Abstract
Asthma’s a problem of inhaling too much Hyperinflation, air trapping, and such. But physicians find themselves just as aghast When a patient finally exhales his last. So which deserves the frowning pout: Breathing in or breathing out? So it is with the mitochondrion, Which is better, fission or fusion? Both out and in make good respiration Ditto for mitochondrial undulation. Roberta A. Gottlieb
Keywords: mitochondria, fission, fusion, mitophagy, biogenesis, metabolism, retrograde signaling, mitochondrial unfolded protein response
I. Introduction
Mitochondria can present many different faces depending on environmental conditions and cellular requirements. They change their structure, their proteome, their metabolism, and even their genome, in response to cellular and environmental cues. As they remodel, they in turn alter cellular programs and capabilities. Mitochondrial remodeling is a component of the pathophysiology of diverse diseases, including neurological, cardiovascular, metabolic and oncologic. In this setting, mitochondrial alterations may be either secondary processes or primary mechanisms due to mutations in genes that regulate mitochondrial remodeling. However, the role of mitochondrial remodeling in normal physiological processes is gaining increased recognition. In this review we will cover structural remodeling, comprising fusion and fission; mitochondrial turnover accomplished by mitophagy and biogenesis; proteomic alterations and posttranslational modifications; metabolic alterations; reactive oxygen species; and retrograde signaling whereby mitochondria modify the nuclear program.
II. Mitochondrial Structural Remodeling: rearranging and recycling
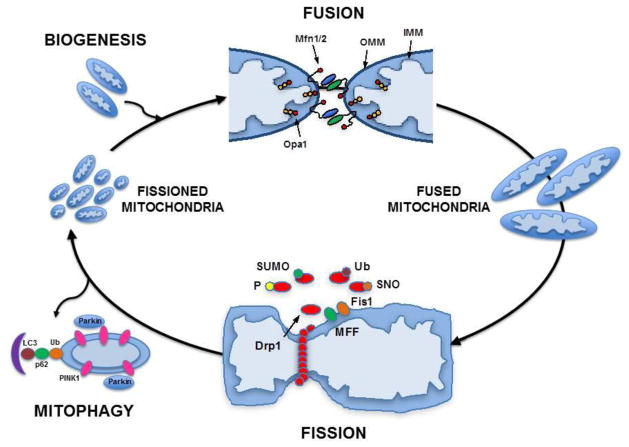
The dynamic nature of mitochondrial morphology was apparent even to the 19th century scientists who first proposed the name: mitos (thread-like) and chondrion (granule-like) (1, 2). Mitochondrial dynamics are those processes that regulate mitochondrial morphology, including: (a) Mitochondrial fission: the process that creates more numerous, smaller and more circular mitochondria; (b) Mitochondrial fusion: the process that creates fewer, larger and more elongated mitochondria; (c) Mitophagy: the process of removing damaged mitochondria; and (d) Mitochondrial biogenesis: the process that increases mitochondrial number and/or volume through increased expression of both mitochondrial and nuclear transcripts. This is illustrated in Fig. 1.
Figure 1. Mitochondrial dynamics.
The opposing processes of mitochondrial fission and fusion are both involved mitochondrial quality control and the maintenance of normal cellular homeostasis. Fission is regulated by interaction of mitochondrial proteins (Fis1 or MPP) with the cytosolic protein Drp1, after its translocation to the mitochondria fraction. Post-translational modifications (phosphorylation, sumoylation, s-nitrosylation and ubiquitination) regulate this process in response to intracellular cues, including alterations in Ca2+. When fission occurs as a result of pathologic stress, mitochondria with low membrane potential are marked for mitophagy by accumulation of Pink1, recruitment of Parkin, and then eliminated through autophagic sequestration, lysosomal fusion and degradation. Fusion is regulated by the interaction of mitochondrial inner (Opa1) and outer membrane proteins (Mfn1/2). Biogenesis is the process that results in synthesis of new mitochondrial components and is regulated by the PPARγ coactivator (PGC) family of transcriptional coactivators, PGC-1α, PGC-1β, and the PGC-related PRC. (Based in part on Archer et al. NEJM 2013 (91)).
A. Mitochondrial fission is mediated by dynamin-1-like protein (Drp1), a member of the dynamin superfamily, consisting of a GTPase and GTPase effector domain separated by a helical segment. Fission occurs when Drp1 is translocated from the cytosol to the outer mitochondrial membrane (OMM) leading to a complex, and as yet still not fully understood, interaction with three integral OMM proteins, Fis1, Mff, and MIEF1.
Fis-1. Fis1 is the most evolutionarily conserved pathway, and is a major regulator of fission in yeast. In yeast cells, two adapter proteins (Mdiv1 and Caf4) link Drp1 and Fis1, although similar adapters have not been found in mammalian cells. The role of Fis1 in fission in mammalian cells is still controversial, with some studies supportive (overexpression of Fis1 induces fragmentation; knockdown of Fis1 results in elongation (3–5)), yet others arguing against a role (the absence of bridging proteins; Fis1 is uniformly distributed on the OMM, whereas when Drp1 is translocated, it localizes to punctate structures; altering the level of Fis1 does not affect the distribution of Drp1 on the OMM (5, 6); ablation of Fis1 does not alter mitochondrial morphology or block fission (7)). It is possible, therefore, that Fis1 may play a role in recruitment of Drp1 in certain cell types and under certain stressors, but not others (8, 9). Some of the strongest evidence confirming the role of Fis1 in mammalian cells comes from studies showing that a peptide designed to specifically inhibit the Drp1-Fis1 interaction can block fission in both cardiomyocytes and neuronal cells, attenuating ischemic injury or neurodegeneration, respectively (10–12).
Mff is an OMM protein not found in yeast. Knockdown of Mff leads to increased mitochondrial fusion whereas overexpression leads to fragmentation. However, Mff and Fis1 are located in different complexes on the OMM (13) and Mff-mediated fission is independent of Fis1 (7, 14), suggesting that these two proteins play different roles or activate fission in response to different stressors.
MIEF1 (mitochondrial elongation factor 1). Also known as MiD51 and MiD49 (mitochondrial dynamic proteins), MIEF1 is co-localized with Drp1 in punctate structures on the OMM. Overexpression of MIEF1 recruits Drp1 but, intriguingly, induces mitochondrial elongation rather than fragmentation; similarly, knockdown of MIEF1 leads to fragmentation. As well, the recruitment of Drp1 by MIEF1 is independent of Drp1 phosphorylation or its GTPase activity (15). The current paradigm is that although MIEF1 recruits Drp1, it sequesters it from interaction with Fis1, thus inhibiting its activity. MIEF1 also associates with Fis1 in a Drp1-independent manner (15), competing for MIEF1 binding with Drp1, and thus attenuating the inhibitory effects of MIEF1 on Drp1.
Post-translational modifications (PTMs) are key to the regulation of mitochondrial dynamics. Drp1 is activated/deactivated via several post-translational modifications (16, 17): Phosphorylation at Ser637 by PKA inhibits fission, whereas phosphorylation at Ser616 by the cell cycle regulator cyclin B1/Cdk1 activates fission (18). Intracellular Ca2+ modulates fission at two sites: calcineurin-mediated dephosphorylation (pro-fission) at Ser637 (19, 20) and CaMK phosphorylation (pro-fission) at Ser616 (21). Additional PTMs regulating fission include Drp1 sumoylation (pro-fission) by SUMO1, MAPL and SENP5 (22–24); and S-nitrosylation by nitric oxide (25). The ubiquitin-proteasomal system also plays a role in regulation of fission, with Drp1 ubiquitinated by Parkin (26) and by RING-CH protein MARCH5 (27, 28).
B. Mitochondrial fusion is mediated by the coordinated interaction of mitofusins on the OMM and OPA1 on the inner mitochondrial membrane (IMM).
Mitofusin exists in two isoforms, Mfn1 and Mfn2, anchored in the OMM by two transmembrane domains and containing an N terminal GTPase domain. Mitofusins form homo- or heterodimers that tether together adjacent mitochondria, promoting fusion, a process requiring activation of the GTPase and the presence of Opa1. The roles of the two isoforms are not identical, as knockout of Mfn1 results in more fragmentation than does Mfn2. Knockout of both results in embryonic lethality (29). Mfn2 is also critical for the tethering of mitochondrial to the sarcoplasmic reticulum in cardiomyocytes, enhancing mitochondrial Ca2+ signaling (30).
Opa1 (Mgm1p in yeast) is a dynamin-related GTPase located in the IMM, containing a mitochondrial targeting sequence (MTS) at its N-terminal which is cleaved after import. There are eight different isoforms, both long and short, with the short form lacking the transmembrane anchor. Processing of OPA1 between long and short forms helps to regulate the balance between fusion and fission. Two IMM peptidases OMA1 and YME1L, convert long OPA1 into short isoforms, with fusion dependent on long OPA1, and fission dependent on the short isoforms (31–33). Opa1 activity requires Mfn1, but not Mfn2. In yeast, Ugo1p acts as an anchor between the Mfn equivalent (Fzo1p) and Opa1 equivalent (Mgm1p), however, no such homologue has been found in mammalian cells. Knockout of Opa1 in cultured cells results in fragmentation, whereas knockout in mice is embryonic lethal (33).
MIEF1(mitochondrial elongation factor 1): In addition to inhibiting Drp1 (above), MIEF1 promotes fusion, even in the absence of Mfn2, and overexpression of MIEF1 can reverse the fragmentation associated with Mfn2 deficiency (34).
C. Mitophagy is a process for clearing damaged mitochondria, and a critical component of normal mitochondrial quality control (35). Traditionally, mitochondrial fragmentation has been associated with pathologic insults such as ischemia (36, 37) or diabetes (38), and thought to be deleterious, due to the propensity for fragmented mitochondria to produce greater reactive oxygen species (ROS) and have lower mitochondrial membrane potential (Δψm) (10, 39). In this setting, fission is asymmetrical, segregating portions of mitochondria with low membrane potential (Δψm) from those with normal Δψm. The low Δψm daughter mitochondrion is then marked for elimination by mitophagy (40) through accumulation of PINK1 and recruitment of Parkin (41, 42), resulting in autophagic sequestration, lysosomal fusion and degradation.
In addition to the role of PINK1 and Parkin in regulating mitophagy, both proteins play a role in mitochondrial functional regulation by directing the localized translation of nuclear-encoded respiratory chain complex mRNAs on the OMM. This process, mediated by displacement of translational repressors, links dynamic regulation of mitochondrial quality control and oxidative phosphorylation (OXPHOS) (43, 44). Parkin-dependent mitophagy has also been implicated in metabolic programming, e.g. that which occurs in the heart during the transition from fetal (carbohydrate-based) to post-natal (fatty acid-based) metabolism (45).
