Abstract
Across a range of taxa, hormones regulate suites of traits that influence survival and reproductive success; however, the mechanisms by which hormone-mediated traits evolve are still unclear. We hypothesized that phenotypic divergence might follow from differential regulation of genes encoding key steps in hormone biosynthesis and thus the rate of hormone production. We tested this hypothesis in relation to the steroid hormone testosterone by comparing two subspecies of junco (Junco hyemalis) in the wild and in captivity. These subspecies have diverged over the last 10-15k years in multiple testosterone-mediated traits, including aggression, ornamentation, and body size. We show that variation in gonadal gene expression along the steroid biosynthetic pathway predicts phenotypic divergence within and among subspecies, and that the more androgenized subspecies exhibits a more prolonged time-course of elevated testosterone following exogenous stimulation. Our results point to specific genes that fulfill key conditions for phenotypic evolution because they vary functionally in their expression among individuals and between populations, and they map onto population variation in phenotype in a common garden. Our findings therefore build an important bridge between hormones, genes, and phenotypic evolution.
Keywords: testes, individual variation, steroidogenic acute regulatory protein, cytochrome p450 side-chain cleavage, cytochrome p450 17α-hydroxylase, 3-β-hydroxysteroid-dehydrogenase
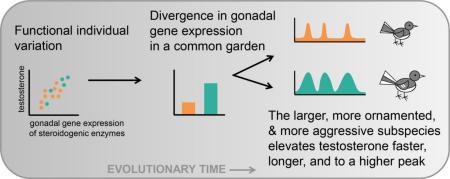
Graphical Abstract
Steroid hormones are chemical messengers that link environmental stimuli with the expression of a variety of traits across a range of taxa, including growth and immunity in plants (Oklestkova et al., 2015), color and size in insects (Oostra et al., 2011), and many social and sexual behaviors in vertebrates (Adkins-Regan, 2005; O'Connell and Hofmann, 2012). Because these effects are often mediated via hormonal activation of gene activity, hormones can influence how genotype is translated into phenotype, generating the potential for hormones to be proximate mediators of phenotypic evolution (Ketterson and Nolan, 1999; Zera et al., 2007). In vertebrates, the sex steroid testosterone (T) has been experimentally linked with survival, reproductive success, and many components of phenotype (Ketterson et al., 1992; Reed et al., 2006; Sinervo et al., 2000). Artificial selection on circulating T levels can influence hormone-mediated phenotypes and fitness (Mills et al., 2012; Robison et al., 1994; Walker et al., 2004), and interspecific differences in T profiles correspond to divergence in other traits as well (Dijkstra et al., 2012; Goymann et al., 2007). Thus, changes in circulating T have the potential to bring about phenotypic evolution (Hau, 2007; Ketterson et al., 2009; Wingfield, 2012); however, the underlying mechanisms by which these changes occur remain unclear.
There are several reasons for this lack of clarity. First, like many hormones, T is not a direct gene product, but rather the product of a multi-enzyme pathway and complex endocrine cascade. The hypothalamo-pituitary-gonadal (HPG) axis is activated when an environmental cue, such as day length or a conspecific, stimulates the hypothalamus to secret gonadotropin-releasing hormone (GnRH). GnRH acts on the pituitary to secrete gonadotropins, including luteinizing hormone (LH), which stimulates the gonad to produce T, which is derived from cholesterol via several intermediates. Further complexity arises via feedback along the HPG axis, variation in hormone receptor densities in target tissues, and other factors that exhibit plasticity in response to the environment (Ball and Balthazart, 2008). This plasticity poses a challenge for evolutionary biologists seeking to understand how hormone-mediated traits evolve because plasticity can obscure otherwise consistent individual variation that represents the raw material of evolutionary change (Whitehead and Crawford, 2006).
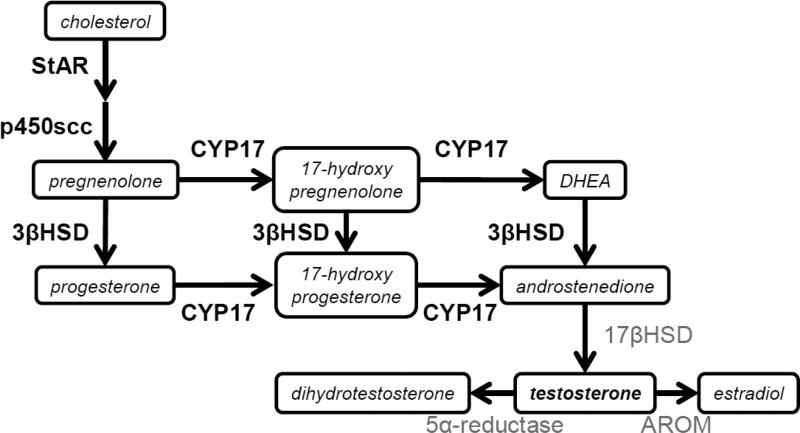
Here, we sought to identify mechanistic sources of variation in T production, within and among populations, concentrating on gene expression related to the biosynthesis of T in the gonad (Figure 1). Empirical and theoretical research on other biosynthetic pathways suggests that genes whose products are located early in a pathway or those catalyzing multiple, branching steps ought to be major regulators of flux through these pathways (Rausher, 2013; Wright and Rausher, 2010). As a consequence, we focused on steroidogenic acute regulatory protein (StAR), cytochrome p450 side-chain cleavage (p450scc), 3-β-hydroxysteroid-dehydrogenase (3βHSD), and cytochrome p450 17α-hydroxylase (CYP17), due to their position in the pathway to produce T. In addition, all have been linked with variation in T secretion and T-mediated phenotypes in other contexts, e.g. during ascent to social dominance in a cichlid fish (Huffman et al., 2012) or in association with hormonal disorders in humans (LaVoie and King, 2009; Payne and Youngblood, 1995). However, we are not aware of any study that has identified these candidate genes as potential sources of within-sex individual differences in T. We hypothesized that differences in the expression of these genes would underlie individual and population variation in the regulation of T and the expression of T-mediated traits.
Figure 1. A simplified overview of testosterone synthesis.
Genes of interest are highlighted in bold. StAR= steroidogenic acute regulatory protein. P450scc= Cytochrome P450 side-chain cleavage; CYP17= Cytochrome P450 17α-hydroxylase; 3βHSD= 3β-hydroxysteroid dehydrogenase/isomerase; 17βHSD= 17β-hydroxysteroid dehydrogenase. DHEA= dehydroepiandrosterone. AROM=aromatase.
