Summary
A hallmark of the well-studied vertebrate class Va myosin is its ability to take multiple steps on actin as a single molecule without dissociating, a feature called “processivity”. Thus it was surprising when kinetic and single molecule assays showed that human Myosin Vc (MyoVc) was not processive on single actin filaments [1–3]. We explored the possibility that MyoVc is processive only under conditions that resemble its biological context. Recently, it was shown that zymogen vesicles are transported on actin “superhighways” composed of parallel actin cables nucleated by formins from the plasma membrane [4]. Loss of these cables compromises orderly apical targeting of vesicles. MyoVc has been implicated in transporting secretory vesicles to the apical membrane [5]. We hypothesized that actin cables regulate the processive properties of MyoVc. We show that MyoVc is unique in taking variable size steps, which are frequently in the backward direction. Results obtained with chimeric constructs implicate the lever arm/rod of MyoVc as being responsible for these properties. Actin bundles allow single MyoVc motors to move processively. Remarkably, even teams of MyoVc motors require actin bundles to move continuously at physiological ionic strength. The irregular stepping pattern of MyoVc, which may result from flexibility in the lever arm/rod of MyoVc, appears to be a unique structural adaptation that allows the actin track to spatially restrict the activity of MyoVc to specialized actin cables in order to co-ordinate and target the final stages of vesicle secretion.
Keywords: myosin Vc, Myo5c, processivity, actin cables, secretory granules
Results and Discussion
Single MyoVc motors do not move processively on actin
Here we test the hypothesis that MyoVc can move processively as a single molecule when assayed under conditions that mimic its cellular environment during transport. We expressed a truncated, constitutively active dimeric human MyoVc construct, using the baculovirus/Sf9 cell expression system (Figure 1A and S1A). The C-terminus of the heavy chain has a biotin tag for attachment to streptavidin-quantum dots (Qdots). MyoVc was co-expressed with calmodulin that does not bind calcium (CaMΔall) [6] because it produces a more homogeneous protein preparation by reducing calmodulin dissociation from the lever arm. To further ensure full occupancy of the lever arm, excess CaMΔall was present throughout the protein preparation and in single molecule assay solutions. Total internal reflection fluorescence (TIRF) microscopy was used to track single motors with high spatial (6 nm) and temporal (33 ms) resolution [7].
Figure 1.
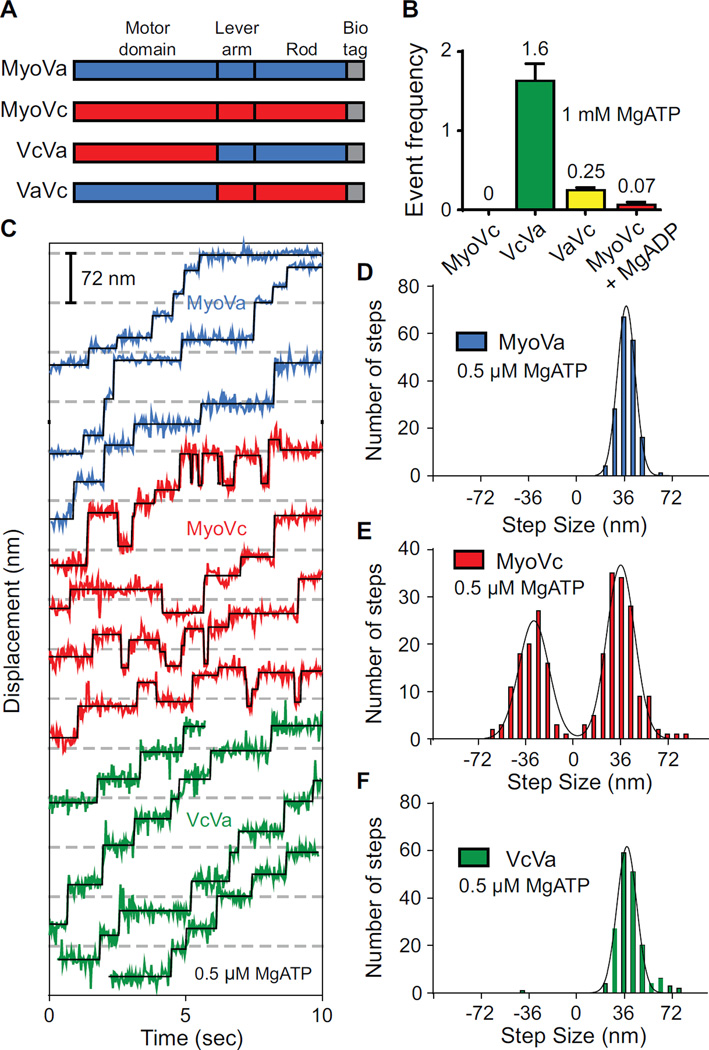
Stepping patterns of MyoVc, MyoVa, and a chimeric motor on single actin filaments.
