstert.2008.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungheim ES, Schoeller EL, Marquard KL, Louden ED, Schaffer JE, Moley KH. Diet-induced obesity model: abnormal oocytes and persistent growth abnormalities in the offspring. Endocrinology. 2010;151:4039–4046. doi: 10.1210/en.2010-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungheim ES, Schon SB, Schulte MB, DeUgarte DA, Fowler SA, Tuuli MG. IVF outcomes in obese donor oocyte recipients: a systematic review and meta-analysis. Hum Reprod. 2013;28:2720–2727. doi: 10.1093/humrep/det292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Fairbanks LA, Bailey JN, Jorgensen MJ, Wilson M, Zhang L, et al. Characterization and heritability of obesity and associated risk factors in vervet monkeys. Obesity (Silver Spring) 2007;15:1666–1674. doi: 10.1038/oby.2007.199. [DOI] [PubMed] [Google Scholar]
- Kundu MC, May MC, Chosich J, Bradford AP, Lasley B, Gee N, et al. Assessment of luteal function in the vervet monkey as a means to develop a model for obesity-related reproductive phenotype. Syst Biol Reprod Med. 2013;59:74–81. doi: 10.3109/19396368.2012.752547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lujan ME, Krzemien AA, Reid RL, Van Vugt DA. Caloric restriction inhibits steroid-induced gonadotropin surges in ovariectomized rhesus monkeys. Endocrine. 2005;27:25–31. doi: 10.1385/ENDO:27:1:025. [DOI] [PubMed] [Google Scholar]
- Luke B, Brown MB, Missmer SA, Bukulmez O, Leach R, Stern JE. The effect of increasing obesity on the response to and outcome of assisted reproductive technology: a national study. Fertil Steril. 2011;96:820–825. doi: 10.1016/j.fertnstert.2011.07.1100. [DOI] [PubMed] [Google Scholar]
- Metwally M, Cutting R, Tipton A, Skull J, Ledger WL, Li TC. Effect of increased body mass index on oocyte and embryo quality in IVF patients. Reprod Biomed Online. 2007;15:532–538. doi: 10.1016/s1472-6483(10)60385-9. [DOI] [PubMed] [Google Scholar]
- Motola S, Popliker M, Tsafriri A. Response of follicle cells to ovulatory stimuli within the follicle and in primary culture. Mol Cell Endocrinol. 2008;282:26–31. doi: 10.1016/j.mce.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Musicki B, Aten RF, Behrman HR. Inhibition of protein synthesis and hormone-sensitive steroidogenesis in response to hydrogen peroxide in rat luteal cells. Endocrinology. 1994;134:588–595. doi: 10.1210/endo.134.2.7507829. [DOI] [PubMed] [Google Scholar]
- Pazol K, Wilson ME, Wallen K. Medroxyprogesterone acetate antagonizes the effects of estrogen treatment on social and sexual behavior in female macaques. J Clin Endocrinol Metab. 2004;89:2998–3006. doi: 10.1210/jc.2003-032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotsky AJ, Hailpern SM, Skurnick JH, Lo JC, Sternfeld B, Santoro N. Association of adolescent obesity and lifetime nulliparity–the Study of Women’s Health Across the Nation (SWAN) Fertil Steril. 2010;93:2004–2011. doi: 10.1016/j.fertnstert.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttabyatappa M, Brogan RS, Vandevoort CA, Chaffin CL. EGF-like ligands mediate progesterone’s anti-apoptotic action on macaque granulosa cells. Biol Reprod. 2013;88:18. doi: 10.1095/biolreprod.112.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg V, Seshadri S, Sunkara SK, Sobaleva S, Oteng-Ntim E, El-Toukhy T. Effect of body mass index on IVF treatment outcome: an updated systematic review and meta-analysis. Reprod Biomed Online. 2011a;23:421–439. doi: 10.1016/j.rbmo.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Rittenberg V, Sobaleva S, Ahmad A, Oteng-Ntim E, Bolton V, Khalaf Y, et al. Influence of BMI on risk of miscarriage after single blastocyst transfer. Hum Reprod. 2011b;26:2642–2650. doi: 10.1093/humrep/der254. [DOI] [PubMed] [Google Scholar]
