Abstract
Morphine and its pharmacological derivatives are the most prescribed analgesics for moderate to severe pain management. However, chronic use of morphine reduces pathogen clearance and induces bacterial translocation across the gut barrier. The enteric microbiome has been shown to play a critical role in the preservation of the mucosal barrier function and metabolic homeostasis. Here, we show for the first time, using bacterial 16s rDNA sequencing, that chronic morphine treatment significantly alters the gut microbial composition and induces preferential expansion of gram-positive pathogenic and reduction in bile-deconjugating bacterial strains. A significant reduction in both primary and secondary bile acid levels was seen in the gut, but not in the liver with morphine treatment. Morphine induced microbial dysbiosis and gut barrier disruption was rescued by transplanting placebo-treated microbiota into morphine-treated animals, indicating that microbiome modulation could be exploited as a therapeutic strategy for patients using morphine for pain management.
Introduction
Despite being the predominant drug of choice for moderate to chronic pain management, morphine treatment results in severe co-morbidities due to peripheral side effects. In the past few years, several groups including ours have been actively working on understanding the phenomenon and delineating the mechanism underlying peripheral effects of morphine on immune cells and its role in exacerbating co-morbidities associated with its use or abuse. The emerging consensus from all these studies conclusively demonstrate that opioid drugs cause adverse effects, including increased pre-disposition to infection, exacerbating pathogenesis, impairing endotoxin tolerance, and more recently inducing gut barrier disruption and bacterial translocation1–5. The mammalian gut houses a robust and resilient microbiota that maintains a high degree of species diversity- the most important defense against pathogen virulence and invasion6. In diseased states, the microbial balance that favors homeostasis is perturbed, resulting in a loss in the richness and diversity of the bacterial components. Any significant shift in the composition of the microbiota (“dysbiosis”) favors the appearance of distinct pathogens and has been implicated in the pathogenesis of diverse illnesses, such as obesity, type 2 diabetes, inflammatory bowel disease, and cardiovascular disease7–9. However, until now there have been no studies investigating how morphine treatment modulate the gut microbiota and its contribution to morphine induced pathology such as microbial translocation and systemic immune activation. Furthermore, a cyclical relationship exists between gut microbial homeostasis and healthy hepato-enteric circulation of host metabolites particularly bile acid. Change in gut microbial composition (microbial dysbiosis) leading to bile acid changes has been correlated with gut barrier disruption and inflammation10–12. Systemic insults like opioid use/abuse, which elevate systemic inflammation, would be expected to alter the gut microbial composition (microbial dysbiosis) and induce bile acid changes. The linearity and order of events needs to be determined for designing any kind of clinical intervention. Furthermore, pro-inflammatory environment and infiltration of immune cells in the gut tissues is strongly correlated to the maintenance of the pathogenic phenotype.
In this study, we establish a link between the two phenomena, namely gut barrier compromise and dysregulated bile acid metabolism. We show for the first time that morphine fosters significant gut microbial dysbiosis and disrupts cholesterol/bile acid metabolism. Changes in the gut microbial composition is strongly correlated to disruption in host inflammatory homeostasis13,14 and in many diseases (e.g. cancer/HIV infection), persistent inflammation is known to aid and promote the progression of the primary morbidity. We show here that chronic morphine, gut microbial dysbiosis, disruption of cholesterol/bile acid metabolism and gut inflammation; have a linear correlation. This opens up the prospect of devising minimally invasive adjunct treatment strategies involving microbiome and bile acid modulation and thus bringing down morphine-mediated inflammation in the host.