(A) Schematic showing truncated MyoVa-HMM and MyoVc-HMM constructs used here. MyoVa-HMM (MyoVa, blue) and MyoVc-HMM (MyoVc, red) contain three domains: motor domain, lever arm and rod. The chimeric construct, VcVa contains the motor domain of MyoVc and the lever arm and rod of MyoVa. VaVc contains the motor domain of MyoVa and the lever arm and rod of MyoVc. Constructs contain a C-terminal biotin tag (gray) for attachment to streptavidin-Qdots for visualizing movement on different actin tracks.
(B) Event frequencies (µM myosin*µm actin*sec)−1 of MyoVc (0), VcVa (green, 1.6 ± 0.22), VaVc (yellow, 0.25 ± 0.033) and MyoVc + 200 µM MgADP (red, 0.067 ± 0.032) on single actin filaments. Single motor motility was done at 1 mM MgATP and 25 mM KCl.
(C) Representative displacement versus time data for MyoVa (blue), MyoVc (red) and the VcVa chimera (green) at 0.5 µM MgATP. Steps were fitted to the raw data using a step-finding algorithm (black lines). Displacements were determined to be steps only if they persisted for at least two frames (67 ms).
(D–F) Step size histograms of (D) MyoVa, (E) MyoVc and (F) a VcVa chimeric construct. Histograms were fit to single or multiple Gaussian distributions to determine the average step sizes ± SD for each construct; MyoVa, 37nm ± 6.8 (N = 173); MyoVc, 33nm ± 10.6 and -33nm ± 11.4 (N = 247); VcVa, 38nm ± 7.5 (N = 177).
See also Figures S1–S4 and Movie S1.
Consistent with previous studies [1–3], MyoVc did not move processively on individual actin filaments at 1 mM MgATP (Figure 1B). Occasionally, Qdots associated with individual actin filaments for 1.0 ± 1.4 seconds (mean ± SD), but gave no net displacement (Table 1 and Movie S1). Binding of any of three prevalent human cytoplasmic tropomyosins (Tpm3.1, Tpm1.8, and Tpm4.2) to actin did not convert MyoVc into a processive motor. We previously showed that tropomyosin was essential for a class V myosin from budding yeast (Myo2p) to move processively [8].
Table 1.
Summary of single molecule parameters
| Construct | Track | [KCl] (mM) | Run length (µm)a | Speed (µm/sec)b | N |
|---|---|---|---|---|---|
| Single motor motilityc | |||||
| MyoVc | Actin | 25 | No runs | No runs | 0 |
| MyoVa | Actin | 25 | 0.48 ± 0.02 | 0.48 ± 0.21 | 182 |
| VcVa chimera | Actin | 25 | 0.18 ± 0.004 | 0.54 ± 0.21 | 182 |
| MyoVc | Actin bundles | 25 | 0.49 ± 0.10 | 0.44 ± 0.23 | 207 |
| MyoVc, 37°C | Actin bundles | 25 | 1.26 ± 0.02 | 0.54 ± 0.34 | 206 |
| Multiple motor motilityd | |||||
| MyoVc | Actin | 25 | 0.26 ± 0.02 | 0.13 ± 0.10 | 108 |
| MyoVc | Actin | 150 | No runs | No runs | 0 |
| MyoVc | Actin bundles | 150 | 0.35 ± 0.02 | 0.16 ± 0.13 | 203 |
The characteristic run length (λ) was obtained by fitting the cumulative frequency histogram to an exponential function. Error is in standard error of the fit. No runs detected, with a minimum detectable run length of 120 nm.
Mean ± SD.
Mixing ratios of 1 myosin dimer per 5–10 Qdots ensures that the majority of Qdots are bound by a single motor. Experiments were performed at 1 mM MgATP and 22°C unless specified.
Mixing ratios of 10 myosin dimers per 1 Qdot promotes the recruitment of multiple MyoVc motors per Qdot.
The MyoVc motor domain supports processive motion when coupled to the lever arm/rod of MyoVa
Processive motion requires that one motor domain remains bound to the actin track at all times. This requires that the motor has a high duty cycle, i.e. remains tightly bound to actin for more than half of its ATPase cycle. Transient kinetic studies of a single-headed MyoVc inferred a duty cycle <50%, implying that the kinetics of the MyoVc motor domain are incompatible with processivity [1–3].