PINK1 (PTEN-induced putative kinase 1) is a OMM serine/threonine kinase with an N-terminal mitochondrial targeting sequence (MTS), a transmembrane domain and a cytosolic-facing C-terminal kinase domain. However, PINK1 is also located in the cytoplasm and its recruitment to the OMM is thought to be a mechanism by which cells distinguish functional vs. dysfunctional mitochondria. Healthy mitochondria maintain Δψm, which enhances the import of PINK1 to the IMM, where it is cleaved by PARL and cleared. When mitochondria are stressed or damaged, Δψm decreases, leading to decreased PINK1 import and accumulation on the OMM, where it can recruit Parkin. PINK1 may also function as a vesicular trafficking pathway, segregating oxidized and damaged proteins for lysosomal degradation, another mechanism by which it regulates mitochondrial quality control (46).
Parkin is a cytosolic E3 ubiquitin ligase, which translocates to damaged mitochondria when PINK1 accumulates on the OMM, ubiquitinating OMM proteins and inducing engulfment of mitochondria by autophagosomes (mitophagy) through p61 and LC3 (41, 42). Parkin also regulates mitochondrial dynamics by the ubiquitination and degradation of other fission and fusion proteins: Mfn1/2, Drp1 and Fis1(26, 47–49). Knockdown of either PINK1 or Parkin leads to mitochondrial fragmentation (50, 51).
Other proteins such as Nix and Bnip3 (BH-3 pro-apoptotic proteins), can also function as autophagy receptors by binding to LC3 on the autophagosome and targeting mitochondria to autophagosomes (52, 53).
D. Mitochondrial biogenesis
Mitochondrial biogenesis is controlled by the PPARγ coactivator (PGC) family of transcriptional coactivators, most importantly PGC-1α, PGC-1β, and the PGC-related coactivator PRC. PGC-1α works in tandem with nuclear respiratory factor 2 (NRF-2) to co-activate NRF-1 (54). NRFs direct transcription of nuclear encoded mitochondrial proteins, the mitochondrial protein import machinery, and co-factors required for assembly of the respiratory chain complexes, as well as the regulatory factors required for mitochondrial DNA transcription and translation, most importantly mitochondrial transcription factor A (Tfam), which is also responsible for maintenance of mtDNA copy number (55). Overexpression of PGC-1α is sufficient to drive mitochondrial biogenesis (56). While acetylation of PGC1α suppresses its transcriptional coactivator activity, histone deacetylase sirtuin 1 (Sirt1) can activate PGC1α and mito-biogenesis (57). PGC-1α also responds to a variety of exogenous cues including ROS/RNS, cold exposure, endurance exercise, and intracellular signaling downstream of cAMP.
Mitochondrial biogenesis requires coordinated synthesis and assembly of nuclear-encoded and mitochondrial-encoded subunits of the OXPHOS complexes. Excess or unincorporated subunits are degraded by matrix proteases, and the resulting peptides have been shown (in yeast and C. elegans) to trigger the mitochondrial unfolded protein response (see Section VIIA). Nuclear-encoded mRNAs are translated on ribosomes in cytosol or associated with the mitochondrial outer membrane, and in most cases, are co-translationally imported through the TOM (translocase outer membrane) and TIM (translocase inner membrane) complexes. Import of some, but not all, proteins requires adequate mitochondrial membrane potential, ensuring that newly-synthesized proteins are not delivered to senescent mitochondria (58). Information contained in the 3′UTR and in the mitochondria-targeting sequences of the protein-encoding portion of the mRNA is sufficient to target the mRNA or nascent protein to the mitochondria. Additional information, usually in a cleavable pre-sequence, directs the protein to the outer membrane, the intermembrane space, the inner membrane, or the matrix. Peptidases may cleave the signal sequence and additional chaperone machinery assists with protein folding and insertion of prosthetic groups such as FAD, Fe-S clusters, and heme groups (59). Codon usage in mitochondria differs from cytosolic ribosomes, and mitochondria encode their own tRNAs as well as rRNAs. There are 13 mitochondria-encoded proteins that are essential subunits of Complexes I, III, IV, and V. Mitochondrial mRNAs are generated by a mitochondrial RNA polymerase; translation of individual mRNAs is governed by specific and rate-limiting translational activators which may govern several subunits of a Complex (60). The resulting mRNAs are translated on ribosomes associated with the inner mitochondrial membrane Specific assembly factors for OXPHOS complexes are nuclear-encoded and facilitate the assembly of the various nuclear and mitochondrial-encoded subunits into functional complexes (61–63). The complexes are then arranged into supercomplexes or respirasomes, for instance I+III2+IV (64).
There is also post-transcriptional control of mitochondrial biogenesis. Nuclear-encoded mRNAs destined for the mitochondria are associated with ribosomes located in proximity to the mitochondrial outer membrane. Many mitochondrial-targeted mRNAs have a half-life in excess of 24h (65) and may be stored on mitochondria-associated polysomes, in stress granules, or cytosolic processing bodies (P-bodies) (66). Stress granules and P-bodies are known for their role in RNA degradation; the related GW-bodies are important role for mRNA storage (67). Interestingly, P-bodies have been described to interact frequently with mitochondria (68) and more recently, both miRNAs and Ago2, a key protein in P-bodies, have been identified in mitochondria and mitoplasts, where they have been shown to regulate mitochondrial mRNA translation (69). During muscle differentiation, muscle-specific miR-1 enhanced translation of mitochondrial transcripts. Protein levels of ND1 and COX1 increased during differentiation without a significant increase in mRNA levels or mtDNA copy number. In the absence of the P-body scaffold protein GW182, miRNAs interacting with Ago2 can enhance translation by binding to an mRNA target lacking a 5′cap and 3′poly-A tail, features characteristic of mitochondrial transcripts. As GW182 has not been detected in mitochondria, it seems plausible that miRNAs present in mitochondria may enhance rather than suppress translation of mitochondrial transcripts.
E. The balance between fission and fusion in the mitochondrial response to stress
That both fission and fusion are critical to normal cellular function is highlighted by the lethality induced by knockout of Mfn1/2, Opa1 or Drp1 (70–76) and the severe diseases caused by mutations in these proteins (77–79). Mitophagy is also critical for metabolic remodeling (45). The effects of inhibiting fission, however, have been variable, depending on the method used and cell type. Cardiac-specific Drp1 knockouts show preservation of mitochondrial function, but increased mitophagy and eventual heart failure due to opening of the mitochondrial permeability transition pore (MPTP) (71). In contrast, the Drp-1 inhibitor Mdivi decreases mitophagy and rescues heart failure induced by pressure overload (80). Others have shown that disruption of fission decreases mitophagy but results in the accumulation of dysfunctional mitochondria (35, 50, 81). Thus, any disruption of baseline dynamics appears to be deleterious. However, as all of these studies have relied on genetic manipulation of dynamic regulators, the role of fission in normal “healthy” physiologic adaptation is still unknown (82).
Mitochondrial dynamics may also be coupled to respiratory chain function by altering the structure of OXPHOS supercomplexes, controversial protein superstructures that are thought by some to play a role in vivo regulating respiratory function (83). For example, ablation of the IMM fusion regulator Opa1 results in disruption of normal cristae architecture, impairs the assembly of respiratory chain supercomplexes, and alters mitochondrial metabolism (84). The formation of respiratory supercomplexes may themselves alter Opa1 oligomerization, for example during starvation, resulting in narrowing of the cristae, and increasing ATP synthase activity (85).
F. Mitochondrial dynamics and human disease
Pathologic alterations in mitochondrial dynamics can result in impaired bioenergetics and mitochondrial-mediated cell death, and are associated with a wide spectrum of pathologies, including ischemic cardiomyopathy, diabetes, pulmonary hypertension, Parkinson’s and Huntington’s diseases and skeletal muscle atrophy (37, 39, 86–92). Diseases where specific mutations of genes encoding mitochondrial dynamic regulators have been found include Mfn2 in Charcot-Marie-Tooth neuropathy (93) and OPA1 in autosomal optic atrophy (94). Alzheimer’s disease has been associated with amyloid-β mediated S-nitrosylation of Drp1 (SNO-Drp1) and resultant mitochondrial fragmentation. Neurons are particularly sensitive to changes in mitochondrial function due to their limited capacity for glycolysis and high level of energetic use. Cardiomyocytes, which have one of the highest volume fractions of mitochondria due to the high energy requirements of rhythmic contraction, are also uniquely sensitive to mitochondrial dynamic alterations. The compact organization of the cardiomyocyte sarcomeric structure has led some to suggest that mitochondrial morphologic changes are minimal in the heart, and that dynamic regulators are mostly important for maintaining quality control (95, 96). Although the degree of mitochondrial fragmentation is less in the cardiomyocyte compared to many other cell types, there is still abundance evidence for active mitochondrial structural dynamics in the heart (37, 97–100). In normal physiology, fission plays a role in the post-natal closure of the ductus arteriosus, where redox sensing in the arterial smooth muscle wall is dependent on Drp1 activation (101). Mitochondrial dynamic alterations also play a role in many cancers, where fusion can reduce the susceptibility of cells to apoptotic signaling, and inducing aerobic glycolysis (the Warburg effect) (102, 103). These alterations are thought to be a direct effect of cancer-causing mutations, a secondary effect of tumor hypoxia, or a potentially a mechanism to enhance cancer cell growth by reducing apoptotic cell death.
G. Alterations in mitochondrial dynamics as a normal physiologic response to stress
Studies in skeletal muscle during exercise suggest a role for mitochondrial fragmentation in maintaining energetic homeostasis, evidenced by an increase in Fis1 (104, 105). During recovery, fusion is activated (increased Mfn1/2), and with chronic exercise, mitochondrial biogenesis (PGC-1α) (106–108). However, as none these studies have examined mitochondrial morphology or function, the data remain incomplete. Similar cycles of fission and fusion induce mitochondrial metabolic reprogramming, as seen after starvation and re-feeding (109). Cardiac mitochondrial biogenesis has been demonstrated in chronic exercise conditioning (110, 111). Our preliminary data in the heart suggest that mitochondrial fission may be a component of the normal adaptation to increased energetic demand, such as occurs during exercise, a process we call “physiologic fragmentation” (112). In contrast to pathologic fragmentation, exercise-induced fragmentation is associated with normal or enhanced, rather than degraded, mitochondrial function, maintenance of normal Δψm and ROS production, and repression, rather than activation, of mitophagy.