Morphine induces global changes in gut microbiota
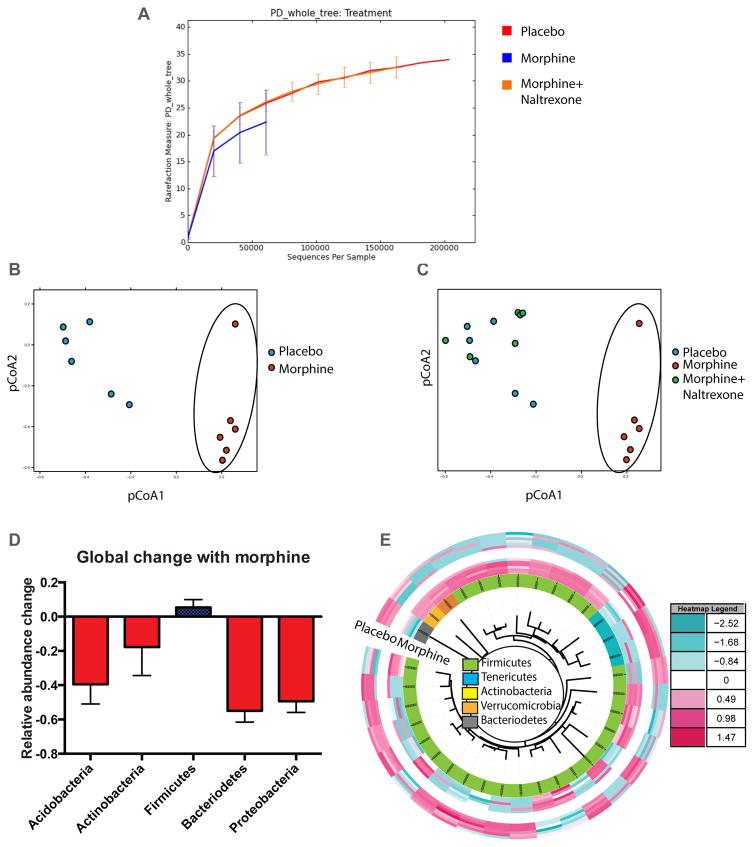
We have previously shown that bacterial translocation due to the gut mucosal barrier compromise are derived from the gut, mostly from commensal flora3,15. In this report, morphine (~1μM serum concentration) was administered into C57Bl6/j mice in analgesic doses and intestinal fecal contents were collected 72 hours later and analyzed for microbial composition. While non-significant changes were observed in α-diversity among treatment groups (Fig. 1A), a significant shift in gut microbial composition between placebo and morphine treated animals was observed (Principal Coordinate analysis, pCoA; Fig. 1B). Microbiome of the animals treated with both morphine and naltrexone (μ-opioid receptor antagonist), clustered with the placebo-treated animals (Fig. 1C). Analysis of the significant changes in β-diversity due to morphine shows a net expansion of gram-positive Firmicutes compared to all other major phyla of gut bacteria (Fig 1D). Of the 5,585 cumulative OTUs detected among placebo and morphine-implanted animals, 117 OTUs (within 30 families) were found to be significantly different between the groups. Top OTUs from each family with higher (red) or lower (blue) relative abundance were used to compute an idealized tree using taxonomic classifications (Fig 1E). In this heat map, 28 out of 34 OTUs belong to phylum Firmicutes with a net preferential expansion in the morphine treated animals. Reduction in comparative abundance was observed for phylum Bacteroidetes in the morphine treated animals, thus reducing the Bacteroidetes/Firmicutes ratio in those animals (Figure S1). Similar changes in the Bacteroidetes/Firmicutes ratio is correlated with increased systemic inflammation in obesity and aging9,16,17. Additionally, elevated abundance of bacterial families Enterococcaceae, Staphylococcaceae, Bacillaceae, Streptococcaceae and Erysipelotrichaceae was seen in the morphine-implanted animals, all belonging to phylum Firmicutes. In this context, we and others have shown that chronic morphine increases the susceptibility18,19 and rate of pathogenesis18,20 of mucosal infections, leading to sepsis and septic shock5,21,22 with significant contribution of gram-positive bacteria in morphine-induced polymicrobial sepsis23.
Figure 1. Morphine induced global changes in the microbiome.
Phylogenetic Diversity (PD) among the 16s rDNA content from animals treated with placebo, morphine and morphine+naltrexone exhibits a trend (non-significant) towards reduced species richness for the morphine treated animals (A). Principle co-ordinate analysis (pCoA) of 16s rDNA content from WT animals implanted with Placebo or Morphine (circled) pellets show distinct clustering of microbiome (B). Microbial composition of animals co-implanted with morphine and naltrexone pellets show high similarity with Placebo implanted animals, distinct from morphine-implanted animals (circled; C). Ratio of relative abundance of annotated OTUs between placebo and morphine implanted animals show a net expansion of phylum Firmicutes among 5 major commensal phyla (D). Top OTUs from each family, with significant difference between placebo and morphine groups, was rendered as a heat map using interactive tree of life (iToL; itol.embl.de). In this heat map, 28 out of 34 OTUs belong to phylum Firmicutes with a net preferential expansion in the morphine treated animals (E). n=6 for pCoA analyses (See also Figure S1 and S2).