This inference was tested with a chimera composed of the MyoVc motor domain joined to the lever arm/rod of processive MyoVa (VcVa) (Figure 1A). This chimera moved processively on single actin filaments at 1 mM MgATP with a run length of 0.18 µm and average speed of 0.5 µm/sec (Table 1, Figure 1B, and S1B). This result has two important implications. First, the ability of single molecules to move processively implies that the MyoVc motor domain has a duty cycle >50%. The VcVa chimeric construct has a shorter run length than MyoVa (Table 1 and Figure S1B). Run length increases with duty cycle, and thus the MyoVc motor domain likely has a duty cycle less than the >70% duty cycle of MyoVa [9, 10], which supports a run length of 0.48 µm under comparable conditions (Table 1 and Figure S1B). The VcVa chimera moves with speeds that are similar to MyoVa, consistent with the similar maximal actin-activated ATPase rates (Vmax) for MyoVa, VcVa and MyoVc (10.9 sec−1,14.2 sec−1 and 11.2 sec−1 respectively) (Figure S2). Interestingly, the actin concentration at half-maximal ATPase (Kactin) is less for VcVa than for MyoVc. This observation implies that the MyoVa lever arm/rod either promotes binding of the second head to actin, or enhances communication between the two heads, properties that would enhance run length in single molecule assays. These experiments suggest that features in the lever arm and/or rod domains of MyoVc preclude its ability to move processively, perhaps by introducing flexibility in the molecule and diminishing inter-head coupling. Processive run lengths are enhanced when the two actin-bound heads are coupled via strain, which slows ADP release from the leading head and keeps it attached until the trailing head swings forward and rebinds to actin [11, 12]. Because of intramolecular strain, the effective duty cycle of a two-headed construct, which is predictive of processivity, cannot be determined solely from a single-headed construct [3].
The lever arm of class V myosins has six IQ motifs (consensus sequence IQxxxRGxxxR), each of which bind calmodulin. A 3 amino acid insert (KAI) in the MyoVc lever arm increases the spacing between IQ motifs 2 and 3 compared with MyoVa and MyoVb (Figure S3A). When these 3 amino acids were deleted to restore typical IQ spacing, MyoVcΔKAI remained non-processive on single actin filaments.
Kinetic studies showed that MyoVc has a high affinity for MgADP, and simulations predicted that its duty ratio could be elevated to the more than 50% needed to support processive motion in the presence of MgADP [1]. Addition of 200 µM MgADP to the single molecule motility assay produced a few runs, but they were ~20-fold less frequent than the VcVa chimera (Figure 1B). Thus MgADP is not likely to be the physiologic switch for processivity.
Is the Vc lever arm/rod sufficient to abolish processivity of MyoVa? Processive events of VaVc, a chimeric construct containing the known high duty cycle motor domain of MyoVa, and the lever arm/rod domains of MyoVc, were rare and ~7-fold reduced in run frequency compared with the VcVa chimera (Figure 1B). We conclude that MyoVc is non-processive primarily because of elements in the lever arm/rod.
The lever arm/rod of MyoVc is responsible for back steps
The stepping dynamics of MyoVc and MyoVa at low MgATP concentrations were compared to gain mechanistic information about how structural features in the rod/lever arm of MyoVc impact coordinated stepping on actin. 0.5 µM MgATP is necessary to resolve individual step displacements, and allows MyoVc to take multiple steps because nucleotide binding becomes rate limiting. Center-of-mass displacements were monitored with a Qdot bound to the C-terminus of the heavy chain. The average step size was determined by fitting a Gaussian to the step size histogram. MyoVa takes only forward steps with an average step size of 37 nm ± 6.8 (mean ± S.D.) (Figure 1C,D), consistent with previous studies under unloaded conditions [7, 13, 14]. Strikingly, we find that under the same conditions, MyoVc takes ~40% back steps (Figure 1C,E). The average size of the back steps (-33 nm ± 11.4) are approximately the same size as forward steps (33 nm ± 10.6), suggesting a reversal of the powerstroke. Similar stepping properties were observed with a MyoVc construct containing a biotin tag on the N-terminus for Qdot attachment to the motor domain (Figure S3B).
The standard deviations of the forward and back steps of MyoVc (11 nm) are larger than for MyoVa (7 nm), indicating that MyoVc has a more variable stepping pattern. Additional flexibility in the lever arm/rod may allow the motor to access multiple actin binding sites along the filament. MyoVcΔKAI, which lacks the 3 amino acid insert in the lever arm, has a step size distribution identical to that of MyoVc (Figure S3C). In contrast, the stepping pattern of the VcVa chimera was identical to that of MyoVa, with no back steps and an average step size of 38 nm ± 7.5 (Figure 1C, F). The standard deviation of the step size distribution of the VcVa chimera is similar to MyoVa (Figure 1F). The step size variability of MyoVc and its back stepping behavior thus result from structural features in the lever arm/rod, and not the properties of the motor domain per se.