III. Post-translational modification (PTM) of the mitochondrial proteome (redecorating): regulation of both mitochondrial function and dynamics
A. Role of calcium in mediating mitochondrial dynamics and function
Ca2+ is a major regulator of both ATP production and mitochondrial dynamics (113, 114). In organs like the heart, where energy needs can rapidly change, β1-adrenergic receptors (ARs) increase [Ca2+]i in response to increased workload, mediated through activation of L-type Ca2+ channels, increasing extracellular Ca2+ import and also by post-translational modification of proteins that control exchange of Ca2+ across the sarcoplasmic reticulum (e.g. ryanodine receptors [RyR] and the sarcoplasmic reticulum calcium transport ATPase [SERCA]). Changes in [Ca2+]i are rapidly sensed by mitochondria, the consequence of peri-mitochondrial microdomains where local [Ca2+] can be many-fold higher than the rest of the cytosol, although this relationship varies dramatically between cell types (115, 116). In excitable cells such as cardiomyocytes, Mfn2 tethers mitochondria to the sarcoplasmic reticulum (30), the microdomain where Ca2+-induced Ca2+ release is localized. Additional mechanisms for Ca2+-mediated alterations in mitochondrial structure and function include through PKA-AKAP, Ca2+-induced post-translational modifications and the mitochondrial Ca2+ uniporter (MCU) (113, 114, 117, 118) (see below).
Mitochondria also serve as a major buffer for intracellular Ca2+, protecting cells against Ca2+ overload, and as a mechanism for Ca2+ compartmentalization within cellular microdomains (115). In pancreatic acinar cells, for example, mitochondria prevent the cell-wide propagation of Ca2+ signaling away from the apical region (119). Mitochondrial Ca2+ uptake occurs through the MCU, but given the low affinity of this uniporter, uptake is dependent on localization of mitochondria near high Ca2+ microdomains. Uptake through the MCU is also highly dependent on Δψm, which is in turn driven by mitochondrial respiration, and is inhibited by magnesium, and by the OXPHOS uncouplers dinitrophenol and carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (120). Mitochondrial Ca2+ uptake is also mediated by the Letm1 Ca2+/H+ antiporter, coupling Ca2+ uptake to mitochondrial alkalinization, and functioning at much lower (nanomolar) concentrations than the MCU (121).
Mitochondrial release of Ca2+ is mediated in excitable (but not in non-excitable) cells, by the mitochondrial Na+-Ca2+ exchanger, NCX (122), although in pathological conditions, such as ischemia, this exchanger may operate in reverse mode, increasing mitochondrial Ca2+ overload (123, 124). Ca2+ is also extruded via opening of the MPTP (125), which is usually associated with mitochondrial stress and the initiation of apoptotic cell death. Although ablation of the MCU does not alter mitochondrial morphology, it does reduce exercise capacity (114), suggesting that altering mitochondrial Ca2+ does affect oxidative function, at least under stress conditions. Increased Ca2+ enhances mitochondrial function by activating pyruvate (126), isocitrate and oxoglutarate dehydrogenases (127). As well, increasing Ca2+ increases mitochondrial fragmentation (128) through calcineurin (removing an inhibitory phosphate on Drp1) or CaMK (phosphorylating Ser600) (21, 129). This process, although generally associated with pathologic stress, may also play a role in the response to normal physiologic stresses (see below (112)). In contrast, excessive Ca2+ opens the MPTP and can trigger cell death (36), highlighting the danger of excessive Ca2+, a consequence of chronic β-AR signaling in diseases such as heart failure (130).
B. Post-translational modifications of the mitochondrial proteome
These have been described during both physiologic (fasting) and pathologic (ischemia/reperfusion) stress. As described above, PTMs of Drp1 are a major regulator of the balance between fission and fusion (19, 21, 24, 131). PTM of mitochondrial proteins differs between low vs. high level stresses, e.g. normal levels of ROS act as a signaling second messenger, but excessive ROS leads to oxidative PTMs (oxPTMs) (132). Phosphorylation is also a regulator of mitochondrial function, including complexes I, IV and V, VDAC and ANT, targeted by β-AR-regulated kinases, e.g. PKA, PKC and GSK3β. Modern proteomic techniques, e.g. multiple-reaction monitoring (133–135), have identified >200 different phospho-transitions of proteins in the TCA cycle, and pyruvate dehydrogenase and branched-chain α-keto acid dehydrogenase complexes (118, 133, 135–138). Bioinformatic site analysis suggests the involvement of PKA, PKC, casein kinase II, DNA-dependent kinase, tyrosine kinase, Src kinase, GSK-3, ERK1/2, and EGFR (118). Several of these kinases and phosphatases are either tethered to or are translocated to the OMM where they target membrane proteins such as Bcl-2, BAD, VDACs 1–3 and TOM70 (118). For example, the A kinase anchoring protein (AKAP) family of scaffolding proteins localize cAMP-dependent PKA to the OMM (139). Finally, there are many mitochondrial proteins with oxPTMs, some of which can modulate the phosphorylation status and activity of target proteins, such as ATP synthase (140–143).
PTM of subunits of ATP synthase (complex V) have also been shown to play a role in enhancing mitochondrial respiration. Many of these PTMs are altered during the metabolic remodeling that is associated with diseases such as heart failure, leading to an increase in anaerobic metabolism and a shift from fatty acid utilization to glucose utilization. Whether this shift represents a component of the more global “recapitulation of fetal pathways” (144) or is a mechanism to improve energy economy (145) is still being debated, and there are even contradictory studies showing an increase, not a decrease, in fatty acid oxidation in human heart failure (146, 147). Phosphorylation of the E1 subunit of the pyruvate dehydrogenase complex by pyruvate dehydrogenase kinase (PDK) results in its inactivation, one of the first demonstrations of the role of PTMs in mitochondrial energetics (148). Pyruvate dehydrogenase phosphatase, which mediates activation of this complex, is regulated by intracellular Ca2+ (149). Other Ca2+-mediated phosphatases, targeted to mitochondria, include regulators of branched-chain amino acid catabolism (150).
Post-translational control of mitochondrial function may also be mediated directly by intramitochondrial PKA. Activation of soluble adenylyl cyclase directly in the mitochondrial matrix by [Ca2+]mt has been shown to increase mitochondrial cAMP formation, which in turn increases mitochondrial ATP production (60–63). However, the role of direct mitochondrial PKA in these processes is still debated. The identification of true mitochondrial-associated signaling systems is complicated by the difficulty in separating mitochondrial membranes from mitochondrial-associated membranes containing endoplasmic reticulum (ER)-associated signaling molecules, and by conflicting results, as has been the case for the putative mitochondrial angiotensin system (64, 151).
Therapeutic maneuvers can reverse many disease-specific deleterious PTMs. In heart failure, cardiac resynchronization therapy (CRT) induces dramatic changes in the mitochondrial proteome, including increases in expression and PTM of most respiratory chain enzymes, and in metabolic pathway enzymes responsible for replenishing Krebs cycle intermediates, including pyruvate carboxylase, pyruvate dehydrogenase, α-keto acid dehydrogenase, ferredoxin and fatty acid-binding protein (142). Phosphorylation of the ATP synthase-β subunit has been shown to be a major regulator of complex V activity. Additional pathways that were upregulated included proteins that mediate mitochondrial protein synthesis and import, mitochondrial membrane channels, and ROS scavenging proteins such as thioredoxin-dependent peroxide reductase (142).
IV. Metabolic alterations in fuel utilization
Mitochondria, as the major site of ATP production, both respond to fuel availability and determine the extent and type of fuel utilization in the cell. Mitochondria can utilize glucose, fatty acids, amino acids, and ketone bodies for fuel, and there are complex inter-regulatory mechanisms that determine fuel choices. These choices will result in global metabolic alterations to the cell, with far-reaching consequences.
A. Glucose utilization
Glucose may be metabolized through anaerobic glycolysis, siphoned off for the hexose monophosphate shunt, or directed into oxidative phosphorylation in the mitochondria. When oxidative phosphorylation is inhibited (e.g., by oligomycin), the cell rapidly switches to anaerobic glycolysis, resulting in the buildup of lactate. When glucose is directed by the rate-controlling enzyme glucose-6-phosphate dehydrogenase (G6PD) into the cytosol-located hexose monophosphate shunt (also known as the pentose phosphate pathway), it generates reducing equivalents in the form of NADPH and ribose 5-phosphate, an essential precursor for nucleotide biosynthesis. G6PD is activated by NADP+ and inhibited by acetyl-CoA. G6PD is also activated by deacetylation mediated by SIRT2. Hexokinase II plays a key role in glucose utilization: when this enzyme is associated with the mitochondrial outer membrane, glucose is utilized for glycolysis; when it is free in the cytosol, glucose is stored as glycogen (152). A key enzyme complex linking glycolysis to mitochondrial oxidative phosphorylation is the pyruvate dehydrogenase complex, responsible for converting pyruvate into acetyl-CoA in the mitochondrial matrix, where it is used in the tricarboxylic acid cycle for oxidative phosphorylation. The pyruvate dehydrogenase complex is regulated by phosphorylation mediated by pyruvate dehydrogenase kinase, which inactivates it. Calcium affects the rate of mitochondrial oxidative phosphorylation through multiple sites including dehydrogenases, the cytochrome chain, the F(1)F(o) ATPase, and substrate delivery to the mitochondrial matrix (153).
B. Fatty acid oxidation
As mitochondria can generate ATP from a variety of fuels, the ability to dynamically tune fuel utilization will increase cellular adaptability and is known as metabolic flexibility. The pyruvate dehydrogenase complex and its regulatory kinases and phosphatases are key determinants of metabolic flexibility. Phosphorylation of PDC will shift the mitochondria towards fatty acid oxidation, whereas its dephosphorylation will restore glucose oxidation. The generation of malonyl-CoA from acetyl CoA by acetyl CoA carboxylase will prevent fatty acid oxidation through inhibition of carnitine palmitoyltransferase I, the major shuttle for fatty acids into the mitochondria (154). Thus this critical mitochondrial enzyme complex influences glycolysis, glucose oxidation, fatty acid oxidation, lipid storage, and redox poise.
V. Metabolic output
Production of ATP is the prime metabolic output. Additional consequences of glycolysis and oxidative phosphorylation include shifts in pH and the redox poise. Reactive oxygen species, a byproduct of oxidative phosphorylation will be discussed in the next section.
A. ATP production
Approximately 80% of the high-energy phosphates (ATP and creatine phosphate) in the heart are derived from oxidative phosphorylation of fatty acids, with the remainder coming from glucose oxidation, to a small extent, amino acid oxidation. In ischemia, glycolysis may be called into play, utilizing stored glycogen and generating lactic acid (more on this later). Early reports suggested that amino acid supplementation might be beneficial in the ischemic and reperfused heart (155); replenishing amino acid precursors of Krebs’ cycle intermediates (e.g., glutamate, aspartate) was felt to be needed to ensure more effective oxidative metabolism to produce energy for cellular repair and subsequent mechanical function (156); subsequent work showed that glutamate could be metabolized anaerobically to contribute up to 20% of the ATP in the ischemic dog heart (157). A subsequent study using NMR phosphate spectroscopy concluded that supplementation with aspartate/glutamate did not improve myocardial bioenergetics (158). However, the aspartate/glutamate-supplemented group had higher intracellular pH at every time point; while they did not find it to be statistically significant in their small sample size (two pig hearts per group), it may hint at a degree of lactate sparing that could have been shown to be beneficial in a larger sample size. Thus the potential benefit of glutamate in ischemic metabolism might be under-appreciated. More pertinent to the present review, cell metabolism that is well-adapted to support anaerobic metabolism of amino acids could demonstrate greater benefit from amino acid availability. Thus metabolic flexibility is important to ischemic tolerance of the myocardium.