The subject of this research is the dark-eyed junco (Junco hyemalis), a songbird that has been used extensively in studies of evolutionary and behavioral endocrinology (Ketterson et al., 2009; Ketterson and Nolan, 1992). Juncos are thought to have rapidly diverged into several phenotypically distinct subspecies since the last glaciation (Mila et al., 2007), and we compare two subspecies that differ in a number of T-mediated traits. White-winged juncos (J. h. aikeni), which breed only in the Black Hills of South Dakota, are significantly larger, more ornamented, and more aggressive than the Carolina subspecies (J. h. carolinensis) (Bergeon Burns et al., 2013; Nolan et al., 2002), which breed in the Appalachian mountains of Virginia. The subspecies do not differ in average T levels when sampled in the field (Bergeon Burns et al., 2013) or when sampled in a common aviary environment 30 min after standardized HPG axis activation (Bergeon Burns et al., 2014). Despite these similar T levels measured in prior research, captive VA males appear to be more sensitive to LH stimulation of the gonad (more LH receptor mRNA), and they also may be more sensitive to negative feedback at the top of the HPG axis (more hypothalamic AR mRNA), compared to SD males (Bergeon Burns et al., 2014). These findings led us to hypothesize that the populations may differ in temporal regulation of T secretion, despite not differing significantly in the magnitude of circulating T at the sampling points used in earlier research. Critically, individual male juncos are also repeatable in how much T they produce in response to exogenous injection of GnRH (Jawor et al., 2006), and this individual variation is remarkably similar to the amount of T produced in response to a standardized LH injection (Bergeon Burns et al., 2014). Thus, our past work suggested that individual differences in T production may lie primarily downstream of this LH signal (i.e. at the level of the gonad).
In this study, we examined gonadal tissues from free-living male juncos that were sampled on their breeding territories in the early spring, as well as captive males held in a common aviary environment. We measured T levels and expression of genes whose products have central roles in gonadal steroidogenesis. We asked whether the two phenotypically divergent populations varied in gonadal gene expression in the wild, and whether any differences persisted in a common garden. We investigated which genes, if any, predict individual differences in T levels. We predicted that males with higher T would have greater gonadal gene expression for these steroidogenic enzymes. We further predicted that males from the larger, more ornamented, and more aggressive population would have a greater molecular capacity to produce T, and that when the HPG axis was stimulated, these males would elevate T more rapidly and sustain that elevation for a longer period of time.
Material and Methods
Study 1: Testosterone and gonadal gene expression in the field
Male juncos were captured on their breeding territories in the spring near Custer, South Dakota (“SD,” 43°46′N 103°36'W, n=17, dates: 14 to 22 May 2010) and Mountain Lake, Virginia (“VA,” 37°22'N, 80°32'W, n=17, dates: 1 May to 5 June 2010), as described in prior publications on aggressive behavior and neural gene expression in these same birds (Bergeon Burns et al., 2014; Rosvall et al., 2012a). All animals experienced a 6 min simulated territorial intrusion (“STI”) by a live same-sex conspecific between 0600-1200 local time. Males were captured shortly thereafter (4.7±0.8 min after STI), and immediately killed by overdose of isoflurane, followed by decapitation. Trunk blood was collected and stored on ice in the field, and plasma was stored at −20°C. Gonads were dissected from the body using RNAse-free techniques, flash frozen on dry ice, and stored at −80°C until processing. Exact breeding stage was not known for most males, but males were reliably defending a territory, and in some cases we observed females building nests and incubating, suggesting that the timing of collection was after territory establishment, in the early- to mid- breeding period. Enlarged gonads were typical of full reproductive condition (see below).
The short STIs are not expected to have affected T levels because juncos do not elevate T in response to STIs under these conditions (Rosvall et al., 2014; Rosvall et al., 2012b). In addition, appreciable transcription of all but immediate early genes is thought to require more time (Herdegen and Leah, 1998), and it is thought to require at least 60 min for appropriate upstream hormonal stimuli to affect our genes of interest (LaVoie and King, 2009). Accordingly, we anticipate that expression of genes measured here represents a close approximation of each male's constitutive expression prior to the short STI, rather than a genomic response to the STI itself. Consistent with this view, latency to capture was not correlated with expression of any gene p>0.12, |rs|<0.28; we did not include this covariate in final analyses.
Study 2: Testosterone and gonadal gene expression in a common aviary environment
Male juncos were captured as juveniles in the late summer in SD (n=19) and VA (n=26) and brought into captivity at Indiana University (IU) (Bergeon Burns et al., 2014; Rosvall et al., 2013). Birds from SD were studied in 2009-10 and birds from VA in 2010-11 using exactly the same rooms, protocols, light advancement schedule, and days of the year. Birds were held overwinter in a mixed sex flock on their native light cycle in an indoor aviary. On March 2, before either population had reached a photo-stimulatory day length, birds were transferred to individual cages and lights were advanced by 1h every other day, until 16L:8D on March 10. This schedule was designed to override subtle population differences in stimulatory day length. We later recorded gonad size to confirm that the populations were at equivalent reproductive stages. Three weeks after reaching 16L:8D, we began hormone challenges to explore population variation along the HPG axis (Bergeon Burns et al., 2014): one LH injection to assess transient T production and two GnRH injections (one to assess T, and another to assess LH production) per male, one per day every 5th day, randomized and counterbalanced for order. Prior work demonstrates that T returns to baseline within about 2h of these injections (Jawor et al., 2006), and we found with these males that prior injections had no influence on T levels measured in later injections (Bergeon Burns et al., 2014). Here, we focus on T levels following the widely used ‘30 min GnRH challenge’ because this is a well-established method for assessing repeatable individual differences in ability to produce T in a range of vertebrates (Apfelbeck and Goymann, 2011; Gleason et al., 2012; Jawor et al., 2006). Males that produce more T in response to exogenous GnRH also have higher T following ~30-min STIs administered while their mates were re-nesting (McGlothlin et al., 2008). Thus, although recent work suggests that T elevation in response to a male rival is either absent or highly context-specific in this species (occurring only when mates are fertile (Rosvall et al., 2014; Rosvall et al., 2012b)), the GnRH challenge remains a highly useful tool for assaying a biologically relevant hormonal phenotype because it predicts T response to at least one naturally occurring life history transition (McGlothlin et al., 2008), as well as many components of male phenotype and fitness (McGlothlin et al., 2007; McGlothlin et al., 2010).
Each individual was removed from its cage and an initial blood sample (≤140μL) was collected. Each individual was then injected in the pectoral muscle with 1.25μg of GnRH (Chicken LH–RH, American Peptide #54-8-23) dissolved in 50μL PBS. Thirty min later, we collected a second blood sample. Blood was stored on ice, centrifuged, and plasma was frozen at −20°C. Five days after the final hormone challenge, each male was removed from its cage and immediately euthanized with an overdose of isoflurane followed by decapitation. Gonads were dissected from the body, as above. All procedures occurred between 0600 and 1200.