One reason for the unusually high percentage of back steps for MyoVc is the frequent occurrence of back–forward and forward–back steps with ~10-fold faster lifetimes than typical MyoVc or MyoVa stepping (Figure S4A–D). Their short lifetimes likely explain why they were not observed previously [3]. The slow rate of ATP binding [1, 2] at 0.5 µM MgATP (~0.5 sec−1) suggests that these displacements are not ATP-dependent. Back steps are also observed for MyoVa under a resistive load [15], or when it steps short of its preferred actin binding site under no load [16]. These missteps are thought to result in an off-axis component to the resistive load that weakens the actomyosin bond causing leading head detachment. The broad step size distribution of MyoVc indicates that the motor frequently falls short of this preferred 36 nm pseudo-repeat of the actin filament. Back stepping under unloaded conditions may thus result from off-axis strain.
MyoVc moves processively as a single molecule on actin bundles
MyoVc appears to function predominantly in the trafficking of large (~0.5 µm) exocrine secretory granules [5, 17, 18]. Interestingly, this transport occurs on specialized ~4µm actin cables composed of multiple parallel actin filaments that emanate from the apical side of epithelial cells [4] (Figure 3F). The broad step size distribution of MyoVc suggested that it may move better on actin bundles that provide additional lateral actin binding sites, which enhance the probability of at least one head remaining bound to actin. We imaged single MyoVc motors bound to a Qdot moving on actin-fascin bundles, which are composed of multiple parallel actin filaments separated by ~9 nm [19]. Remarkably, on actin bundles, MyoVc showed long processive runs as a single molecule at 1 mM MgATP, with a characteristic run length (λ) of 0.49 µm and a speed of ~0.5 µm/sec (Figure 2A–B, Table 1, and Movie S2).
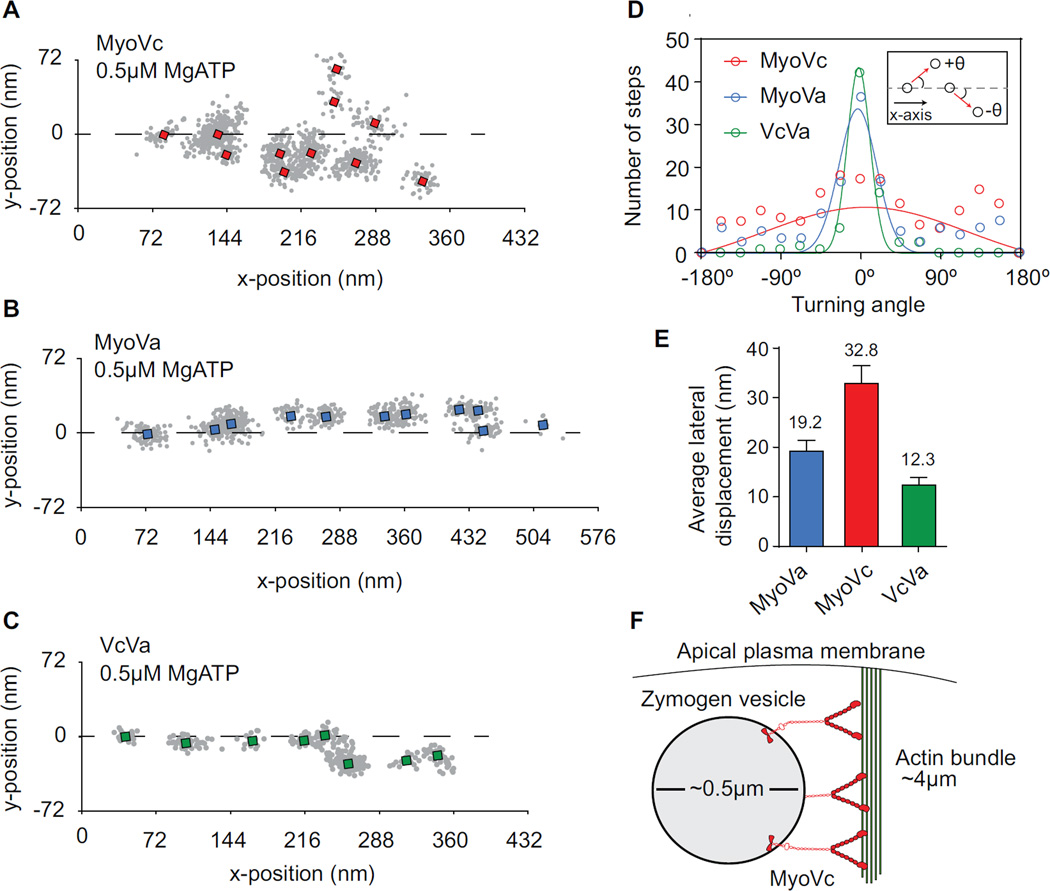
Figure 3.