Role of TLR2 and μ-opioid receptor in morphine-induced microbial dysbiosis
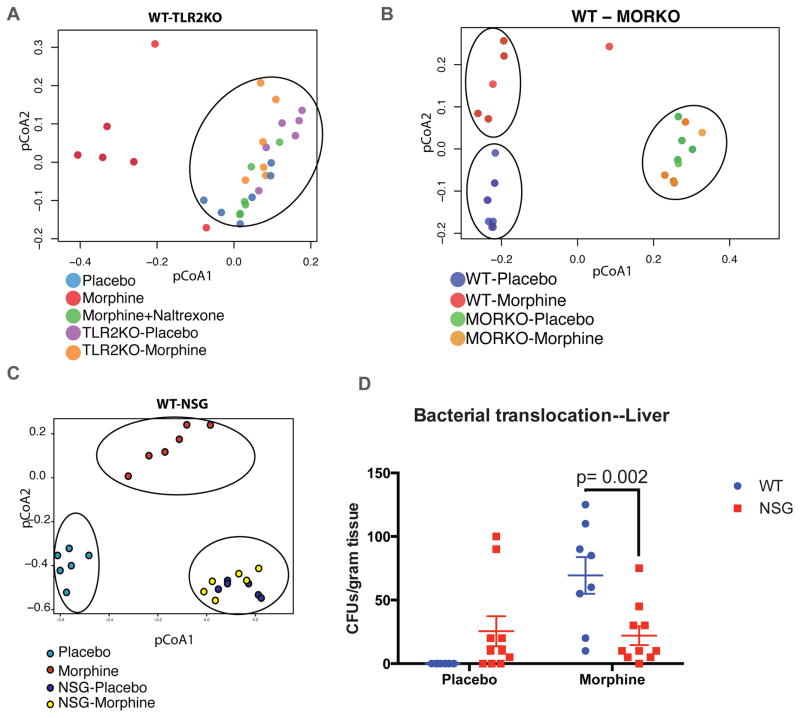
Literature5,22, including our own published works2–4,19, show that morphine mediated gut microbial dysbiosis and mucosal barrier compromise results in bacterial translocation (predominantly gram-positive) leading to localized gut and systemic inflammation. Interestingly, the translocated bacteria, serotyped from the liver of morphine treated animals3, belonged to the classes Staphylococcaceae,, Enterococcaceae and Bacillaceae, all part of the commensal flora and the same classes which exhibit preferential expansion in the gut microbiome upon morphine treatment (Fig 1D and Figure S2). Hence, we wanted to verify in TLR-2 knockout (TLR2KO) and μ-opioid receptor knockout (MORKO) animals, whether morphine effects are influenced by these receptors. TLR2KO animals treated with placebo or morphine were compared to WT animals as above (Fig. 2A). pCoA analysis of the five groups indicates WT-morphine as the only group that shows separation from the others, implying that TLR2KO-morphine animals are protected from the microbial shift.
Figure 2. Dependence of morphine induced microbial changes on TLR2, μ-opioid receptor, and host immune system.
TLR2KO animals, implanted with placebo or morphine pellets, exhibit similar microbial composition as placebo and morphine+naltrexone implanted WT animals in pCoA analysis (circled), distinct from morphine implanted WT animals (A). μ-Opioid receptor knockout (MORKO) animals, although exhibiting a distinct microbial composition compared to WT animals, do not show morphine-mediated microbial changes (B). Similar to MORKO animals, severely immune-compromised NSG animals exhibit a distinct microbial composition compared to WT animals, with no morphine-mediated changes in the microbiota (C). [Individual groups forming a distinct cluster circled in each pCoA plot. N=6 for each group]. Liver lysates of WT and NSG animals, implanted with placebo or morphine pellets for 72 hours were plated on sheep-blood agar plates and total CFUs counted as a surrogate of gut barrier compromise and bacterial translocation. While WT-morphine animals exhibited robust translocation of bacteria, NSG animals failed to exhibit morphine-mediated exacerbation in bacterial translocation (D). [n=5; lysate from each animal plated in duplicate].