B. Consequences of fuel selection on pH and intracellular Ca2+
Under conditions of normal blood flow, fuel utilization carries little consequence and is likely related to the mix of available substrates. In the failing or borderline ischemic heart, fuel utilization is thought to shift to glucose oxidation as the oxygen cost for ATP production is lower than for fatty acids. However, anaerobic glycolysis comes at a high price beyond its inefficiency (2 ATP per molecule of glucose vs. 30 ATP via aerobic metabolism): lactic acid must be eliminated from the cell; the protons are removed via the Na+/H+ exchanger, but the increased intracellular Na+ then competes with Ca2+ for extrusion via the Na+/Ca2+ exchanger, resulting in a steady rise in intracellular Ca2+. Calcium doesn’t have to rise by much in order to wreak havoc, as acidosis increases the calcium sensitivity of some enzymes. For instance, the actin capping protein gelsolin binds phosphatidylinositol 4,5-bisphosphate (PIP2) in the presence of microM Ca2+, but the Ca2+ requirement is reduced further as the pH drops (159). Similarly, phospholipase C activity is increased at low pH in the absence of Ca2+ (58). An alternate method for extruding H+ is via the vacuolar proton ATPase, which can pump the protons into autophagolysosomes or across the plasma membrane. Although it consumes ATP, it is Ca2+-sparing. Ischemic preconditioning, which induces autophagy, activates the vacuolar proton ATPase and limits acidification and calcium overload (160, 161). To the extent that mitochondrial dysfunction (.e.g, in the face of hypoxia) forces the cell to rely on glycolysis, there is the likelihood that calcium overload may ensue. Mitochondria play an important role in calcium homeostasis, as discussed above.
C. Redox poise
Mitochondria determine the relative balance of reducing equivalents in the cell through conversion of NADH to NAD+ in Complex I. The mitochondria govern redox poise (NAD+/NADH, NADP+/NADPH) and acetyl-CoA levels; in this way they indirectly regulate countless processes through histone acetyltransferases (HATs) and deacetylases (HDACs) that modify cytosolic enzymes to control their activity, transcription factors such as PGC-1α, and histones whose acetylation status determines chromatin structure and access of transcription factors to specific regions (162). Interestingly, there is a close relationship between histone acetylation and intracellular pH: as intracellular pH decreases, histones are globally deacetylated by HDACs, and the acetate anions are exported along with protons through monocarboxylate transporters. As intracellular pH rises, histone acetylation also increases. Interestingly, inhibition of HDACs or monocarboxylate transporters limits acetate export and results in intracellular acidification, a potentially problematic side effect of HDAC inhibitors (163). Availability of acetyl-CoA will determine the extent of histone acetylation; however, the activity of acetyl-CoA carboxylase will determine whether acetyl-CoA is directed into fatty acid biosynthesis versus leaving it available for histone acetylation (164). The HDACs are influenced by both the ratio and absolute levels of NAD+; therefore its biosynthesis, particularly the activity of the rate-limiting enzyme nicotinamide phosphoribosyl transferase (NAMPT), is an important determinant of HDAC activity. NAMPT expression can increase by 2–3 fold with fasting or caloric restriction (165).
VI. Reactive oxygen species
Byproducts of oxidative phosphorylation are reactive oxygen species (ROS), predominatly superoxide, which is rapidly dismuted to hydrogen peroxide by superoxide dismutase and thence to water by the action of catalase. Hydrogen peroxide represents 1–2% of oxygen consumed in the process of oxidative phosphorylation.
A. Mitochondria generate ROS but have antioxidant defenses
Mitochondria are both a source and a target of ROS, which are generated in the mitochondria at Complex I, Complex II, and Complex III (166–168). Under ordinary circumstances, mitochondrial antioxidant defenses (MnSOD, catalase, glutathione peroxidase) convert the superoxide to hydrogen peroxide and then to water, but this may be impaired in certain settings. First, the production of superoxide may be high enough to overwhelm normal defenses. Catecholamine overstimulation increases mitochondrial ROS production due to signaling through protein kinase A (169) or Ca2+ (170). Secondly, normal defenses comprising glutathione and glutathione S-transferase P (GSTP) may be attenuated. Polymorphisms of human GSTP are associated with increased susceptibility to doxorubicin cardiomyopathy (171). A variety of stressors can cause depletion of glutathione, or the failure to regenerate reduced glutathione (GSH) from its oxidized counterpart (GSSG). Importantly, there is a dynamic interplay of ROS, GSH, and mitochondrial Ca2+ handling. In the absence of Ca2+-induced stimulation of the tricarboxylic acid cycle, NADH and NADPH become more oxidized and are unable to regenerate the GSH antioxidant system. This leads to an unstable system wherein small increases in hydrogen peroxide result in exaggerated ROS emission with increased workload (172).
B. Oxidative damage to mitochondrial proteins
Mitochondria undergo reversible S-nitrosylation, in which nitric oxide modifies the thiol moiety of cysteine. S-nitrosylation of mitochondrial proteins has been demonstrated in the setting of cardioprotection where it often inhibits OXPHOS components (173, 174). Interestingly, S-nitrosylation increases the activity of parkin, the E3 ubiquitin ligase important for mitophagy (175). Other forms of oxidative protein modifications such as thiol oxidation to sulfonic or sulfinic acid are less reversible. Nitration of tyrosine and carbonylation of cysteine are additional oxidative posttranslational modifications of proteins, many of which are detected in mitochondria. These lesions are repaired through distinct enzymatic systems including denitrosylating enzymes and mitochondrial proteases (AAA proteases including Lon). Alternatively, the entire organelle may also be eliminated via mitophagy, a process discussed above.
C. Lipid peroxidation
Lipid peroxidation is also a common consequence of mitochondrial ROS generation, often resulting in hydroxyl-alkenals such as 4-hydroxy-2-nonenal (HNE), commonly derived from linoleic acid and arachidonic acid, 4-hydroxyhexenal (HHE), and malondialdehyde (MDA). HHE can trigger mitochondrial permeability transition pore opening. These aldehydes can form covalent adducts on proteins by reacting with lysine amino groups, cysteine sulfhydryl groups, and histidine imidazole groups (176). As a result, they can modify protein structure and in many instances can trigger mitochondrial dysfunction. These aldehydes can form adducts on ethanolamine phospholipids, and unsaturated lipids can undergo lipid peroxidation, thereby altering membrane fluidity with myriad effects on cellular processes.
D. Oxidized mitochondrial DNA
Mitochondrial DNA is a common target of oxidative damage, resulting in oxidation of guanine residues (8-oxoG) at levels 2–3 fold higher than nuclear DNA (nDNA) (177). These are recognized and removed by oxoguanine glycosylase (OGG1), one of the few DNA repair enzymes localized to mitochondria (178). Sirt3 is also reported to interact with OGG1. Deacetylation by Sirt3 prevents degradation of OGG1 protein and controls its incision activity (179). Thus, oxidative stress elicits a coordinated repair response including both protein folding and DNA base excision. Failure to clear 8-oxoG lesions will result in mispairing with A, resulting in a G:C→A:T transversion (180). When mtDNA was isolated from fibroblasts of humans of various ages, mutations in the control region were found to be more common in older individuals, although the relationship was far from linear (181). It is unknown whether the age-related mtDNA mutations can be attributed to cumulative damage, to age-related deterioration in DNA repair processes, increased production of ROS, or age-related impaired autophagy. If the mtDNA mutations affect mitochondrial function, they would be cleared by mitophagy. Mitochondrial quality control (fission, fusion, mitophagy, and biogenesis) has been suggested to be sufficient for mitochondrial homeostasis even in the presence of ongoing mtDNA damage and limited repair (182). However, autophagy diminishes with age, which could contribute to the increase in mtDNA mutations with age (183, 184). Dysfunctional mitochondria left behind by inadequate mitophagy might also explain the increased ROS production seen with aging. Thus the extent of mtDNA damage seen with increased age might be a consequence of diminished autophagy in the face of a constant burden of DNA damage over a lifespan. There is abundant evidence linking autophagy to longevity (185, 186). Recently CISD2, a protein localized to mitochondria (187) has been linked to longevity through autophagy and regulation of Ca2+ homeostasis (188, 189). One also wonders whether the age:mtDNA damage relationship makes sense, since the very oldest individuals have probably survived that long in part because they had effective quality control mechanisms; one might expect the very oldest patients to exhibit less mtDNA damage. A more comprehensive investigation of the age-relatedness of mtDNA mutations, covering both coding and the control regions of the genome, may reveal additional insights.
VII. Nuclear–mitochondrial cross-talk
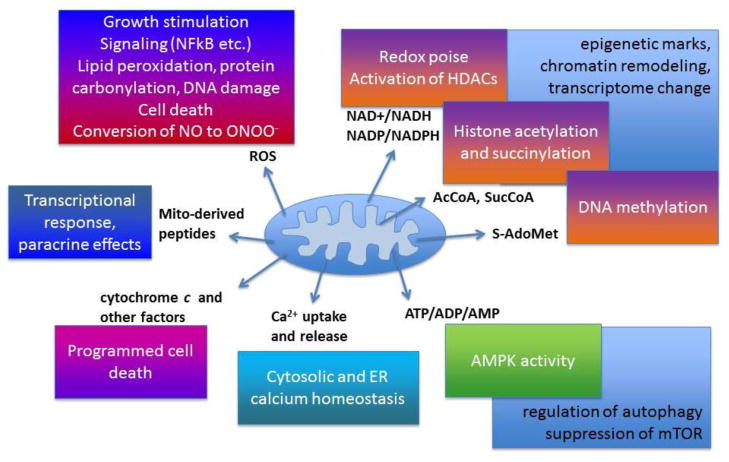
Nuclear instructions for mitochondrial biogenesis primarily driven by PGC-1α represent the best instance of nucleus-to-mitochondria signaling. Mitochondria “talk back” to the nucleus, a phenomenon called retrograde signaling, in which mitochondrial signals alter nuclear gene expression. This can occur through the mitochondrial unfolded protein response (UPRmt), through mitochondrial-derived peptides, and through metabolic milieu shifts. In addition to nuclear signaling, mitochondria alter the cellular and metabolic milieu in multiple ways as outlined above and diagrammed in Figure 2.