Study 3: Time-course of testosterone production in a common garden
Adult male juncos were captured between May 2010 and May 2012 (n=15 VA, 7 SD) and held in an aviary at IU until this study in spring 2014, which used the same rooms, cages, protocols, light advancement, and dates as Study 1. We injected each male with GnRH once every 5 days to construct a 6-point time-course of T production, sampling at 0 min (pre-injection) and 15, 30, 45, 60, and 90 min post-injection. To obtain all time-points while maintaining a safe blood draw volume, each male was bled twice per GnRH challenge, with time-points randomized and counterbalanced among 3 GnRH challenges. Our prior work administering hormone challenges once every 5 days shows that males are highly repeatable in their T production (Bergeon Burns et al., 2014). We tested for an effect of order on T levels for each of the 6 time-points (i.e. whether data were collected from the first, second, or third GnRH challenge), and we found no significant effect (F<1.54, p>0.24).
Testosterone enzyme immunoassay
We quantified T levels in 20μL plasma using enzyme immunoassay kits (Enzo #901-065, Farmingdale, NY, USA) that have been used extensively in this species (Clotfelter et al., 2004). All samples were spiked with a small, uniform amount of tritiated T and extracted twice with diethyl ether prior to being assayed. Data were corrected for incomplete recoveries (average = 91%). Hormone samples from studies 1, 2, and 3 were assayed separately, with average intra-assay CVs of 9.7%, 7.4%, and 3.6%, and inter-assay CVs of 19.2%, 19.9%, and 5.8%, respectively. Within studies 1 and 2, we used a plate correction factor to correct for interplate variability (Jawor et al., 2006); we did not do this for study 3 due to low interplate variability.
Molecular methods to assess gonadal gene expression
For all 79 males in studies 1 and 2, we extracted RNA from one testis using Trizol (Invitrogen, Carlsbad, CA, USA), treated 1μg of RNA with DNAse (Promega, Madison, WI, USA), and reverse transcribed it to cDNA with Superscript III (Invitrogen). All qPCR reactions were run on Strategene MX3000P using MxPro software (v.4.10, Agilent, Santa Clara, CA, USA) using PerfeCta SYBR Green SuperMix with low ROX (Quanta Biosciences, Gaithersburg, MD, USA). Samples were run in duplicate for each gene of interest (GOI) and a reference gene. We used the comparative Ct method (2−ΔΔCt) that represents transcript abundance for each GOI as the fold difference in expression compared to a pooled standard (i.e. calibrator) and normalized by the reference gene (Livak and Schmittgen, 2001). The calibrator sample was derived from juncos collected during earlier pilot work. We used ribosomal protein L13A (RPL13A) as the reference gene because it has been reported to be one of the 10 best housekeeping genes for qPCR (de Jonge et al., 2007), and we confirmed that the populations did not differ in expression of this gene in the gonad. Further details on primers and our qPCR protocol can be found in ESM.
Gonad size
We weighed and measured one frozen testis from a subset of our subjects (n=19 SD captive, 15 SD field, 26 VA captive, 11 VA field). Mass was recorded to the nearest 0.01 g and length and width to the nearest 0.1 mm. We calculated volume as 4/3πa2b, where a is half the width and b is half the length. Gonads were measured for all captive birds and a subset of field birds.
Statistical analyses
Statistical analyses were performed using JMP (v. 12.1.0, SAS Institute, Cary, NC, USA) unless otherwise noted. All tests are two-tailed, and we report results as mean ± one standard error, and η2 effect size estimates as the ratio of the effect variance (SSeffect) to the total variance (SStotal). We used Shapiro-Wilks to test for normality, and we natural log-transformed T data and log2-transformed gene expression data, when necessary.
To compare gonad mass and volume among populations, we used a two-way ANOVA with fixed effects of population and environment (field vs. captive), as well as their interaction. To compare gene expression between the populations, we used a single MANOVA with population, environment, and their interaction. One captive VA male was excluded from this analysis due to a missing value for p450scc that stemmed from a poor replicate in qPCR amplification. Post-hoc ANOVAs were used to highlight gene-level results, using FDR correction for multiple comparisons (Benjamini and Hochberg, 2000; Pike, 2011).
We used linear mixed models to test the effect of each gene in predicting variation in T among males while controlling for population. For field samples, the dependent variable was T measured in trunk blood; for captive samples, we separately tested baseline and GnRH-stimulated T (30 min after injection) as dependent variables. Genes were correlated in their expression to some degree in the wild (0.32<r<0.73, p's range from 0.06 to <0.0001) and to a lesser degree in captivity (ESM Table 2), but low variance inflation factors justified inclusion of all four genes in a single model.
We compared populations in the time-course of T elevation (study 3) using a repeated-measures mixed-model, with individual as a random repeated factor, and fixed effects of population and time. We estimated area under the curve of T vs. time using R statistical package (v. 3.2.2, R Development Team, 2015). Calculations used the trapezoidal rule, which calculates area as where i is a time-point at which blood was sampled and i + 1 is the subsequent time-point, measured in minutes, and Ti and Ti+1 are the corresponding testosterone levels. We compared these measures of integrated T among populations using a Welch's ANOVA.
Results
Population comparisons in the wild and in a common garden
Gonad mass and volume did not significantly differ among populations, but gonads were larger and heavier in the field than in captivity for both populations (studies 1 and 2: n=71, Mass: R2adj=0.34, overall: F=12.85, p<0.0001, environment: η2=0.36, F=37.5, p<0.001; population: η2=0.00, F=0.0, p=0.94, pop*env: η2=0.00, F=0.24, p=0.62; Volume: R2adj=0.34, overall: F=13.11, p<0.0001, environment: η2=0.35, F=37.00, p<0.0001, population: η2=0.00, F=0.04, p=0.83, pop*env: η2=0.01, F=1.52, p=0.22). As previously reported, T levels in these same birds did not differ between populations in the field (Bergeon Burns et al., 2013), or in captivity before or 30 min after GnRH challenge (Bergeon Burns et al., 2014). Baseline T levels in captivity were lower than in the field (averages of 2.4 and 3.7 ng/mL, respectively), although captive males were capable of marked T elevation (study 2, average T 30 min after GnRH challenge = 7.7 ng/mL, ranging from 4.4 to 15.9 ng/mL).
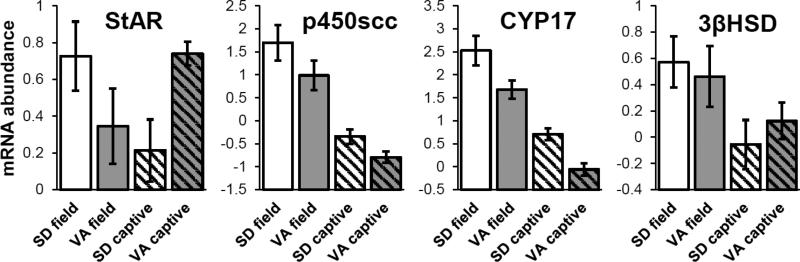
MANOVA including all 4 genes showed significant overall effects of population and environment (n=78, Wilks’ λ=0.34, F=7.88, p<0.0001; population: F=4.51, p=0.0026, environment: F=21.33, p<0.0001, pop*env: F=2.16, p=0.083). Post-hoc ANOVAs revealed significant main effects of population and environment for p450scc and CYP17, significant effects of environment only for 3βHSD, and a significant pop*env interaction for StAR (Table 1, Figure 2).