MyoVc motion on actin bundles.
(A-C) Single molecule tracking of (A) MyoVc, (B) MyoVa and (C) VcVa chimera moving on an actin bundle at 0.5 µM MgATP. Run trajectories are rotated to align with the axis of the bundle (dashed line). Single x,y positions were classified manually and averaged (squares).
(D) Histogram comparing the turning angles of MyoVc (red), MyoVa (blue) and VcVa chimera (green). Turning angles (θ) were made relative to the axis of the bundle. Positive values for θ are defined as stepping to a leftward adjacent filament within the bundle. Negative values are defined as stepping to a rightward adjacent filament (inset). The average turning angle ± SD was determined for MyoVc, MyoVa and VcVa chimera by fitting the histograms to a Gaussian distribution; MyoVa, -0.8° ± 24, N = 156; MyoVc, 8.3° ± 128, N = 203; VcVa 3.6° ± 14. Standard deviations of these distributions represent variability in the motors turning angle.
(D) Average lateral displacement (nm) per step for MyoVa (blue), MyoVc (red) and VcVa chimera (green) moving on a bundle; MyoVa, 16.73 nm ± 1.30 (N = 93); MyoVc, 24.38 nm ± 2.38 (N = 76); VcVa, 12.3 ± 1.60 (N = 83).
(E) Model for actomyosin transport of secretory vesicles to the apical membrane of acinar cells.
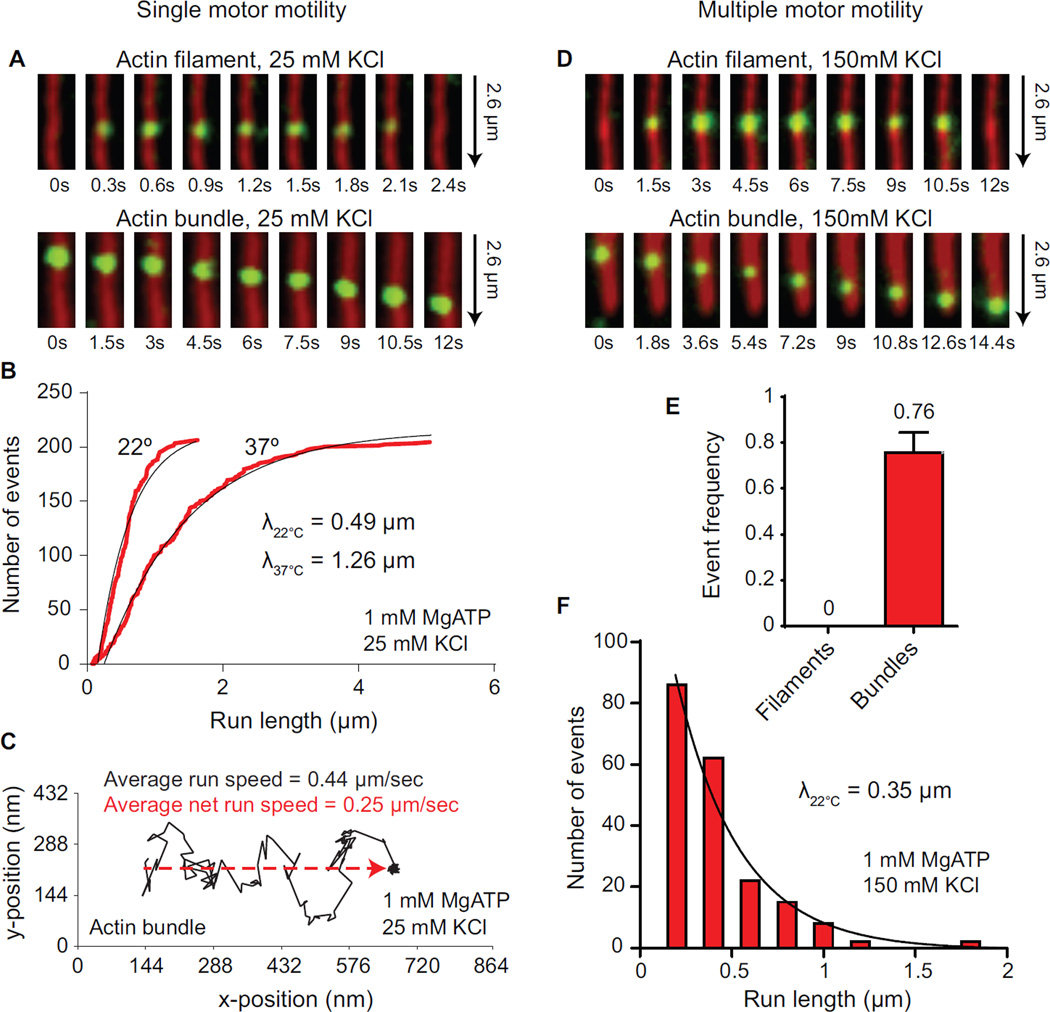
Figure 2.