Figure 2. How mitochondria alter the cellular milieu.
Mitochondrial ROS production at low levels stimulates proliferation, but at high levels triggers damage to lipids, proteins, and DNA, triggering cell death. Superoxide can interact with NO produced locally or in neighboring cells, generating peroxynitrite and impairing endothelial function. Metabolites including NAD+, NADP, Acetyl-CoA, and S-adenosyl-methionine affect HDAC activity, histone acetylation and succinylation, and DNA methylation, resulting in chromatin remodeling and transcriptional shifts. The ratio of ATP to AMP regulates AMPK which in turn activates autophagy and suppresses mTOR (when AMP is high). Mitochondria take up Ca2+ rapidly and release it more slowly, supporting ER homeostasis and helping to buffer changes in cytosolic Ca2+. Mitochondria can release cytochrome c to trigger programmed cell death. Mitochondrial-derived peptides such as humanin elicit transcriptional responses and can have broad paracrine effects. Thus mitochondria impact broad signaling pathways with profound consequences.
A. The mitochondrial unfolded protein response
This pathway was first characterized in yeast and C. elegans and is under active investigation in mammals. Mitochondria possess machinery for responding to an imbalance between imported nuclear-encoded proteins and mitochondrial-encoded proteins. The correct stoichiometry of OXPHOS subunits is essential for assembly of functional respiratory complexes. Excess or unincorporated subunits are degraded to short peptides by matrix proteases of the AAA+ family (ATPases associated with a variety of cellular activities); Lon protease is responsible for the largest share of intramitochondrial protein degradation. In C. elegans, the peptide fragments generated by the related protease ClpP are extruded into the cytosol through the transporter Haf1. The mammalian homolog is thought to be ABCme10. The extruded peptides are recognized by transcription factors CHOP, C/EBPβ, and cJun/AP1, which direct expression of mitochondrial chaperone hsp60 and proteases ClpP, Yme1L1, and MPPβ. Upregulation of these factors reduces protein aggregation in the mitochondrial matrix. Additional proteins upregulated in the UPRmt include Tim17A, NDUFB2, and EndoG, but not ER stress proteins Bip, Grp94, calreticulin, or calnexin (190). PKR (double-stranded RNA-activated protein kinase) phosphorylates eIF2α and cJun, with the resulting suppression of translation and import of nuclear-encoded mitochondrial proteins (reviewed in CM Haynes and D Ron)(191). There is much less known about the significance of the UPRmt in mammalian systems or its connection to mitophagy and mitochondrial biogenesis. However, agents such as chloramphenicol and rapamycin, which induce mitophagy/autophagy, are also reported to be potent inducers of UPRmt (192). Chloramphenicol inhibits mitochondrial protein synthesis, which would result in an excess of nuclear-encoded subunits over mitochondrial subunits and would therefore trigger UPRmt. In contrast, rapamycin, which suppresses cytosolic protein synthesis via inhibition of mTOR, would result in an excess of mitochondrial subunits over nuclear-encoded proteins. Thus an imbalance in either direction is sufficient to trigger UPRmt, which may also be linked to activation of mitophagy. Interestingly, over a decade ago we identified mitochondrial elongation factor Tu (mtEF-Tu) as a major target of phosphorylation during ischemic preconditioning (193). In bacteria, phosphorylation of EF-Tu suppresses protein synthesis. Subsequent large-scale proteomic analyses documented phosphorylation of mtEF-Tu at Thr273 and Ser312 (194). Phosphorylation of these residues in the tRNA-binding region is believed to destabilize tRNA binding and thereby inhibit protein synthesis at the elongation step (195, 196). Thus a kinase signaling pathway activated during preconditioning converges on mitochondrial protein synthesis and the UPRmt. mtEF-Tu is also a target of acetylation at K88 and K256 (197). While the significance of acetylation of mtEF-Tu is unknown, mitochondrial SIRT3 deacetylates mitochondrial ribosomal protein L10 (MRPL10) to suppress mitochondrial protein synthesis (197).
B. Apoptotic signaling
Although this topic has been covered by many reviews (188, 189), it is important to mention the release of cytochrome c (190) and endonuclease G (191) as two proteins released from the intermembrane space and matrix, respectively, that trigger programmed cell death. Apoptosis Inducing Factor 1 (AIFM1) is an NADH oxidoreductase in the intermembrane space which also triggers DNA fragmentation, activates caspase-7, and inhibits protein synthesis through interaction with EIF3G (192–196). Permeabilization of the outer membrane to release intermembrane space factors (cytochrome c and AIFM1) is governed by the balance of pro-and anti-apoptotic members of the Bcl-2 family (reviewed in (197–199)). Calcium overload can activate calpains resulting in proteolytic activation of proapoptotic factors including Bid and AIFM1 (200, 201). Programmed cell death can also be accomplished via the mitochondrial permeability transition pore (MPTP), a large-conductance channel of the inner membrane that can be activated by Ca2+ overload or excessive ROS. Recently MPTP opening was shown to involve dimerization of the F0F1 ATPase and regulated by cyclophilin D (202). MPTP opening results in matrix swelling, loss of Ca2+ homeostasis, depletion of ATP and pyridine nucleotides, and outer membrane rupture with release of proapoptotic factors (203).
C. Mitochondrial-derived peptides
These short mitochondrial-encoded peptides represent an additional mechanism by which mitochondria communicate with the nucleus. MDPs are short peptide sequences hidden within the rRNA sequence of mtDNA. Two such peptides, humanin and MOTS-c, have been identified to date, and more may exist within the rRNA sequences (187). These peptides can act on intracellular targets including Bax (198) to suppress apoptosis, but can also act as paracrine factors; reports differ as to the specific receptors responsible (199) but in the case of humanin, signaling through STAT3 has been implicated (200). Within the mitochondria, the a humanin analog has been reported to reduce hydrogen peroxide production (201). MOTS-c has been shown to regulate metabolism and mitigate insulin resistance (202, 203). Both mitochondrial derived peptides are associated with longevity (204, 205).
D. Shifts in the metabolic milieu
Metabolic alterations represent a profound and global mechanism by which mitochondria accomplish retrograde signaling. By altering substrate utilization and electron flow, mitochondria can profoundly alter NADH/NAD+, or acetyl-CoA, thereby activating sirtuins/HDACS or HATS, respectively, with corresponding epigenetic alterations and global transcriptional responses. Mitochondria are responsible for the lion’s share of ROS production, and also regulate Ca2+ uptake/release, thereby accomplishing global shifts in various signaling pathways.
E. Impact of differing mitochondrial genomes
Differences in mitochondrial genome remodel the cell through altered metabolic milieu eliciting global changes in the nuclear-encoded transcriptome. Recent work exploring mitochondrial haplogroups (ethno-ancestrally distinct patters of SNPs in mtDNA) in an isogenic nuclear background has revealed the profound importance of relatively modest differences in mtDNA. Different mitochondrial haplogroups have been shown to exhibit differences in mitochondrial oxygen consumption, membrane potential, and mitochondrial number (206, 207). These haplogroup differences also result in broad changes in the transcriptome (208, 209). Moreover, mitochondrial haplogroups also modify susceptibility to disease (210–218). This has been elegantly dissected in a mouse model in which the mitochondrial genome of C3H/HeN mice has been placed into the C57Bl/6J nuclear background and vice versa (219). Using these mitochondrial-nuclear exchange (MNX) mice, Ballinger’s group was able to show that the mitochondrial genome determined susceptibility to the pathological stress of cardiac volume overload. Thus, mtDNA, which is maternally transmitted, plays an important role in complex multigenic diseases. It seems likely that the future of precision medicine will be required to take mitochondrial function into account in order to optimize patient-specific therapy.
VIII. Conclusions
Mitochondria adapt to a wide range of cellular requirements, adjusting fuel utilization, meeting ATP requirements, regulating ROS output, and titrating the redox poise of the cell. Each metabolic shift results in a corresponding shift in nuclear chromatin, epigenetic marks, the transcriptome, and ultimately, the proteome. Short-term alterations in energy requirements may be met by post-translational modifications or reorganization of mitochondrial structure by fission/fusion, while long-term alterations, especially those due to diseases such as myocardial ischemia or diabetes, are accomplished by mitophagy to eliminate mitochondria ill-suited to the new requirements, followed by biogenesis which often involves alterations in the mitochondrial proteome, accomplished by changes in expression of nuclear-encoded genes. These responses are coordinated across genomes to accomplish a synchronized metabolic remodeling. Although not discussed here, this includes circadian alterations as well as developmental changes or those elicited in response to pathologic conditions. While some may argue that the mitochondrial genome is a tiny tail to wag the dog, one should recall there are ~1000 mitochondria per cardiomyocyte that may all be wagging in synchrony.
Highlights.