Table 1.
Post-hoc ANOVAs comparing gene expression between two populations, in two environments (field and captivity).
| population | environment | pop × env | |
|---|---|---|---|
| StAR | F = 0.22, p = 0.77 | F = 0.15, p = 0.77 | F = 8.90, p = 0.011* |
| p450scc | F = 5.58, p = 0.038* | F = 60.33, p < 0.0001* | F = 0.27, p = 0.77 |
| CYP17 | F = 16.62, p = 0.0004* | F = 81.34, p < 0.0001* | F = 0.05, p = 0.78 |
| 3βHSD | F = 0.03, p = 0.78 | F = 6.77, p = 0.025* | F = 0.61, p = 0.69 |
FDR adjusted p-values (i.e. q-values) are denoted as significant when in bold with asterisk.
Figure 2.
Gonadal gene expression in the field and in captivity (studies 1 and 2). SD= South Dakota (J. h. aikeni). VA= Virginia (J. h. carolinensis). mRNA abundance is expressed on a log2 scale, where a difference of 1 unit represents a two-fold difference in expression. See MANOVA in text and Table 1 for statistics.
Among individuals
In the wild (study 1), when controlling for population, StAR and p450scc were significant predictors of individual variation in T (Table 2). In captivity (study 2), none of our genes of interest were significant predictors of baseline T levels (Table 2), but CYP17 gene expression was weakly but significantly related to GnRH-induced T levels (Table 2).
Table 2.
Predictors of individual variation in testosterone.
| F | p | VIF | |
|---|---|---|---|
| Field: Testosterone (Study 1) | |||
| Overall R2adj=0.48 | 6.98 | 0.0003 | |
| Population | 0.53 | 0.47 | 1.37 |
| StAR | 5.83 | 0.023* | 1.88 |
| p450scc | 15.14 | 0.0006* | 2.21 |
| CYP17 | 3.35 | 0.078 | 2.80 |
| 3bHSD | 0.086 | 0.77 | 2.11 |
| Captivity: Baseline testosterone (Study 2) | |||
| Overall R2adj=0.06 | 1.56 | 0.20 | |
| Population | 0.0022 | 0.96 | 1.62 |
| StAR | 0.16 | 0.69 | 1.32 |
| p450scc | 1.38 | 0.25 | 1.43 |
| CYP17 | 1.55 | 0.22 | 1.81 |
| 3bHSD | 0.00 | 1.00 | 1.15 |
| Captivity: GnRH-induced testosterone (Study 2) | |||
| Overall R2adj=0.07 | 1.62 | 0.18 | |
| Population | 0.72 | 0.40 | 1.62 |
| StAR | 0.98 | 0.33 | 1.32 |
| p450scc | 0.34 | 0.563 | 1.43 |
| CYP17 | 5.58 | 0.023* | 1.81 |
| 3bHSD | 2.81 | 0.10 | 1.15 |
Significant effects are in bold with asterisk. VIF = variance inflation factors.
Time-course of T elevation following GnRH challenge
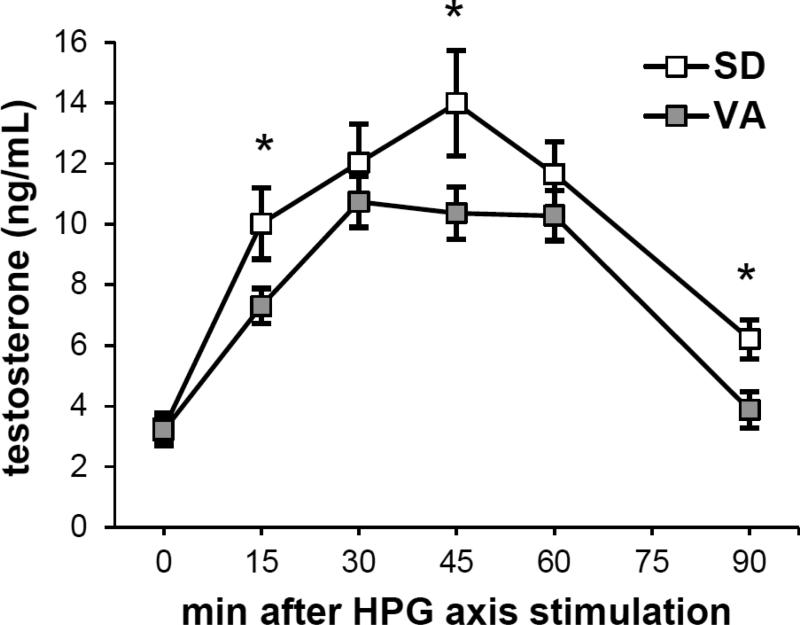
Repeated measures mixed models showed significant effects of population (study 3: n=15 VA, n=7 SD, F=5.24, p=0.033; Figure 3) and time-point (n=6 time-points per male, F=47.5, p<0.0001). Post-hoc comparisons demonstrated higher T for SD vs. VA at 15, 45, and 90 min (F>4.38, p<0.049), but not at 0, 30, or 60 min (F<0.95, p>0.34). The area under the curve of T vs. time was significantly larger for SD vs. VA (Welch's ANOVA, F=4.91, p=0.047).
Figure 3.
VA and SD males differ in the time-course of T production following HPG axis stimulation (study 3). Asterisks indicate time-points that significantly differ between populations.
DISCUSSION
These studies reveal robust population differences in gonadal gene expression for proteins that play a central role in steroid synthesis. Most notably, the two cytochrome enzymes (p450scc and CYP17) were more highly expressed in SD than VA, both in the field and in a common aviary environment, suggesting stable gonadal differences between populations that are consistent with our prediction that the larger, more aggressive, and more ornamented subspecies (SD) would have a greater molecular capacity to produce T. Although not all genes in the steroidogenic pathway followed this same pattern, and there are surely additional factors that influence T levels in circulation, our direct comparison of the time-course of T elevation showed that SD males indeed elevate T more rapidly, to a higher peak, and sustain T levels above baseline for longer than VA males, following HPG axis stimulation. This pattern ought to represent greater tissue exposure to T over time in SD vs. VA. Among free-living males, we found that StAR and p450scc gene expression were significant predictors of an individual's current T level – collectively accounting for nearly half of the variation in T levels. These findings suggest that gene expression for key proteins regulating steroidogenesis may be significant contributors to functional individual differences in hormone levels, as well as associated variation in hormone-mediated morphology or behavior. Our findings also lend support to the view that the gonad itself may be a much more important contributor to phenotypic variation than previously thought (Kitano et al., 2011; Lynn et al., 2015; McGuire et al., 2013). Below, we discuss these and other implications for understanding the proximate mechanisms of hormone-mediated phenotypic evolution.