Similar to TLR2KO profile, MORKO animals exhibited a pCoA distribution unperturbed by morphine (Fig. 2B). While this was expected, one major difference seen here is a distinct clustering of the MORKO (both placebo and morphine implanted) animals away from the WT-placebo animals. This indicates that unconditional absence of μ-opioid receptor has changed the basic composition of the microbiome in these animals, which is different from pharmacological inhibition of μ-opioid signaling with naltrexone. Morphine-induced changes in the microbiome might be mediated by μ-opioid signaling in the peripheral immune cells and could be due to an altered tutoring effect, by now a well established phenomenon17,24. To test this, we implanted placebo and morphine pellets into Non-obese diabetic, severe combined immune-deficient (NOD-SCID) with IL-2 receptor gamma knockout (NSG; Jackson Laboratories) animals, with impaired innate and adaptive immune compartments. Principle coordinate analysis of NSG animals shows a very similar profile as MORKO animals; where there are fundamental differences in the basic microbial composition between WT and NSG, but morphine mediated changes are completely abolished (Fig. 2C). Additionally, bacterial translocation due to morphine treatment, a hallmark of gut barrier compromise3,15, was significantly abrogated in the NSG-morphine animals (Fig. 2D). We have previously shown that bacterial translocation due to morphine is significantly diminished in the TLR2KO mice and completely abolished in the MORKO animals3,15. Absence of morphine-mediated bacterial translocation and microbial changes in NSG animals strongly indicates a linear correlation between μ-opioid receptor and TLR2 signaling in the mucosal immune cells influencing a focused change in the microbial composition of the animals. Inhibition/knockout of either of these receptors, irrespective of the baseline changes in the microbiome due to their absence, rescues the morphine-induced microbial dysbiosis and its physiological fallout with respect to bacterial translocation and gut barrier homeostasis.
Microbial transplant influences commensal flora and gut morphology
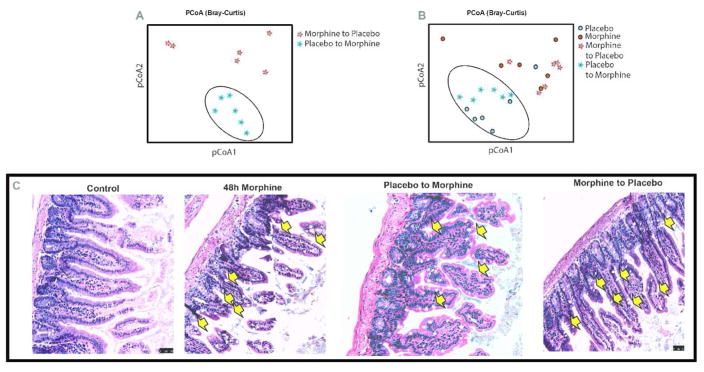
Fecal transplant has been successfully used clinically, especially for treating C. difficile infection25–28. With our expanding knowledge of the central role of microbiome in maintenance of host immune homeostasis17, fecal transplant is gaining importance as a therapy for indications resulting from microbial dysbiosis. There is a major difference between fecal transplant being used for the treatment of C. difficile infection and the conditions described in our studies. The former strategy is based on the argument that microbial dysbiosis caused by disproportionate overgrowth of a pathobiont can be out-competed by re-introducing the missing flora by way of a “normal microbiome” transplant. This strategy is independent of host factors and systemic effects on the microbial composition. Here, we show that microbial dysbiosis caused due to morphine can be reversed by transplantation of microbiota from the placebo-treated animals. There is a distinct clustering of the transplanted microbiome (Fig. 3A) and that “Placebo to placebo (PP)” and “placebo to morphine (PM)” groups cluster together, which is distinct from “morphine to morphine (MM)” and “morphine to placebo (MP)” groups (Fig. 3B). This indicates that the reciprocally transplanted microbiome tend to cluster together with the donor microbiome rather than the treatment condition. As an extension of the transplant experiment, we asked whether morphine-induced dysbiotic microbiome is capable of recapitulating gut pathologies in WT animals. Donor mice (48 hours morphine) exhibit morphine-induced hallmark distortion of gut morphology as described earlier4 and early signs of morphological changes are evident in the recipient animals as well (Fig. 3C). This data indicates that if not all, a significant measure of gut pathology and resultant systemic inflammation is mediated by morphine-induced microbial dysbiosis, which can be partially restored with “normal” microbial transplant.
Figure 3. Donor microbiome predominates treatment condition and influences gut physiology.