Mitochondrial dynamics: fission and fusion
Mitochondrial turnover: mitophagy and biogenesis
Metabolism
Reactive oxygen species and mitochondria
Mitochondrial unfolded protein response
Retrograde signaling
Acknowledgments
NIH P01 HL112730-03 (to RAG); and NIH R01 HL123655 and HL117083-01 (to DB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Lewis MR, Lewis WH. Mitochondria (and other cytoplasmic structures) in tissue cultures. Amer J Anatomy. 1915;17:339–401. [Google Scholar]
- 2.Lewis MR, Lewis WH. Mitochondria in Tissue Culture. Science. 1914;39:330–333. doi: 10.1126/science.39.1000.330. [DOI] [PubMed] [Google Scholar]
- 3.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 5.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 6.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Scimia MC, Wilkinson D, Trelles RD, Wood MR, Bowtell D, Dillin A, Mercola M, Ronai ZA. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaddour-Djebbar I, Choudhary V, Brooks C, Ghazaly T, Lakshmikanthan V, Dong Z, Kumar MV. Specific mitochondrial calcium overload induces mitochondrial fission in prostate cancer cells. Int J Oncol. 2010;36:1437–1444. doi: 10.3892/ijo_00000629. [DOI] [PubMed] [Google Scholar]
- 10.Disatnik MH, Ferreira JCB, Campos JC, Gomes KS, Dourado PMM, Qi X, Mochly-Rosen D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. Journal of the American Heart Association. 2013;2:e000461. doi: 10.1161/JAHA.113.000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J Clin Invest. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X, Qvit N, Su YC, Mochly-Rosen D. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J Cell Sci. 2013;126:789–802. doi: 10.1242/jcs.114439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santel A, Frank S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- 17.Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 19.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO reports. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 21.Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Romero C, Iniguez-Lluhi JA, Stadler J, Chang CR, Arnoult D, Keller PJ, Hong Y, Blackstone C, Feldman EL. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23:3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, Liu Y, Kohlhaas M, Meiser M, Bergem S, et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circ Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wai T, Garcia-Prieto J, Baker MJ, Merkwirth C, Benit P, Rustin P, Ruperez FJ, Barbas C, Ibanez B, Langer T. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science. 2015;350:aad0116. doi: 10.1126/science.aad0116. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J, Lendahl U, Nister M. Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates. Cell Mol Life Sci. 2013;70:951–976. doi: 10.1007/s00018-012-1066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–99. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu T, Sheu SS, Robotham JL, Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc Res. 2008;79:341–351. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi X, Disatnik MH, Shen N, Sobel RA, Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase C{delta} under oxidative stress conditions in vivo. Molecular biology of the cell. 2011;22:256–265. doi: 10.1091/mbc.E10-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crescenzo R, Lionetti L, Mollica MP, Ferraro M, D’Andrea E, Mainieri D, Dulloo AG, Liverini G, Iossa S. Altered skeletal muscle subsarcolemmal mitochondrial compartment during catch-up fat after caloric restriction. Diabetes. 2006;55:2286–2293. doi: 10.2337/db06-0312. [DOI] [PubMed] [Google Scholar]
- 41.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015;21:95–108. doi: 10.1016/j.cmet.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesnik C, Golani-Armon A, Arava Y. Localized translation near the mitochondrial outer membrane: An update. RNA Biol. 2015;12:801–809. doi: 10.1080/15476286.2015.1058686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 48.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 53.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest JID - 7802877. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scarpulla RC. Transcriptional Paradigms in Mammalian Mitochondrial Biogenesis and Function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 56.Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, et al. A Role for the Transcriptional Coactivator PGC-1α in Muscle Refueling. Journal of Biological Chemistry. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- 57.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 58.Schrey MP, Montague W. Phosphatidylinositol hydrolysis in isolated guinea-pig islets of Langerhans. Biochem J. 1983;216:433–441. doi: 10.1042/bj2160433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersson DC, Fauconnier J, Yamada T, Lacampagne A, Zhang SJ, Katz A, Westerblad H. Mitochondrial production of reactive oxygen species contributes to the beta-adrenergic stimulation of mouse cardiomycytes. J Physiol. 2011;589:1791–1801. doi: 10.1113/jphysiol.2010.202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zippin JH, Chen Y, Nahirney P, Kamenetsky M, Wuttke MS, Fischman DA, Levin LR, Buck J. Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains. FASEB J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- 61.Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9:265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katona D, Rajki A, Di Benedetto G, Pozzan T, Spat A. Calcium-dependent mitochondrial cAMP production enhances aldosterone secretion. Mol Cell Endocrinol. 2015;412:196–204. doi: 10.1016/j.mce.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. Mitochondrial Ca(2)(+) uptake induces cyclic AMP generation in the matrix and modulates organelle ATP levels. Cell Metab. 2013;17:965–975. doi: 10.1016/j.cmet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Astin R, Bentham R, Djafarzadeh S, Horscroft JA, Kuc RE, Leung PS, Skipworth JR, Vicencio JM, Davenport AP, Murray AJ, et al. No evidence for a local renin-angiotensin system in liver mitochondria. Sci Rep. 2013;3:2467. doi: 10.1038/srep02467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 66.Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonne R, Filipowicz W, Bertrand E, Rage F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patel PH, Barbee SA, Blankenship JT. GW-Bodies and P-Bodies Constitute Two Separate Pools of Sequestered Non-Translating RNAs. PLoS One. 2016;11:e0150291. doi: 10.1371/journal.pone.0150291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, Dautry F, Weil D. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011;286:24219–24230. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cougot N, Cavalier A, Thomas D, Gillet R. The dual organization of P-bodies revealed by immunoelectron microscopy and electron tomography. J Mol Biol. 2012;420:17–28. doi: 10.1016/j.jmb.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 70.Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116:264–278. doi: 10.1161/CIRCRESAHA.116.303356. [DOI] [PubMed] [Google Scholar]
- 71.Song M, Mihara K, Chen Y, Scorrano L, Dorn GW., 2nd Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–285. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem. 2003;278:17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 74.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 75.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, Rossignol R. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 76.Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, et al. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol. 2015;35:211–223. doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res. 2012;111:1012–1026. doi: 10.1161/CIRCRESAHA.112.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gottlieb RA, Gustafsson AB. Mitochondrial turnover in the heart. Biochimica et biophysica acta. 2011;1813:1295–1301. doi: 10.1016/j.bbamcr.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PLoS One. 2012;7:e32388. doi: 10.1371/journal.pone.0032388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shields LY, Kim H, Zhu L, Haddad D, Berthet A, Pathak D, Lam M, Ponnusamy R, Diaz-Ramirez LG, Gill TM, et al. Dynamin-related protein 1 is required for normal mitochondrial bioenergetic and synaptic function in CA1 hippocampal neurons. Cell Death Dis. 2015;6:e1725. doi: 10.1038/cddis.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottlieb RA, Bernstein D. METABOLISM. Mitochondria shape cardiac metabolism. Science. 2015;350:1162–1163. doi: 10.1126/science.aad8222. [DOI] [PubMed] [Google Scholar]
- 83.Enriquez JA. Supramolecular Organization of Respiratory Complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 84.Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 87.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Obeso JA, Rodriguez-Oroz MC, Goetz CG, Marin C, Kordower JH, Rodriguez M, Hirsch EC, Farrer M, Schapira AH, Halliday G. Missing pieces in the Parkinson’s disease puzzle. Nat Med. 2010;16:653–661. doi: 10.1038/nm.2165. [DOI] [PubMed] [Google Scholar]
- 89.Büeler H. Mitochondrial dynamics, cell death and the pathogenesis of Parkinson's disease. Apoptosis : an international journal on programmed cell death. 2010;15:1336–1353. doi: 10.1007/s10495-010-0465-0. [DOI] [PubMed] [Google Scholar]
- 90.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Archer SL. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 92.Ryan JJ, Marsboom G, Fang YH, Toth PT, Morrow E, Luo N, Piao L, Hong Z, Ericson K, Zhang HJ, et al. PGC1alpha-mediated mitofusin-2 deficiency in female rats and humans with pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:865–878. doi: 10.1164/rccm.201209-1687OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 94.Olichon A, Guillou E, Delettre C, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Daloyau M, Hamel C, Amati-Bonneau P, et al. Mitochondrial dynamics and disease, OPA1. Biochim Biophys Acta. 2006;1763:500–509. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Song M, Dorn GW., 2nd Mitoconfusion: noncanonical functioning of dynamism factors in static mitochondria of the heart. Cell Metab. 2015;21:195–205. doi: 10.1016/j.cmet.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dorn GW, 2nd, Kitsis RN. The mitochondrial dynamism-mitophagy-cell death interactome: multiple roles performed by members of a mitochondrial molecular ensemble. Circ Res. 2015;116:167–182. doi: 10.1161/CIRCRESAHA.116.303554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ong SB, Hausenloy DJ. Mitochondrial morphology and cardiovascular disease. Cardiovascular Research. 2010;88:16–29. doi: 10.1093/cvr/cvq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beraud N, Pelloux S, Usson Y, Kuznetsov AV, Ronot X, Tourneur Y, Saks V. Mitochondrial dynamics in heart cells: very low amplitude high frequency fluctuations in adult cardiomyocytes and flow motion in non beating Hl-1 cells. J Bioenerg Biomembr. 2009;41:195–214. doi: 10.1007/s10863-009-9214-x. [DOI] [PubMed] [Google Scholar]
- 99.Sun CN, Dhalla NS, Olson RE. Formation of gigantic mitochondria in hypoxic isolated perfused rat hearts. Experientia. 1969;25:763–764. doi: 10.1007/BF01897616. [DOI] [PubMed] [Google Scholar]
- 100.Schaper J, Froede R, Hein S, Buck A, Hashizume H, Speiser B, Friedl A, Bleese N. Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation. 1991;83:504–514. doi: 10.1161/01.cir.83.2.504. [DOI] [PubMed] [Google Scholar]
- 101.Hong Z, Kutty S, Toth PT, Marsboom G, Hammel JM, Chamberlain C, Ryan JJ, Zhang HJ, Sharp WW, Morrow E, et al. Role of dynamin-related protein 1 (Drp1)-mediated mitochondrial fission in oxygen sensing and constriction of the ductus arteriosus. Circ Res. 2013;112:802–815. doi: 10.1161/CIRCRESAHA.111.300285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeong SY, Seol DW. The role of mitochondria in apoptosis. BMB Rep. 2008;41:11–22. doi: 10.5483/bmbrep.2008.41.1.011. [DOI] [PubMed] [Google Scholar]