Population divergence in steroidogenic gene expression
Free-living South Dakota males expressed marginally more StAR mRNA and significantly more CYP17 and p450scc mRNA than Virginia males. StAR is found in steroid-producing Leydig cells, and along with several co-factors is responsible for shuttling cholesterol into the inner mitochondria where the first committed step of steroid synthesis begins, when p450scc converts cholesterol to pregnenolone. CYP17, in turn, catalyzes reactions to convert pregnenolone and progesterone into the androgens dehydroepiandrosterone (DHEA) and androstenedione, respectively. StAR, p450scc, and CYP17 have been suggested as rate-limiting steps in steroid synthesis (Huffman et al., 2012; LaVoie and King, 2009; Payne and Youngblood, 1995). To the extent that these transcript-level patterns extend to protein, our data suggest that SD males have a greater capacity to produce steroids in the gonad, compared to VA males at the same breeding stage. Interestingly, the populations did not differ in gonadal abundance of 3βHSD mRNA, demonstrating that there has not been integrated divergence along the entire biosynthetic pathway. Thus, at least for gonadal steroidogenesis, different components of the endocrine system may be able to change independently of others, a pattern that may influence the rate at which hormone levels can evolve (Ketterson et al., 2009).
A key question is the degree to which population differences in gonadal gene expression represent heritable differences among populations vs. plasticity due to the different environments of Virginia and South Dakota. Comparisons of field and captive gonads show clear environmental effects on gene expression. For example, averaged across populations, 3βHSD, CYP17, and p450scc were 0.5, 3.4, and 3.7 times more highly expressed in the wild than in captivity, perhaps related to the observation that animals often do not reach their full reproductive potential in captivity (Calisi and Bentley, 2009) and in keeping our finding that captive gonads were roughly 75% of the size of field gonads. Even so, captive males studied here were capable of robust T production that differed markedly in its time-course between populations (elaborated below).
Similar to the field results, the captive study revealed significant population differences in expression of StAR, p450scc, and CYP17 mRNA, but not 3βHSD. Surprisingly, the direction of the population effect in StAR reversed signs from the wild to captivity, with VA captive males having more StAR mRNA than SD, despite SD still having higher expression of p450scc and CYP17. One interpretation is that the populations differ in some mechanism regulating reproductive timing (Ketterson et al., 2015), indicating that perhaps we did not compare them at equivalent stages of reproductive readiness. This potential confound is not supported, however, because the populations did not differ in gonad size, and the GnRH-induced T levels observed in captives were on par with those observed in the field for both populations (Jawor et al., 2006) and Bergeon Burns, unpubl. data). Why StAR was greater in VA captives is something we cannot currently explain; however, the observation that captive SD males can nonetheless produce more T over time (Figure 3) suggests that StAR, or the availability of hormone precursor, may not necessarily cause differences in T levels, but rather the rate of conversion to active steroids may be more functionally important for T output (e.g. abundance of p450scc and CYP17). Indeed, StAR is regulated by an unusually complex set of signaling pathways and cofactors, only some of which overlap with those regulating the cytochrome enzymes that showed consistently higher mRNA levels in SD vs. VA in the wild and in captivity (Stocco et al., 2005).
There are many ecological factors that could differ between these populations and influence gonadal steroidogenesis, including diet, climate, or the social environment (Goymann, 2009; Lynn et al., 2015). In SD, for example, territories seem to be larger, the environment is more extreme, and the breeding season is shorter than in VA (Bergeon Burns, unpubl. data), such that it may be adaptive for males to have higher ‘integrated T’ to capitalize upon more limited mating opportunities. On the other hand, population variation in steroidogenesis also may be shaped by founder effects, particularly since drift is thought to have played a role in the post-glacial diversification of the junco (Mila et al., 2007). Genetic differentiation of T production via divergence in gene activity is just one interpretation of our findings; greater experimentation is needed to distinguish among alternative hypotheses underlying the marked population differences described here. For example, early developmental effects might influence T or phenotype directly (e.g. Cameron et al., 2008; Pfannkuche et al., 2011), and if the populations differ genetically in other traits, there also may be dynamic feedbacks that affect steroidogenesis, e.g. mediated via stress signaling, ornamentation, or behavior (Deviche et al., 2012; Safran et al., 2008). Regardless of the drivers of these proximate differences, our common garden results clearly demonstrate robust differences in gonadal gene expression when food, social interactions, day length, and temperature are standardized, suggesting stable population divergence at the level of the gonad.
Population divergence in the time-course of testosterone elevation
Not only do VA and SD males differ in the gonad's apparent molecular capacity to produce T, but they also differ in the time-course over which they elevate T after a standardized dose of GnRH in a common aviary environment. Males from the two populations did not differ in baseline T levels (i.e. prior to GnRH injection), but at 15, 45, and 90 min post-GnRH injection, SD males had higher T levels than VA males, such that SD males had a greater integrated T, or area under the curve of T over time. By focusing on this time-course of T production, we can now support the hypothesis of evolution via divergence in hormone signal strength (i.e. phenotypic integration) (Ketterson et al., 2009), whereas our previous tests of this hypothesis in this system, which focused on only 2 time-points (0 and 30 min), did not identify these differences (Bergeon Burns et al., 2014; Bergeon Burns et al., 2013). Although the magnitude of an endocrine response is likely related to its duration, it is clear that important information can be missed when individuals or populations are compared at a single time-point. These findings highlight the importance of quantifying the dynamic nature of hormone signaling when asking questions about how hormone-mediated traits evolve.
We propose that population divergence in the time-course of T production stems in part from divergence in gonadal gene expression for steroidogenic enzymes, and that p450scc and CYP17 are likely to play a key role in differentiating the two populations. In previous work, we showed that SD males have lower hypothalamic AR mRNA than VA males, suggesting that SD males also may be less sensitive to negative feedback, which may also be contributing to this apparent population divergence in the time-course of T elevation (Bergeon Burns et al., 2014). Thus, multiple components of the HPG axis may evolve in concert. Other components of endocrine signaling also may contribute to population differences in the rate of T production, including divergence in the rate of GnRH release or cross-talk with the HPA axis. This greater exposure to T over time in SD may provide one mechanism that differentiates SD males from VA males in a range of T-mediated phenotypes. Some of these effects are likely to be activational, particularly with respect to behavior. Although the populations may also differ in sensitivity to T in target tissues, previous work demonstrates SD males do not have uniformly greater sensitivity to T in the brain (Bergeon Burns et al. 2013), further suggesting that peripheral T is an important source of phenotypic divergence.