- Rizzo A, Roscino MT, Binetti F, Sciorsci RL. Roles of reactive oxygen species in female reproduction. Reprod Domest Anim. 2012;47:344–352. doi: 10.1111/j.1439-0531.2011.01891.x. [DOI] [PubMed] [Google Scholar]
- Santoro N, Lasley B, McConnell D, Allsworth J, Crawford S, Gold EB, et al. Body size and ethnicity are associated with menstrual cycle alterations in women in the early menopausal transition: The Study of Women’s Health across the Nation (SWAN) Daily Hormone Study. J Clin Endocrinol Metab. 2004;89:2622–2631. doi: 10.1210/jc.2003-031578. [DOI] [PubMed] [Google Scholar]
- Sawada M, Carlson JC. Superoxide radical production in plasma membrane samples from regressing rat corpora lutea. Can J Physiol Pharmacol. 1989;67:465–471. doi: 10.1139/y89-074. [DOI] [PubMed] [Google Scholar]
- Shah DK, Missmer SA, Berry KF, Racowsky C, Ginsburg ES. Effect of obesity on oocyte and embryo quality in women undergoing in vitro fertilization. Obstet Gynecol. 2011;118:63–70. doi: 10.1097/AOG.0b013e31821fd360. [DOI] [PubMed] [Google Scholar]
- Svensson EC, Markstrom E, Shao R, Andersson M, Billig H. Progesterone receptor antagonists Org 31710 and RU 486 increase apoptosis in human periovulatory granulosa cells. Fertil Steril. 2001;76:1225–1231. doi: 10.1016/s0015-0282(01)02891-6. [DOI] [PubMed] [Google Scholar]
- Warren WC, Jasinska AJ, Garcia-Perez R, Svardal H, Tomlinson C, Rocchi M, et al. The genome of the vervet (Chlorocebus aethiops sabaeus) Genome Res. 2015;25:1921–1933. doi: 10.1101/gr.192922.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LL, Dunning KR, Yang X, Russell DL, Lane M, Norman RJ, et al. High-fat diet causes lipotoxicity responses in cumulus-oocyte complexes and decreased fertilization rates. Endocrinology. 2010;151:5438–5445. doi: 10.1210/en.2010-0551. [DOI] [PubMed] [Google Scholar]
- Wu LL, Russell DL, Norman RJ, Robker RL. Endoplasmic reticulum (ER) stress in cumulus-oocyte complexes impairs pentraxin-3 secretion, mitochondrial membrane potential (DeltaPsi m), and embryo development. Mol Endocrinol. 2012;26:562–573. doi: 10.1210/me.2011-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu LL, Chura LR, Liang X, Lane M, Norman RJ, et al. Exposure to lipid-rich follicular fluid is associated with endoplasmic reticulum stress and impaired oocyte maturation in cumulus-oocyte complexes. Fertil Steril. 2012;97:1438–1443. doi: 10.1016/j.fertnstert.2012.02.034. [DOI] [PubMed] [Google Scholar]
- Young KA, Hennebold JD, Stouffer RL. Dynamic expression of mRNAs and proteins for matrix metalloproteinases and their tissue inhibitors in the primate corpus luteum during the menstrual cycle. Mol Hum Reprod. 2002;8:833–840. doi: 10.1093/molehr/8.9.833. [DOI] [PubMed] [Google Scholar]
- Young KA, Tumlinson B, Stouffer RL. ADAMTS-1/METH-1 and TIMP-3 expression in the primate corpus luteum: divergent patterns and stage-dependent regulation during the natural menstrual cycle. Mol Hum Reprod. 2004;10:559–565. doi: 10.1093/molehr/gah079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