- 104.Wibom R, Hultman E, Johansson M, Matherei K, Constantin-Teodosiu D, Schantz PG. Adaptation of mitochondrial ATP production in human skeletal muscle to endurance training and detraining. Journal of applied physiology (Bethesda, Md: 1985) 1992;73:2004–2010. doi: 10.1152/jappl.1992.73.5.2004. [DOI] [PubMed] [Google Scholar]
- 105.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feng Z, Bai L, Yan J, Li Y, Shen W, Wang Y, Wertz K, Weber P, Zhang Y, Chen Y, et al. Mitochondrial dynamic remodeling in strenuous exercise-induced muscle and mitochondrial dysfunction: regulatory effects of hydroxytyrosol. Free Radical Biology and Medicine. 2011;50:1437–1446. doi: 10.1016/j.freeradbiomed.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 107.Ding H, Jiang N, Liu H, Liu X, Liu D, Zhao F, Wen L, Liu S, Ji LL, Zhang Y. Response of mitochondrial fusion and fission protein gene expression to exercise in rat skeletal muscle. Biochimica et biophysica acta. 2010;1800:250–256. doi: 10.1016/j.bbagen.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 108.Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of Human Skeletal Muscle Mitochondrial Biogenesis and Quality Control: Effects of Age and Aerobic Exercise Training. The journals of gerontology. Series A, Biological sciences and medical sciences. 2013 doi: 10.1093/gerona/glt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 2010;299:C203–210. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Neill BT, Kim J, Wende AR, Theobald HA, Tuinei J, Buchanan J, Guo A, Zaha VG, Davis DK, Schell JC, et al. A conserved role for phosphatidylinositol 3-kinase but not Akt signaling in mitochondrial adaptations that accompany physiological cardiac hypertrophy. Cell Metab. 2007;6:294–306. doi: 10.1016/j.cmet.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dworatzek E, Mahmoodzadeh S, Schubert C, Westphal C, Leber J, Kusch A, Kararigas G, Fliegner D, Moulin M, Ventura-Clapier R, et al. Sex differences in exercise-induced physiological myocardial hypertrophy are modulated by oestrogen receptor beta. Cardiovasc Res. 2014;102:418–428. doi: 10.1093/cvr/cvu065. [DOI] [PubMed] [Google Scholar]
- 112.Coronado M, Fajardo G, Nguyen K, Zhao M, Bezold Kooiker K, Jung G, Hu D-Q, Reddy S, Sandoval E, Stotland A, et al. Physiologic mitochondrial fragmentation is a cardiac adaptation to increased energy demand. Circ Res. 2015:e123. doi: 10.1161/CIRCRESAHA.117.310725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dorn GW., 2nd Mitochondrial dynamics in heart disease. Biochim Biophys Acta. 2013;1833:233–241. doi: 10.1016/j.bbamcr.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pan X, Liu J, Nguyen T, Liu C, Sun J, Teng Y, Fergusson MM, Rovira II, Allen M, Springer DA, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nature cell biology. 2013;15:1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santo-Domingo J, Demaurex N. Calcium uptake mechanisms of mitochondria. Biochim Biophys Acta. 2010;1797:907–912. doi: 10.1016/j.bbabio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 116.Lawrie AM, Rizzuto R, Pozzan T, Simpson AW. A role for calcium influx in the regulation of mitochondrial calcium in endothelial cells. J Biol Chem. 1996;271:10753–10759. doi: 10.1074/jbc.271.18.10753. [DOI] [PubMed] [Google Scholar]
- 117.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O’Rourke B, Van Eyk JE, Foster DB. Mitochondrial protein phosphorylation as a regulatory modality: implications for mitochondrial dysfunction in heart failure. Congest Heart Fail. 2011;17:269–282. doi: 10.1111/j.1751-7133.2011.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park MK, Ashby MC, Erdemli G, Petersen OH, Tepikin AV. Perinuclear, perigranular and sub-plasmalemmal mitochondria have distinct functions in the regulation of cellular calcium transport. EMBO J. 2001;20:1863–1874. doi: 10.1093/emboj/20.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marchi S, Pinton P. The mitochondrial calcium uniporter complex: molecular components, structure and physiopathological implications. J Physiol. 2014;592:829–839. doi: 10.1113/jphysiol.2013.268235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Carafoli E. The fateful encounter of mitochondria with calcium: how did it happen? Biochim Biophys Acta. 2010;1797:595–606. doi: 10.1016/j.bbabio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 123.Smets I, Caplanusi A, Despa S, Molnar Z, Radu M, VandeVen M, Ameloot M, Steels P. Ca2+ uptake in mitochondria occurs via the reverse action of the Na+/Ca2+ exchanger in metabolically inhibited MDCK cells. Am J Physiol Renal Physiol. 2004;286:F784–794. doi: 10.1152/ajprenal.00284.2003. [DOI] [PubMed] [Google Scholar]
- 124.Jung DW, Baysal K, Brierley GP. The sodium-calcium antiport of heart mitochondria is not electroneutral. J Biol Chem. 1995;270:672–678. doi: 10.1074/jbc.270.2.672. [DOI] [PubMed] [Google Scholar]
- 125.Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S, et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci U S A. 2010;107:436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Denton RM. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta. 2009;1787:1309–1316. doi: 10.1016/j.bbabio.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 127.McCormack JG, Denton RM. The role of Ca2+ ions in the regulation of intramitochondrial metabolism and energy production in rat heart. Mol Cell Biochem. 1989;89:121–125. [PubMed] [Google Scholar]
- 128.Hom J, Yu T, Yoon Y, Porter G, Sheu SS. Regulation of mitochondrial fission by intracellular Ca2+ in rat ventricular myocytes. Biochim Biophys Acta. 2010;1797:913–921. doi: 10.1016/j.bbabio.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang X, Szeto C, Gao E, Tang M, Jin J, Fu Q, Makarewich C, Ai X, Li Y, Tang A, et al. Cardiotoxic and cardioprotective features of chronic β-adrenergic signaling. Circulation Research. 2013;112:498–509. doi: 10.1161/CIRCRESAHA.112.273896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marsboom G, Toth PT, Ryan JJ, Hong Z, Wu X, Fang YH, Thenappan T, Piao L, Zhang HJ, Pogoriler J, et al. Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ Res. 2012;110:1484–1497. doi: 10.1161/CIRCRESAHA.111.263848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sadek HA, Szweda PA, Szweda LI. Modulation of mitochondrial complex I activity by reversible Ca2+ and NADH mediated superoxide anion dependent inhibition. Biochemistry. 2004;43:8494–8502. doi: 10.1021/bi049803f. [DOI] [PubMed] [Google Scholar]
- 133.Zhang P, Kirk JA, Ji W, dos Remedios CG, Kass DA, Van Eyk JE, Murphy AM. Multiple reaction monitoring to identify site-specific troponin I phosphorylated residues in the failing human heart. Circulation. 2012;126:1828–1837. doi: 10.1161/CIRCULATIONAHA.112.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kirk JA, Zhang P, Murphy AM, Van Eyk JE. Troponin I alterations detected by multiple-reaction monitoring: how might this impact the study of heart failure? Expert Rev Proteomics. 2013;10:5–8. doi: 10.1586/epr.12.77. [DOI] [PubMed] [Google Scholar]
- 135.Lam MP, Lau E, Scruggs SB, Wang D, Kim TY, Liem DA, Zhang J, Ryan CM, Faull KF, Ping P. Site-specific quantitative analysis of cardiac mitochondrial protein phosphorylation. J Proteomics. 2013;81:15–23. doi: 10.1016/j.jprot.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lam MP, Scruggs SB, Kim TY, Zong C, Lau E, Wang D, Ryan CM, Faull KF, Ping P. An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. J Proteomics. 2012;75:4602–4609. doi: 10.1016/j.jprot.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Wang Y, Korge P, et al. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.000117. 000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovascular research. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen Q, Lin RY, Rubin CS. Organelle-specific targeting of protein kinase AII (PKAII). Molecular and in situ characterization of murine A kinase anchor proteins that recruit regulatory subunits of PKAII to the cytoplasmic surface of mitochondria. J Biol Chem. 1997;272:15247–15257. doi: 10.1074/jbc.272.24.15247. [DOI] [PubMed] [Google Scholar]
- 140.Wang SB, Foster DB, Rucker J, O’Rourke B, Kass DA, Van Eyk JE. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ Res. 2011;109:750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kane LA, Youngman MJ, Jensen RE, Van Eyk JE. Phosphorylation of the F(1)F(o) ATP synthase beta subunit: functional and structural consequences assessed in a model system. Circ Res. 2010;106:504–513. doi: 10.1161/CIRCRESAHA.109.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Agnetti G, Kaludercic N, Kane LA, Elliott ST, Guo Y, Chakir K, Samantapudi D, Paolocci N, Tomaselli GF, Kass DA, et al. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ Cardiovasc Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang SB, Murray CI, Chung HS, Van Eyk JE. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc Med. 2013;23:14–18. doi: 10.1016/j.tcm.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 145.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 146.Paolisso G, Gambardella A, Galzerano D, D’Amore A, Rubino P, Verza M, Teasuro P, Varricchio M, D’Onofrio F. Total-body and myocardial substrate oxidation in congestive heart failure. Metabolism. 1994;43:174–179. doi: 10.1016/0026-0495(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 147.Lommi J, Kupari M, Yki-Jarvinen H. Free fatty acid kinetics and oxidation in congestive heart failure. Am J Cardiol. 1998;81:45–50. doi: 10.1016/s0002-9149(97)00804-7. [DOI] [PubMed] [Google Scholar]
- 148.Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34:217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- 149.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- 150.Lu G, Ren S, Korge P, Choi J, Dong Y, Weiss J, Koehler C, Chen JN, Wang Y. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev. 2007;21:784–796. doi: 10.1101/gad.1499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Abadir PM, Foster DB, Crow M, Cooke CA, Rucker JJ, Jain A, Smith BJ, Burks TN, Cohn RD, Fedarko NS, et al. Identification and characterization of a functional mitochondrial angiotensin system. Proc Natl Acad Sci U S A. 2011;108:14849–14854. doi: 10.1073/pnas.1101507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Calmettes G, John SA, Weiss JN, Ribalet B. Hexokinase–mitochondrial interactions regulate glucose metabolism differentially in adult and neonatal cardiac myocytes. The Journal of General Physiology. 2013;142:425–436. doi: 10.1085/jgp.201310968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Glancy B, Balaban RS. Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry. 2012;51:2959–2973. doi: 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Abo Alrob O, Lopaschuk GD. Role of CoA and acetyl-CoA in regulating cardiac fatty acid and glucose oxidation. Biochem Soc Trans. 2014;42:1043–1051. doi: 10.1042/BST20140094. [DOI] [PubMed] [Google Scholar]
- 155.Lazar HL, Buckberg GD, Manganaro AJ, Becker H, Maloney JV., Jr Reversal of ischemic damage with amino acid substrate enhancement during reperfusion. Surgery. 1980;88:702–709. [PubMed] [Google Scholar]
- 156.Rosenkranz ER, Okamoto F, Buckberg GD, Robertson JM, Vinten-Johansen J, Bugyi HI. Safety of prolonged aortic clamping with blood cardioplegia. III. Aspartate enrichment of glutamate-blood cardioplegia in energy-depleted hearts after ischemic and reperfusion injury. J Thorac Cardiovasc Surg. 1986;91:428–435. [PubMed] [Google Scholar]
- 157.Wiesner RJ, Deussen A, Borst M, Schrader J, Grieshaber MK. Glutamate degradation in the ischemic dog heart: contribution to anaerobic energy production. J Mol Cell Cardiol. 1989;21:49–59. doi: 10.1016/0022-2828(89)91492-2. [DOI] [PubMed] [Google Scholar]