Evolutionary implications of individual variation in steroidogenic gene expression
If evolution is to act on variation in the regulation of hormone levels to bring about phenotypic change, there must be functional inter-individual variation in some component of the mechanism governing hormone production. Here, we find that gonadal StAR and p450scc gene expression levels are significant predictors of individual variation in T levels in free-living males. This finding is consistent with prior work suggesting that enzymes early in a pathway have disproportionate effects on flux through that pathway, and in some cases, may be more likely to diversify over evolutionary time (Rausher, 2013; Wright and Rausher, 2010). Some males have constitutively higher StAR and p450scc gene expression, and to the degree that mRNA predicts protein abundance, these males may have more cholesterol available to initiate steroidogenesis, as well as more pregnenolone, which is the precursor of all steroids, including T. Social or environmental factors also may influence gene expression, such that some males have higher expression than others simply because their HPG axis has been recently stimulated, e.g. by a mating opportunity (Goymann, 2009; Goymann et al., 2007). Without manipulating StAR and p450scc, we cannot say for certain that these correlations between gene expression and T levels are causal. However, for p450scc in particular, our observation that SD males have consistently higher expression, paired with a wealth of data on p450scc and its role in the dysfunction of steroid production (Payne and Youngblood, 1995), suggests that a causal link is probable.
Again, a key question is whether this inter-individual variation represents plastic or stable, potentially heritable, differences among males. Gene expression is known to have a heritable component (Stamatoyannopoulos, 2004; Whitehead and Crawford, 2006) and to differ consistently among species or strains in ways that suggest genetic control (Edwards et al., 2009). T secretion and presumably steroidogenic gene expression also may be influenced by a variety of non-genetic factors, including age, prior experience, or maternal effects (Kempenaers et al., 2008). Our captive dataset is not well suited to disentangling these effects among individuals, both because many extrinsic factors were held stable, and because gene expression and hormone levels were sampled days apart. Captive males also were euthanized without GnRH stimulation, when T levels were lower than in the field, and these males were yearlings that had never mated nor held a territory. Although age differences in T are not uniformly detected in this species (Deviche et al., 2000; Jawor et al., 2006; Ketterson and Nolan, 1992; McGlothlin et al., 2008), it is possible that experience, age, or another correlated phenotype might modify the direct connection between steroidogenic gene expression and T levels. In sum, there are multiple explanations for the relatively weak effect sizes linking T and gene expression in captivity, compared to the field (R2 = 0.07 and 0.48, respectively), although the strong patterns in the field suggest that functional variation in gonadal gene expression may emerge only when individuals are stimulated.
A key role for peripheral sources of variation
A central conclusion of all of these data is that variation at the level of the gonad maps onto phenotypic variation within and between populations. Recently, behavioral endocrinology has emphasized the brain's contribution to phenotypic variation, including divergence in sensitivity to hormones in the brain (Goncalves et al., 2010; Young et al., 1995) and the role of the hypothalamus in stimulating or suppressing the HPG axis (Tsutsui et al., 2010). Our data add to a growing appreciation that endocrine signaling derived from peripheral tissues also may be an important contributor to phenotypic variation and divergence (Kitano et al., 2011; Lynn et al., 2015; McGuire et al., 2013), and our findings especially highlight steroidogenic enzymes as key players in this process.
Supplementary Material
Highlights.
Virginia and South Dakota junco subspecies differ in testosterone (T)-mediated traits
The larger, more aggressive SD birds elevate T faster, longer, and to a higher peak
In the wild, SD birds have greater gonadal gene expression for steroidogenic enzymes
Subspecies differences in p450scc and CYP17 mRNA abundance persist in a common garden
Gonads harbor important variation for the evolution of hormone-mediated traits
Acknowledgments
All animal procedures were approved by the IU Bloomington Institutional Animal Care and Use Committee under protocols 06-242, 09-037, and 12-050 (valid 2006-2015). Our fieldwork would not have been possible without the University of Virginia Mountain Lake Biological Station, Black Hills National Forest, and numerous field assistants. Our aviary and lab work would not have been possible without M Abolins-Abols, B Duncan, A Fudickar, R Hanauer, MS Izzo, A Kimmitt, L Sloan, and S Slowinski. S Jayaratna is grateful to her Biology Honors Thesis committee KA Rosvall, GT Smith, and GE Demas.
Funding: NIH (F32HD068222, T32HD049336); NSF (GRFP to CMB, DDIG IOS-0909834, and IOS-0820055, CISAB REU program DBI-0851607); Indiana Academy of Sciences; IU Women in Science undergraduate research fellowship; IU Department of Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Adkins-Regan E. Hormones and animal social behavior. Princeton University Press; Princeton, NJ: 2005. [Google Scholar]
- Apfelbeck B, Goymann W. Ignoring the challenge? Male black redstarts (Phoenicurus ochruros) do not increase testosterone levels during territorial conflicts but they do so in response to gonadotropin-releasing hormone. Proceedings of the Royal Society B: Biological Sciences. 2011;278:3233–3242. doi: 10.1098/rspb.2011.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball GF, Balthazart J. Individual variation and the endocrine regulation of behaviour and physiology in birds: a cellular/molecular perspective. Philosophical Transactions of the Royal Society B-Biological Sciences. 2008;363:1699–1710. doi: 10.1098/rstb.2007.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. On the Adaptive Control of the False Discovery Rate in Multiple Testing with Independent Statistics. Journal of Educational and Behavioral Statistics. 2000;25:60–83. [Google Scholar]