Single and multiple MyoVc motors are selective for actin bundles.
(A) TIRF microscopy images showing no movement of a single MyoVc bound to a Qdot (green) on actin filaments (red, top), versus processive motion on actin bundles (red, bottom) at 1 mM MgATP, 25 mM KCl.
(B) Cumulative frequency distribution of run lengths for MyoVc at 22°C (N = 207) and 37°C (N = 206) moving on actin bundles at 1 mM MgATP. Run lengths, λ, were determined by fitting the distributions to an exponential function (black line).
(C) Representative run trajectory (black line) of a single MyoVc motor showing large lateral displacements (movement in the y-axis of the plot) on an actin bundle at 1 mM MgATP. Net distance covered along the axis of the bundle is indicated by the red dashed line. The average speed of MyoVc moving on a bundle at 22°C (0.44 µm/sec ± 0.23, N = 207) is compared to its net speed (0.25 µm/sec ± 0.15, N = 207). Error is SD.
(D) TIRF microscopy images showing no movement of multiple MyoVc motors bound to a Qdot (green) on actin filaments (red, top), versus continuous motion on actin bundles (red, bottom) at 1 mM MgATP, 150 mM KCl.
(E) Event frequencies of MyoVc motor ensembles from a representative experiment comparing motility on single actin filaments and actin bundles at physiological ionic strength. Event frequency was normalized per µM Qdot per µm actin track per s.
(F) Frequency distribution of run lengths for MyoVc motor ensembles moving on actin bundles at 1 mM MgATP, 150 mM KCl. Run length, λ, was determined by fitting the distributions to an exponential function (black line).
We investigated whether temperature could increase MyoVc run length on actin bundles. Kinetic studies showed that the ATP hydrolysis step for MyoVc is shifted to the pre-hydrolyzed form (M.ATP) [1, 2]. Increasing temperature should shift the equilibrium to the M.ADP.Pi state and favor flux through the strong-binding AM.ADP state, thus increasing duty cycle and run length [20, 21]. When temperature was increased to 37°C, MyoVc remained non-processive on single actin filaments, but run length was enhanced ~2.5-fold on actin bundles (Table 1 and Figure 2B). This observation suggests that physiologic temperature enhances the duty ratio of MyoVc, but is not sufficient for MyoVc to move processively on single actin filaments, presumably because the stepping pattern remains too variable.
MyoVc moves on actin bundles with large lateral displacements giving a serpentine-like quality to the x,y run coordinate trajectories (Figure 2C). Run speeds measured along these trajectories are consistent with stepping rates for the VcVa chimera, but the net speed along the axis of the bundle is reduced ~2-fold (Figure 2C, dashed red line), which may contribute to the slow speeds observed for secretory vesicles in MCF-7 cells and primary cultures of pancreatic acini (~30 nm/sec) [4, 5]. MyoVc motility is reminiscent of the somewhat random nature of vesicle trafficking in vivo [5], a likely consequence of back and side steps.
MyoVc motor ensembles also require actin bundles for motility at physiological ionic strength
Secretory vesicles are likely transported by multiple MyoVc motors in the cell [5]. We asked if multiple MyoVc motors showed the same requirement for bundled actin as single motors. At low ionic strength (25 mM KCl), MyoVc motor ensembles are motile on single actin filaments (Table 1), consistent with a recent study which showed that two MyoVc motors coupled via a DNA scaffold moved continuously on single actin filaments at low ionic strength [3]. This study did not explore the effect of ionic strength. We hypothesized that actin bundle track selectivity will persist even if cargoes are moved by multiple motors, provided that the ionic strength is near physiologic levels (150 mM KCl). Consistent with this idea, MyoVc motor ensembles are non-motile on single actin filaments at physiological ionic strength (Figure 2D–E, Table 1, Movie S3) but show robust motility on actin bundles (Figure 2D–F, Table 1, Movie S4) at 1 mM MgATP with a run length of 0.35 µm. Single MyoVc motors showed very few processive events on actin bundles at 150 mM KCl, with a short ~150 nm run length. These data imply that MyoVc has evolved to move in teams on actin bundles which mimic the actin cables in the cell. The localization of cables at the apical surface of the exocrine pancreas may restrict granule transport to the apical membrane by providing the only track suitable for continuous motion.