- 158.Ghomeshi HR, Tian G, Ye J, Sun J, Hoffenberg EF, Salerno TA, Deslauriers R. Aspartate/glutamate-enriched blood does not improve myocardial energy metabolism during ischemia-reperfusion: A 31p magnetic resonance spectroscopic study in isolated pig hearts. The Journal of Thoracic and Cardiovascular Surgery. 1997;113:1068–1080. doi: 10.1016/s0022-5223(97)70294-0. [DOI] [PubMed] [Google Scholar]
- 159.Lin KM, Wenegieme E, Lu PJ, Chen CS, Yin HL. Gelsolin binding to phosphatidylinositol 4,5-bisphosphate is modulated by calcium and pH. J Biol Chem. 1997;272:20443–20450. doi: 10.1074/jbc.272.33.20443. [DOI] [PubMed] [Google Scholar]
- 160.Karwatowska-Prokopczuk E, Nordberg JA, Li HL, Engler RL, Gottlieb RA. Effect of vacuolar proton ATPase on pHi, Ca2+, and apoptosis in neonatal cardiomyocytes during metabolic inhibition/recovery. Circ Res. 1998;82:1139–1144. doi: 10.1161/01.res.82.11.1139. [DOI] [PubMed] [Google Scholar]
- 161.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Jr, Gottlieb RA. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 163.McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, et al. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Galdieri L, Vancura A. Acetyl-CoA Carboxylase Regulates Global Histone Acetylation. The Journal of Biological Chemistry. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Muller FL, Liu Y, Van Remmen H. Complex III Releases Superoxide to Both Sides of the Inner Mitochondrial Membrane. Journal of Biological Chemistry. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 167.Quinlan CL, Treberg JR, Perevoshchikova IV, Orr AL, Brand MD. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radic Biol Med. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Perevoshchikova IV, Quinlan CL, Orr AL, Gerencser AA, Brand MD. Sites of superoxide and hydrogen peroxide production during fatty acid oxidation in rat skeletal muscle mitochondria. Free Radic Biol Med. 2013;61:298–309. doi: 10.1016/j.freeradbiomed.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Robinson AD, Ramanathan KB, McGee JE, Newman KP, Weber KT. Oxidative stress and cardiomyocyte necrosis with elevated serum troponins: pathophysiologic mechanisms. Am J Med Sci. 2011;342:129–134. doi: 10.1097/MAJ.0b013e3182231ee3. [DOI] [PubMed] [Google Scholar]
- 170.Jian C, Hou T, Yin R, Cheng H, Wang X. Regulation of superoxide flashes by transient and steady mitochondrial calcium elevations. Sci China Life Sci. 2014;57:495–501. doi: 10.1007/s11427-014-4628-z. [DOI] [PubMed] [Google Scholar]
- 171.Aplenc R, Blanco J, Leisenring W, Davies S, Relling M, Robison L, Sklar C, Stovall M, Bhatia S. Polymorphisms in candidate genes in patients with congestive heart failure (CHF) after childhood cancer: A report from the Childhood Cancer Survivor Study (CCSS). In. ASCO Annual Meeting Proceedings; 2006. p. 9004. [Google Scholar]
- 172.Gauthier Laura D, Greenstein Joseph L, O’Rourke B, Winslow Raimond L. An Integrated Mitochondrial ROS Production and Scavenging Model: Implications for Heart Failure. Biophysical Journal. 2013;105:2832–2842. doi: 10.1016/j.bpj.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun J, Murphy E. Protein S-Nitrosylation and Cardioprotection. Circulation Research. 2010;106:285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Piantadosi CA. Regulation of mitochondrial processes by protein S-nitrosylation. Biochim Biophys Acta. 2012;1820:712–721. doi: 10.1016/j.bbagen.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ozawa K, Komatsubara AT, Nishimura Y, Sawada T, Kawafune H, Tsumoto H, Tsuji Y, Zhao J, Kyotani Y, Tanaka T, et al. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci Rep. 2013;3:2202. doi: 10.1038/srep02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Catalá A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chemistry and Physics of Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 177.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 178.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 179.Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, et al. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 2013;4:e731. doi: 10.1038/cddis.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 181.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-Dependent Large Accumulation of Point Mutations in the Human mtDNA Control Region for Replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 182.Mouli PK, Twig G, Shirihai OS. Frequency and selectivity of mitochondrial fusion are key to its quality maintenance function. Biophys J. 2009;96:3509–3518. doi: 10.1016/j.bpj.2008.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 184.Linton PJ, Gurney M, Sengstock D, Mentzer RM, Jr, Gottlieb RA. This old heart: Cardiac aging and autophagy. J Mol Cell Cardiol. 2015;83:44–54. doi: 10.1016/j.yjmcc.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Grimmel M, Backhaus C, Proikas-Cezanne T. WIPI-Mediated Autophagy and Longevity. Cells. 2015;4:202–217. doi: 10.3390/cells4020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Madeo F, Zimmermann A, Maiuri MC, Kroemer G. Essential role for autophagy in life span extension. J Clin Invest. 2015;125:85–93. doi: 10.1172/JCI73946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY, Liu FC, Yang CW, Wei YH, Hsu MT, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang CH, Kao CH, Chen YF, Wei YH, Tsai TF. Cisd2 mediates lifespan: is there an interconnection among Ca(2)(+) homeostasis, autophagy, and lifespan? Free Radic Res. 2014;48:1109–1114. doi: 10.3109/10715762.2014.936431. [DOI] [PubMed] [Google Scholar]
- 189.Wang CH, Chen YF, Wu CY, Wu PC, Huang YL, Kao CH, Lin CH, Kao LS, Tsai TF, Wei YH. Cisd2 modulates the differentiation and functioning of adipocytes by regulating intracellular Ca2+ homeostasis. Hum Mol Genet. 2014;23:4770–4785. doi: 10.1093/hmg/ddu193. [DOI] [PubMed] [Google Scholar]
- 190.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 192.Mottis A, Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response in mammalian physiology. Mamm Genome. 2014;25:424–433. doi: 10.1007/s00335-014-9525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.He H, Chen M, Scheffler NK, Gibson BW, Spremulli LL, Gottlieb RA. Phosphorylation of mitochondrial elongation factor Tu in ischemic myocardium: basis for chloramphenicol-mediated cardioprotection. Circ Res. 2001;89:461–467. doi: 10.1161/hh1701.096038. [DOI] [PubMed] [Google Scholar]
- 194.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, Singh Y. Interaction of Mycobacterium tuberculosis Elongation Factor Tu with GTP Is Regulated by Phosphorylation. Journal of Bacteriology. 2011;193:5347–5358. doi: 10.1128/JB.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Alexander C, Bilgin N, Lindschau C, Mesters JR, Kraal B, Hilgenfeld R, Erdmann VA, Lippmann C. Phosphorylation of Elongation Factor Tu Prevents Ternary Complex Formation. Journal of Biological Chemistry. 1995;270:14541–14547. doi: 10.1074/jbc.270.24.14541. [DOI] [PubMed] [Google Scholar]
- 197.Koc EC, Koc H. Regulation of mammalian mitochondrial translation by post-translational modifications. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819:1055–1066. doi: 10.1016/j.bbagrm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 198.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 199.Zapala B, Kaczynski L, Kiec-Wilk B, Staszel T, Knapp A, Thoresen GH, Wybranska I, Dembinska-Kiec A. Humanins, the neuroprotective and cytoprotective peptides with antiapoptotic and anti-inflammatory properties. Pharmacol Rep. 2010;62:767–777. doi: 10.1016/s1734-1140(10)70337-6. [DOI] [PubMed] [Google Scholar]
- 200.Hashimoto Y, Suzuki H, Aiso S, Niikura T, Nishimoto I, Matsuoka M. Involvement of tyrosine kinases and STAT3 in Humanin-mediated neuroprotection. Life Sci. 2005;77:3092–3104. doi: 10.1016/j.lfs.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 201.Paharkova V, Alvarez G, Nakamura H, Cohen P, Lee KW. Rat Humanin is encoded and translated in mitochondria and is localized to the mitochondrial compartment where it regulates ROS production. Mol Cell Endocrinol. 2015;413:96–100. doi: 10.1016/j.mce.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 202.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zarse K, Ristow M. A mitochondrially encoded hormone ameliorates obesity and insulin resistance. Cell Metab. 2015;21:355–356. doi: 10.1016/j.cmet.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 204.Fuku N, Pareja-Galeano H, Zempo H, Alis R, Arai Y, Lucia A, Hirose N. The mitochondrial-derived peptide MOTS-c: a player in exceptional longevity? Aging Cell. 2015;14:921–923. doi: 10.1111/acel.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A, Cohen P. IGF-I regulates the age-dependent signaling peptide humanin. Aging Cell. 2014;13:958–961. doi: 10.1111/acel.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 207.Krzywanski DM, Moellering DR, Westbrook DG, Dunham-Snary KJ, Brown J, Bray AW, Feeley KP, Sammy MJ, Smith MR, Schurr TG, et al. Endothelial Cell Bioenergetics and Mitochondrial DNA Damage Differ in Humans Having African or West Eurasian Maternal Ancestry. Circ Cardiovasc Genet. 2016;9:26–36. doi: 10.1161/CIRCGENETICS.115.001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kenney MC, Chwa M, Atilano SR, Pavlis JM, Falatoonzadeh P, Ramirez C, Malik D, Hsu T, Woo G, Soe K, et al. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PLoS One. 2013;8:e54339. doi: 10.1371/journal.pone.0054339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Caceres-del-Carpio J, Nesburn AB, Boyer DS, et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet. 2014;23:3537–3551. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Appleyard GD, Forsyth GW, Kiehlbauch LM, Sigfrid KN, Hanik HL, Quon A, Loewen ME, Grahn BH. Differential mitochondrial DNA and gene expression in inherited retinal dysplasia in miniature Schnauzer dogs. Invest Ophthalmol Vis Sci. 2006;47:1810–1816. doi: 10.1167/iovs.05-0819. [DOI] [PubMed] [Google Scholar]
- 212.Hwang S, Kwak SH, Bhak J, Kang HS, Lee YR, Koo BK, Park KS, Lee HK, Cho YM. Gene expression pattern in transmitochondrial cytoplasmic hybrid cells harboring type 2 diabetes-associated mitochondrial DNA haplogroups. PLoS One. 2011;6:e22116. doi: 10.1371/journal.pone.0022116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lin CS, Lee HT, Lee SY, Shen YA, Wang LS, Chen YJ, Wei YH. High mitochondrial DNA copy number and bioenergetic function are associated with tumor invasion of esophageal squamous cell carcinoma cell lines. Int J Mol Sci. 2012;13:11228–11246. doi: 10.3390/ijms130911228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Earp MA, Brooks-Wilson A, Cook L, Le N. Inherited common variants in mitochondrial DNA and invasive serous epithelial ovarian cancer risk. BMC Res Notes. 2013;6:425. doi: 10.1186/1756-0500-6-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8:e61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tiao MM, Liou CW, Huang LT, Wang PW, Lin TK, Chen JB, Chou YM, Huang YH, Lin HY, Chen CL, et al. Associations of mitochondrial haplogroups b4 and e with biliary atresia and differential susceptibility to hydrophobic bile Acid. PLoS Genet. 2013;9:e1003696. doi: 10.1371/journal.pgen.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Del Carpio JC, Nesburn AB, Boyer DS, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim Biophys Acta. 2014;1842:208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Malik D, Hsu T, Falatoonzadeh P, Caceres-del-Carpio J, Tarek M, Chwa M, Atilano SR, Ramirez C, Nesburn AB, Boyer DS, et al. Human retinal transmitochondrial cybrids with J or H mtDNA haplogroups respond differently to ultraviolet radiation: implications for retinal diseases. PLoS One. 2014;9:e99003. doi: 10.1371/journal.pone.0099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, et al. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455:157–167. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]