- Bergeon Burns CM, Rosvall KA, Hahn TP, Demas GE, Ketterson ED. Examining sources of variation in HPG axis function among individuals and populations of the dark-eyed junco. Hormones and Behavior. 2014;65:179–187. doi: 10.1016/j.yhbeh.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeon Burns CM, Rosvall KA, Ketterson ED. Neural steroid sensitivity and aggression: comparing individuals of two songbird subspecies. Journal of Evolutionary Biology. 2013;26:820–831. doi: 10.1111/jeb.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisi RM, Bentley GE. Lab and field experiments: Are they the same animal? Hormones and Behavior. 2009;56:1–10. doi: 10.1016/j.yhbeh.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Cameron N, Del Corpo A, Diorio J, McAllister K, Sharma S, Meaney MJ. Maternal Programming of Sexual Behavior and Hypothalamic-Pituitary-Gonadal Function in the Female Rat. Plos One. 2008;3 doi: 10.1371/journal.pone.0002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotfelter ED, O'Neal DM, Gaudioso JM, Casto JM, Parker-Renga IM, Snajdr EA, Duffy DL, Nolan V, Ketterson ED. Consequences of elevating plasma testosterone in females of a socially monogamous songbird: evidence of constraints on male evolution? Hormones and Behavior. 2004;46:171–178. doi: 10.1016/j.yhbeh.2004.03.003. [DOI] [PubMed] [Google Scholar]
- de Jonge HJM, Fehrmann RSN, de Bont ESJM, Hofstra RMW, Gerbens F, Kamps WA, de Vries EGE, van der Zee AGJ, te Meerman GJ, ter Elst A. Evidence Based Selection of Housekeeping Genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deviche P, Gao S, Davies S, Sharp PJ, Dawson A. Rapid stress-induced inhibition of plasma testosterone in free-ranging male rufous-winged sparrows, Peucaea carpalis: Characterization, time course, and recovery. General and Comparative Endocrinology. 2012;177:1–8. doi: 10.1016/j.ygcen.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Deviche P, Wingfield JC, Sharp PJ. Year-class differences in the reproductive system, plasma prolactin and corticosterone concentrations, and onset of prebasic molt in male dark-eyed juncos (Junco hyemalis) during the breeding period. General and Comparative Endocrinology. 2000;118:425–435. doi: 10.1006/gcen.2000.7478. [DOI] [PubMed] [Google Scholar]
- Dijkstra PD, Verzijden MN, Groothuis TGG, Hofmann HA. Divergent hormonal responses to social competition in closely related species of haplochromine cichlid fish. Hormones and Behavior. 2012;61:518–526. doi: 10.1016/j.yhbeh.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Ayroles JF, Stone EA, Carbone MA, Lyman RF, Mackay TFC. A transcriptional network associated with natural variation in Drosophila aggressive behavior. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-7-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason ED, Holschbach MA, Marler CA. Compatibility drives female preference and reproductive success in the monogamous California mouse (Peromyscus californicus) more strongly than male testosterone measures. Hormones and Behavior. 2012;61:100–107. doi: 10.1016/j.yhbeh.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves D, Saraiva J, Teles M, Teodosio R, Canario AVM, Oliveira RF. Brain aromatase mRNA expression in two populations of the peacock blenny Salaria pavo with divergent mating systems. Hormones and Behavior. 2010;57:155–161. doi: 10.1016/j.yhbeh.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Goymann W. Social modulation of androgens in male birds. General and Comparative Endocrinology. 2009;163:149–157. doi: 10.1016/j.ygcen.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Goymann W, Landys MM, Wingfield JC. Distinguishing seasonal androgen responses from male-male androgen responsiveness - Revisiting the Challenge Hypothesis. Hormones and Behavior. 2007;51:463–476. doi: 10.1016/j.yhbeh.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Hau M. Regulation of male traits by testosterone: implications for the evolution of vertebrate life histories. Bioessays. 2007;29:133–144. doi: 10.1002/bies.20524. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Research Reviews. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Huffman LS, Mitchell MM, O'Connell LA, Hofmann HA. Rising StARs: Behavioral, hormonal, and molecular responses to social challenge and opportunity. Hormones and Behavior. 2012;61:631–641. doi: 10.1016/j.yhbeh.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Jawor JM, McGlothlin JW, Casto JM, Greives TJ, Snajdr EA, Bentley GE, Ketterson ED. Seasonal and individual variation in response to GnRH challenge in male dark-eyed juncos (Junco hyemalis). General and Comparative Endocrinology. 2006;149:182–189. doi: 10.1016/j.ygcen.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Kempenaers B, Peters A, Foerster K. Sources of individual variation in plasma testosterone levels. Philosophical Transactions of the Royal Society B-Biological Sciences. 2008;363:1711–1723. doi: 10.1098/rstb.2007.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterson ED, Atwell JW, McGlothlin JW. Phenotypic integration and independence: Hormones, performance, and response to environmental change. Integrative and Comparative Biology. 2009;49:365–379. doi: 10.1093/icb/icp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterson ED, Fudickar AM, Atwell JW, Greives TJ. Seasonal timing and population divergence: when to breed, when to migrate. Current Opinion in Behavioral Sciences. 2015;6:50–58. [Google Scholar]
- Ketterson ED, Nolan V. Hormones and life histories - an integrative approach. American Naturalist. 1992;140:S33–S62. doi: 10.1086/285396. [DOI] [PubMed] [Google Scholar]
- Ketterson ED, Nolan V. Adaptation, exaptation, and constraint: A hormonal perspective. American Naturalist. 1999;154:S4–S25. doi: 10.1086/303280. [DOI] [PubMed] [Google Scholar]
- Ketterson ED, Nolan V, Wolf L, Ziegenfus C. Testosterone and avian life histories - effects of experimentally elevated testosterone on behavior and correlates of fitness in the dark-eyed junco (Junco hyemalis). American Naturalist. 1992;140:980–999. [Google Scholar]
- Kitano J, Kawagishi Y, Mori S, Peichel CL, Makino T, Kawata M, Kusakabe M. Divergence in sex steroid hormone signaling between sympatric species of Japanese threespine stickleback. Plos One. 2011;6 doi: 10.1371/journal.pone.0029253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie HA, King SR. Transcriptional Regulation of Steroidogenic Genes: STARD1, CYP11A1 and HSD3B. Experimental Biology and Medicine. 2009;234:880–907. doi: 10.3181/0903-MR-97. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynn SE, Perfito N, Guardado D, Bentley GE. Food, stress, and circulating testosterone: Cue integration by the testes, not the brain, in male zebra finches (Taeniopygia guttata). General and Comparative Endocrinology. 2015;215:1–9. doi: 10.1016/j.ygcen.2015.03.010. [DOI] [PubMed] [Google Scholar]
- McGlothlin JW, Jawor JM, Greives TJ, Casto JM, Phillips JL, Ketterson ED. Hormones and honest signals: males with larger ornaments elevate testosterone more when challenged. Journal of Evolutionary Biology. 2008;21:39–48. doi: 10.1111/j.1420-9101.2007.01471.x. [DOI] [PubMed] [Google Scholar]
- McGlothlin JW, Jawor JM, Ketterson ED. Natural variation in a testosterone-mediated trade-off between mating effort and parental effort. American Naturalist. 2007;170:864–875. doi: 10.1086/522838. [DOI] [PubMed] [Google Scholar]
- McGlothlin JW, Whittaker DJ, Schrock SE, Gerlach NM, Jawor JM, Snajdr EA, Ketterson ED. Natural selection on testosterone production in a wild songbird population. American Naturalist. 2010;175:687–701. doi: 10.1086/652469. [DOI] [PubMed] [Google Scholar]