Myosin Vc has greater access to binding sites in an actin bundle
We pursued how an actin bundle could be a “processivity factor” for MyoVc. Actin bundles moderately enhance the run length of MyoVa [22] by providing additional binding sites which reduce run termination. Access to these lateral binding sites requires considerable flexibility in the motor, presumably at the lever arm-rod junction [23–26]. If MyoVc has additional flexible elements in the lever arm/rod, it should in principle have even greater access to lateral actin binding sites. To test this idea, we compared the stepping dynamics of MyoVc, MyoVa, and the VcVa chimera on actin bundles. Qdots bound to the C-terminus of the heavy chain were tracked with high temporal (33 ms) and spatial (6 nm) resolution. Representative x,y trajectories show how motors explore the actin bundle differently (Figure 3A–C). The trajectories were rotated such that the center axis of the bundle runs parallel to the x-axis. Displacement in the x-direction represents movement along a single actin filament while displacement in the y-direction requires switching to an adjacent actin filament in the bundle. MyoVc shows more lateral displacements and samples more filaments in a bundle compared to MyoVa and VcVa (Figure 3A–C). This was quantified by determining the average x,y position of each step (square) and measuring the turning angle (θ) between successive steps (Figure 3A–D). The average turning angle was determined by fitting the histogram of angles with a Gaussian. The standard deviation of this fit is a measure of the variation in the turning angle of the motor. Motors with larger turning angles more effectively explore multiple filaments in a bundle over a given time period. MyoVa and VcVa have a relatively narrow turning angle distribution (S.D. = 24° and 14° respectively) with the majority of its steps moving straight (mean = -0.8° and 3.6° respectively) (Figure 3D), consistent with previous studies [22]. MyoVc also moves with little right or left bias (mean = 8.3°) but has a very broad turning angle distribution (S.D. = 128°), indicating that the motor side steps with a higher probability compared with MyoVa and VcVa (Figure 3D).
We quantified the average lateral displacement between steps. MyoVc has a 63% longer average lateral displacement compared with MyoVa, and a 73% longer displacement compared with the VcVa chimera (Figure 3E). Taken together, these results indicate that MyoVc is more effective at accessing multiple actin filaments in a bundle than MyoVa. This property is due to the lever arm and rod domains since a chimeric construct of MyoVc containing the MyoVa lever arm/rod steps more like MyoVa on single actin filaments and actin bundles.
Unique features of MyoVc bundle selectivity
Some forced dimers of Myosin 10 (Myo10), a motor found at the tips of filopodia, move preferentially on parallel actin bundles instead of single actin filaments [27–29]. An NMR structure of the coiled-coil region of Myo10 [30] provided structural evidence consistent with bundle selectivity. Myo10 has been hypothesized to use an antiparallel coiled-coil, which could provide a “shoulder” that favors the two heads being optimally attached to adjacent filaments in a bundle, rather than to a single actin filament. We find an alternative mechanism by which MyoVc selectively moves on bundles. MyoVc forms a parallel coiled-coil, and the mechanism for bundle selectivity appears to involve additional flexibility in the lever arm/rod regions of MyoVc, which causes irregular and backward steps that cannot be accommodated on a single actin filament. Thus nature has used two very different mechanisms to achieve the same goal of specifying motor location (filopodia versus apical actin cables) by altering track morphology.
It has been inferred but not proven that MyoVc transports secretory vesicles. Our findings show that MyoVc has been tuned for transport on bundles. Given that actin cables have been recently shown to be necessary for orderly apical targeting of vesicles in vivo [4], this study provides the first supporting biophysical evidence implicating MyoVc in this process. This study also demonstrates for the first time that an actin track not only regulates the properties of single motors, but also the properties of teams of motors, a situation that more accurately reflects cargo transport in the cell [5]. We show that bundle selectivity is retained even with small ensembles of MyoVc, provided that the solvent approximates physiologic ionic strength. This study highlights an elegant but simple mechanism by which transport by a human class V myosin can be spatially localized in the cell.
Supplementary Material
Acknowledgments
The authors thank G. Kennedy for technical assistance, D. Warshaw for use of the TIRF microscope, C. Bookwalter, M. Sckolnick, H. Lu and S. Lowey for helpful discussions. This work was supported by funds from the US National Institutes of Health (GM078097 to K.M.T.).