Reciprocal transplant of microbiota preserves distinctness of microbial composition (A), which is closer to the donor microbiome, rather than the treatment condition (placebo or morphine alone; B). This physiologically translates to gut pathologies associated with morphine induced dysbiotic microbiome, e.g. transplantation of morphine-induced dysbiotic microbiome into healthy WT animals results in “morphine-like” diseased phenotype, very similar to morphine-implanted donor animals, whereas transplantation of “normal” microbiome into morphine treated animals shows distinct improvement in the gut pathology (C). Arrows indicate sites of gut injury.
Microbial transplant influences gut immunity
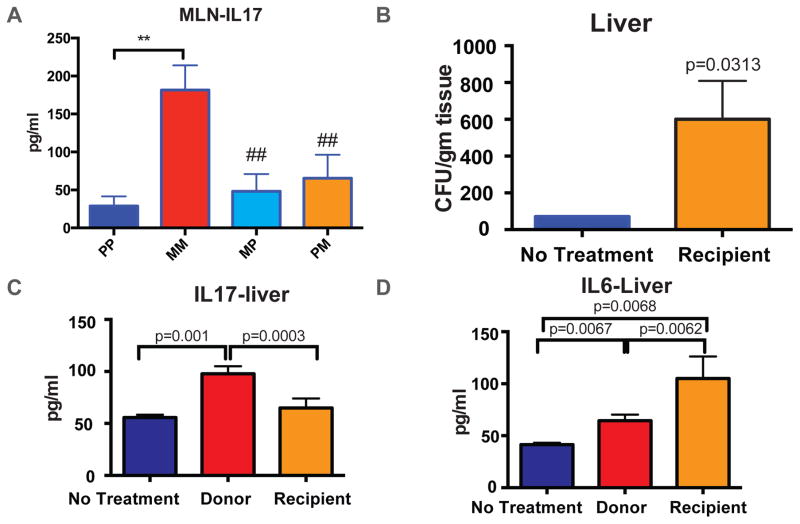
Gut mucosal events leading to host inflammation, follow a cascade, where resident macrophages and dendritic cells within the intestine carry the information to the mesenteric (MLN) and gastric lymph nodes for lymphocyte priming. To study the effect of microbial dysbiosis and host immune response, we performed a multiplexed bead array (CBA) analysis of the MLN homogenate from the animals with fecal transplant from above. Among all cytokines analyzed, only IL17 and IL10 showed significant differences among groups. Elevated levels of IL17 (Fig. 4A) and attenuated levels of IL10 (Figure S3-A) were seen in MM compared to the rest of the four study groups, consistent with the described role of IL17 in host inflammation arising from compromised gut homeostasis, specifically in the context of gram-positive infection29–32. The reciprocally transplanted groups, MP and PM maintained basal levels of the cytokines, comparable with PP. The lack of IL17 or IL10 response in MP or PM animals could be due to compensatory (averaging) effects between microbiome and host immune system. Alternatively, in MP animals, transplanting dysbiotic microbiome may change the general composition of the commensal flora and yet, may not have resulted in severe mucosal barrier compromise to affect systemic immune response within 72 hours. In PM animals, on the other hand, some measure of rescue due to microbial restoration may have resulted in diminished pro-inflammatory response. Additionally, we set out to determine the overall systemic response to microbial dysbiosis by investigating the blood cytokine levels in these animals. No significant differences were found among the WT, TLR2KO and MORKO animals under different treatments (Figure S3-B to D). A significant decrease in IL2 and IL4 levels was observed in the reciprocal fecal transplant animals compared to PP and MM, but no corresponding elevation in pro-inflammatory cytokines was observed (Figure S3-E). This indicates that the 72 hours time point may not be sufficient to elicit pro-inflammatory biomarkers in the systemic circulation; however, localized inflammatory response is initiated within the intestine and mesenteric drainage in immune-competent animals. We have previously shown that morphine exacerbates TLR-mediated inflammation3,4,15 and basal levels of inflammation in the PM animals clearly exhibits protective effect due to restoration of microbial homeostasis with fecal transplant.
Figure 4. Microbial transplant, immune response and rescue.
Morphine treatment results in a robust IL17 response in the Mesenteric Lymph node (MLN) and transplantation of placebo microbiome rescues the inflammatory phenotype (A). Reconstitution of dysbiotic microbiome alone is sufficient to recapitulate most of the morphine induced gut pathologies, including bacterial translocation across the intestinal mucosa (B). Systemic IL17 response was only seen donor animals with morphine treatment (C) and both donors and recipients exhibit a systemic IL6 response indicating pro-inflammatory environment in the host (D). In all cases (A–D), morphine-implanted animals receiving “normal” microbiota show baseline levels of IL17. [**= p<0.05 between indicated groups; ##= p<0.05 compared to MM]. Also See Figure S3.