- McGuire NL, Koh A, Bentley GE. The direct response of the gonads to cues of stress in a temperate songbird species is season-dependent. PeerJ. 2013;1:e139. doi: 10.7717/peerj.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mila B, McCormack JE, Castaneda G, Wayne RK, Smith TB. Recent postglacial range expansion drives the rapid diversification of a songbird lineage in the genus Junco. Proceedings of the Royal Society B-Biological Sciences. 2007;274:2653–2660. doi: 10.1098/rspb.2007.0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SC, Koskela E, Mappes T. Intralocus sexual conflict for fitness: sexually antagonistic alleles for testosterone. Proceedings of the Royal Society B-Biological Sciences. 2012;279:1889–1895. doi: 10.1098/rspb.2011.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan V, Ketterson ED, Cristol DA, Rogers CM, Clotfelter ED, Titus RC, Schoech SJ, Snajdr E. Dark-eyed Junco (Junco hyemalis). In: Poole A, editor. The Birds of North America Online. Cornell Lab of Ornithology; Ithaca, NY. USA: 2002. [Google Scholar]
- O'Connell LA, Hofmann HA. Evolution of a vertebrate social decision-making network. Science. 2012;336:1154–7. doi: 10.1126/science.1218889. [DOI] [PubMed] [Google Scholar]
- Oklestkova J, Rarova L, Kvasnica M, Strnad M. Brassinosteroids: synthesis and biological activities. Phytochemistry Reviews. 2015;14:1053–1072. [Google Scholar]
- Oostra V, de Jong MA, Invergo BM, Kesbeke F, Wende F, Brakefield PM, Zwaan BJ. Translating environmental gradients into discontinuous reaction norms via hormone signalling in a polyphenic butterfly. Proceedings of the Royal Society B: Biological Sciences. 2011 doi: 10.1098/rspb.2010.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AH, Youngblood GL. Regulation of expression of steroidogenic enzymes in Leydig cells. Biology of Reproduction. 1995;52:217–225. doi: 10.1095/biolreprod52.2.217. [DOI] [PubMed] [Google Scholar]
- Pfannkuche KA, Gahr M, Weites IM, Riedstra B, Wolf C, Groothuis TGG. Examining a pathway for hormone mediated maternal effects - Yolk testosterone affects androgen receptor expression and endogenous testosterone production in young chicks (Gallus gallus domesticus). General and Comparative Endocrinology. 2011;172:487–493. doi: 10.1016/j.ygcen.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Pike N. Using false discovery rates for multiple comparisons in ecology and evolution. Methods in Ecology and Evolution. 2011;2:278–282. [Google Scholar]
- Rausher MD. The evolution of genes in branched metabolic pathways. Evolution. 2013;67:34–48. doi: 10.1111/j.1558-5646.2012.01771.x. [DOI] [PubMed] [Google Scholar]
- Reed WL, Clark ME, Parker PG, Raouf SA, Arguedas N, Monk DS, Snajdr E, Nolan V, Ketterson ED. Physiological effects on demography: A long-term experimental study of testosterone's effects on fitness. American Naturalist. 2006;167:667–683. doi: 10.1086/503054. [DOI] [PubMed] [Google Scholar]
- Robison OW, Lubritz D, Johnson B. Realized heritability estimates in boars divergently selected for testosterone levels. Journal of Animal Breeding and Genetics. 1994;111:35–42. doi: 10.1111/j.1439-0388.1994.tb00435.x. [DOI] [PubMed] [Google Scholar]
- Rosvall KA, Bergeon Burns CM, Barske J, Goodson JL, Schlinger BA, Sengelaub DR, Ketterson ED. Neural sensitivity to sex steroids predicts individual differences in aggression: implications for behavioural evolution. Proceedings of the Royal Society B: Biological Sciences. 2012a;279:3547–3555. doi: 10.1098/rspb.2012.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Bergeon Burns CM, Hahn TP, Ketterson ED. Sources of variation in HPG axis reactivity and individually consistent elevation of sex steroids in a female songbird. General and Comparative Endocrinology. 2013;194:230–239. doi: 10.1016/j.ygcen.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Peterson MP, Reichard DG, Ketterson ED. Highly context-specific activation of the HPG axis in the dark-eyed junco and implications for the challenge hypothesis. General and Comparative Endocrinology. 2014;201:65–73. doi: 10.1016/j.ygcen.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Reichard DG, Ferguson SM, Whittaker DJ, Ketterson ED. Robust behavioral effects of song playback in the absence of testosterone or corticosterone release. Hormones and Behavior. 2012b;62:418–425. doi: 10.1016/j.yhbeh.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran RJ, Adelman JS, McGraw KJ, Hau M. Sexual signal exaggeration affects physiological state in male barn swallows. Current Biology. 2008;18:R461–R462. doi: 10.1016/j.cub.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Sinervo B, Miles DB, Frankino WA, Klukowski M, DeNardo DF. Testosterone, endurance, and Darwinian fitness: Natural and sexual selection on the physiological bases of alternative male behaviors in side-blotched lizards. Hormones and Behavior. 2000;38:222–233. doi: 10.1006/hbeh.2000.1622. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. The genomics of gene expression. Genomics. 2004;84:449–457. doi: 10.1016/j.ygeno.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Stocco DM, Wang X, Jo Y, Manna PR. Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: more complicated than we thought. Mol Endocrinol. 2005;19:2647–59. doi: 10.1210/me.2004-0532. [DOI] [PubMed] [Google Scholar]
- Team RDC. A Language and Environment for Statistical. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- Tsutsui K, Bentley GE, Kriegsfeld LJ, Osugi T, Seong JY, Vaudry H. Discovery and evolutionary history of gonadotrophin-inhibitory hormone and kisspeptin: new key neuropeptides controlling reproduction. Journal of Neuroendocrinology. 2010;22:716–727. doi: 10.1111/j.1365-2826.2010.02018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Robison OW, Whisnant CS, Cassady JP. Effect of divergent selection for testosterone production on testicular morphology and daily sperm production in boars. Journal of Animal Science. 2004;82:2259–2263. doi: 10.2527/2004.8282259x. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Molecular Ecology. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Wingfield JC. Regulatory mechanisms that underlie phenology, behavior, and coping with environmental perturbations: an alternative look at biodiversity. Auk. 2012;129:1–7. [Google Scholar]
- Wright KM, Rausher MD. The Evolution of Control and Distribution of Adaptive Mutations in a Metabolic Pathway. Genetics. 2010;184:483–U261. doi: 10.1534/genetics.109.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Nag PK, Crews D. Species-differences in estrogen-receptor and progesterone receptor mRNA expression in the brain of sexual and unisexual whiptail lizards. Journal of Neuroendocrinology. 1995;7:567–576. doi: 10.1111/j.1365-2826.1995.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Zera AJ, Harshman LG, Williams TD. Evolutionary endocrinology: The developing synthesis between endocrinology and evolutionary genetics. Annual Review of Ecology Evolution and Systematics. 2007;38:793–817. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.