Abbreviations
- Qdots
Quantum dots
- TIRF
total internal reflectance fluorescence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T.E.S. and K.M.T. designed experiments. T.E.S. purified proteins, conducted experiments and analyzed the data. E.B.K. cloned, expressed and purified the proteins. T.E.S. and K.M.T. wrote the manuscript.
References
- 1.Takagi Y, Yang Y, Fujiwara I, Jacobs D, Cheney RE, Sellers JR, Kovacs M. Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J Biol Chem. 2008;283:8527–8537. doi: 10.1074/jbc.M709150200. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe S, Watanabe TM, Sato O, Awata J, Homma K, Umeki N, Higuchi H, Ikebe R, Ikebe M. Human myosin Vc is a low duty ratio nonprocessive motor. J Biol Chem. 2008;283:10581–10592. doi: 10.1074/jbc.M707657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunther LK, Furuta K, Bao J, Urbanowski MK, Kojima H, White HD, Sakamoto T. Coupling of two non-processive myosin 5c dimers enables processive stepping along actin filaments. Sci Rep. 2014;4:4907. doi: 10.1038/srep04907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geron E, Schejter ED, Shilo BZ. Directing exocrine secretory vesicles to the apical membrane by actin cables generated by the formin mDia1. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1303796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs DT, Weigert R, Grode KD, Donaldson JG, Cheney RE. Myosin Vc is a molecular motor that functions in secretory granule trafficking. Mol Biol Cell. 2009;20:4471–4488. doi: 10.1091/mbc.E08-08-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE, Trybus KM. Tropomyosin is essential for processive movement of a class v Myosin from budding yeast. Curr Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 11.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 12.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci U S A. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 14.Forkey JN, Quinlan ME, Shaw MA, Corrie JE, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 15.Kad NM, Trybus KM, Warshaw DM. Load and Pi control flux through the branched kinetic cycle of myosin V. J Biol Chem. 2008;283:17477–17484. doi: 10.1074/jbc.M800539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H, Kennedy GG, Warshaw DM, Trybus KM. Simultaneous observation of tail and head movements of myosin V during processive motion. J Biol Chem. 2010;285:42068–42074. doi: 10.1074/jbc.M110.180265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez OC, Cheney RE. Human myosin-Vc is a novel class V myosin expressed in epithelial cells. J Cell Sci. 2002;115:991–1004. doi: 10.1242/jcs.115.5.991. [DOI] [PubMed] [Google Scholar]
- 18.Marchelletta RR, Jacobs DT, Schechter JE, Cheney RE, Hamm-Alvarez SF. The class V myosin motor, myosin 5c, localizes to mature secretory vesicles and facilitates exocytosis in lacrimal acini. Am J Physiol Cell Physiol. 2008;295:C13–C28. doi: 10.1152/ajpcell.00330.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa R, Sakamoto T, Ando T, Higashi-Fujime S, Kohama K. Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. J Neurochem. 2003;87:676–685. doi: 10.1046/j.1471-4159.2003.02058.x. [DOI] [PubMed] [Google Scholar]
- 20.Kodama T. Thermodynamic analysis of muscle ATPase mechanisms. Physiol Rev. 1985;65:467–551. doi: 10.1152/physrev.1985.65.2.467. [DOI] [PubMed] [Google Scholar]
- 21.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 22.Yusuf Ali M, Previs SB, Trybus KM, Sweeney HL, Warshaw DM. Myosin VI has a One Track Mind Versus Myosin Va When Moving on Actin Bundles or at an Intersection. Traffic. 2012 doi: 10.1111/tra.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 24.Shiroguchi K, Kinosita K., Jr Myosin V walks by lever action and Brownian motion. Science. 2007;316:1208–1212. doi: 10.1126/science.1140468. [DOI] [PubMed] [Google Scholar]
- 25.Fujita K, Iwaki M, Iwane AH, Marcucci L, Yanagida T. Switching of myosin-V motion between the lever-arm swing and brownian search-and-catch. Nat Commun. 2012;3:956. doi: 10.1038/ncomms1934. [DOI] [PubMed] [Google Scholar]
- 26.Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 27.Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci U S A. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricca BL, Rock RS. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys J. 2010;99:1818–1826. doi: 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vavra KC, Xia Y, Rock RS. Competition between Coiled-Coil Structures and the Impact on Myosin-10 Bundle Selection. Biophys J. 2016;110:2517–2527. doi: 10.1016/j.bpj.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q, Ye F, Wei Z, Wen Z, Zhang M. Antiparallel coiled-coil-mediated dimerization of myosin X. Proc Natl Acad Sci U S A. 2012;109:17388–17393. doi: 10.1073/pnas.1208642109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.