In the absence of systemic morphine, fecal transplant of dysbiotic microbiome alone was sufficient to induce bacterial translocation to the liver (Fig. 4B). Similar to MM animals in Fig. 4A, fecal transplant induced IL17 response in the liver of the donor mice, but not in the recipient animals (Fig. 4C). There was a significant IL6 response, both in donor and recipient animals (Fig. 4D), indicating that IL17 mediated gut inflammation is contingent upon morphine mediated host immunomodulation, whereas IL6 mediated systemic inflammation is a result of bacterial translocation, common to both donors and recipient animals. This is consistent with our observation that IL17 is an early, whereas IL6 is the sustained host response to morphine-mediated changes in gut homeostasis4, 12.
Morphine-induced cholesterol/bile acid imbalance
Recent reports implicate increase in hydrophobic secondary bile acids (Lithocholic acid; LCA and deoxycholic acid; DCA) and concomitant decrease in hydrophilic secondary bile acids (Ursodeoxycholic acid; UDCA) in gut barrier disruption, bacterial translocation and intestinal inflammation13,14. Since secondary bile acids are produced in entirety through gut microbial fermentation from primary bile acids (Cholic acid; CA and Chenodeoxycholic acid; CDA), secondary bile imbalance is directly correlated, and associated with microbial dysbiosis33. Exclusively within the host liver, primary bile acids are produced from cholesterol catabolism and in fact, one of the major ways of eliminating excess cholesterol from the body34. The secondary bile acids recirculate to the host liver through various bile transporters and also signal through intestinal receptors, constituting the hepato-enteric bile circulation11,35.
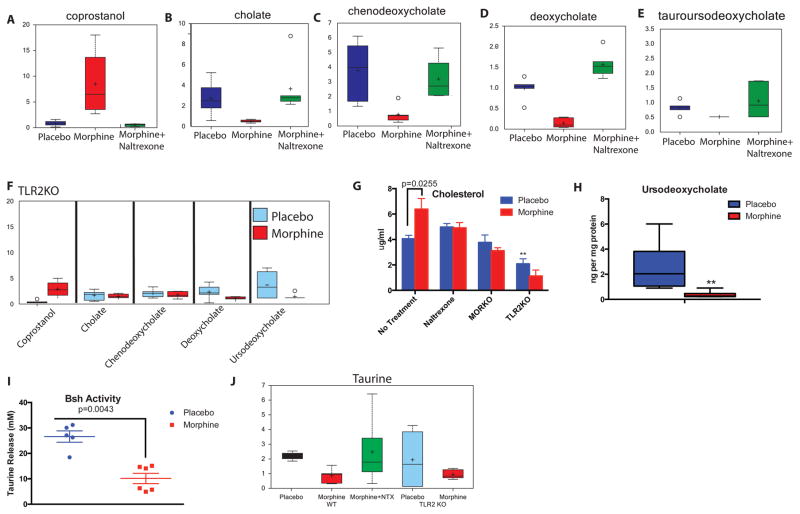
To evaluate if morphine treatment induces metabolic changes in the gut, WT and TLR2KO animals were implanted with placebo or morphine pellet for 72 hours as described above and the fecal content was analyzed with mass-spectrometry for changes in major metabolites. Of 310 compounds tested, lipid metabolites, and especially bile acids (both primary and secondary) exhibited significant changes due to morphine (Figure S4). One of the major observations was significantly high level of coprostanol in the morphine-implanted animals (Fig. 5A). This could be due to abnormal release of cholesterol into the gut, or due to selective expansion of microbes known for this conversion. On the other hand, concentrations of the host-derived primary (Fig. 5B/5C) and microbe-converted secondary bile acids (Fig. 5D/5E) were seen to diminish significantly in the feces of morphine-implanted animals compared to placebo and morphine+naltrexone implanted animals. The morphine-mediated changes were completely abolished in the TLR2KO animals (Fig. 5F).
Figure 5. Morphine induced intestinal and hepatic metabolic changes.
One of the major observations from these studies was that morphine treatment induces the production of significantly high amounts of coprostanol, a direct microbial conversion product of cholesterol in the intestine (A). There is a significant reduction in the abundance of primary bile acids, CA (B) and CDCA (C) and secondary bile acids DCA (D) and UDCA (E) in the fecal contents of morphine treated mice. These affects are reversed in morphine+naltrexone treated (A–E) and TLR2KO animals (F). Morphine induces significant accumulation of cholesterol in the liver, an effect, not seen in naltrexone treated, MORKO and TLR2KO animals (G). These changes in bile/cholesterol levels ultimately result in disrupted hepatoenteric circulation as shown by reduced recovery of UDCA in the liver (H). Bile-salt hydrolase (Bsh), one of the major bacterial enzymes, important for an efficient hepato-enteric circulation shows significantly reduced activity in the microbiome of morphine-treated animals (I). Hence, as expected, a lower level of free taurine was observed in the fecal content of morphine-treated animals, with no significant changes in the TLR2KO animals (J). Also See Figures S4 and S5.
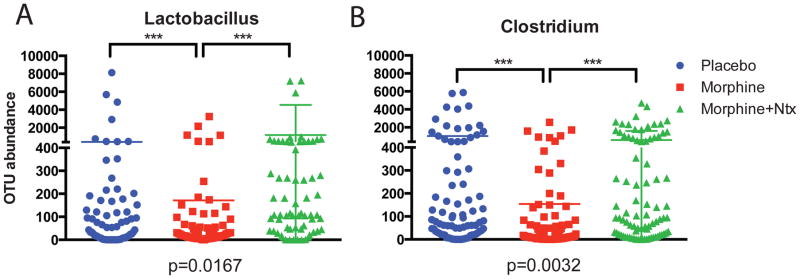
To understand the metabolic status of the host in the context of hepato-enteric circulation and bile metabolism, we subjected the liver lysate of the animals to mass-spectrometric analysis. A significantly elevated level of Cholesterol was seen in morphine-implanted mice (Fig. 5G). These effects are abolished in MORKO animals and with naltrexone. No significant difference were observed between placebo and morphine implanted TLR2KO mice, however, the basal cholesterol levels in these animals were significantly lower than their WT counterparts. Primary bile acid (CA) did not show any significant difference in levels due to morphine treatment (Figure S5-A) and neither did secondary bile acids (DCA, TCA and LCA; Figure S5-B,C and D). Unconjugated Ursodeoxycholate (UDCA), on the other hand, was seen to be significantly down regulated in the morphine implanted animals (Fig. 5H). Since this is a secondary bile acid, its presence in the host liver can only be explained by biliary reabsorption from the intestine. One of the major bacterial enzymes involved in the de-conjugation of secondary bile acids is the Bile salt hydrolase (Bsh) and it’s activity determines the efficacy of hepato-enteric circulation33,36,37. We observed a significant reduction of Bsh activity within the gut bacteria due to morphine treatment (Fig. 5I) and significantly reduced free Taurine in the feces with mass-spectrometry (Fig. 5J). As a direct correlation, among gut bacteria implicated in the de-conjugation of bile acids38,39, we observed significant morphine-mediated lowering in relative abundance of OTUs representing Lactobacillus and Clostridium, (Figure 6). While morphine treatment promotes the expansion of pathogenic/translocating bacteria23 (and Figure S1), it reduces specific bacteria that are responsible for the maintenance of metabolic homeostasis in the gut. Decrease in these bacterial communities may explain the increased cholesterol levels in the liver of morphine-implanted animals due to disruption in hepatoenteric circulation of de-conjugated bile acids and rate limiting steps in cholesterol catabolism. In MORKO and TLR2KO animals (Figure S5-E, F), the levels of secondary bile acids, including UDCA were similar between placebo and morphine implanted mice. CA, the primary bile acid, was significantly diminished in the morphine-implanted TLR2KO animals. This implicates the role of TLR2 in the morphine-mediated modulation of cholesterol hydroxylation in the liver.
Figure 6.
We observed a significant reduction in the abundance of OTUs associated with the genera Lactobacillus and Clostridium in morphine treated mice. These genera have been strongly implicated in de-conjugation of secondary bile acids, enabling their reabsorption through hepatoenteric circulation.
Discussion
Currently there is no alternative to morphine (and its derivatives) for efficient pain management in medical practice. Off-target effects of morphine, especially on the peripheral immune system, however remain a concern in several disease conditions3,5. Dysbiotic microbiome has been independently shown to promote a constant state of inflammation, which becomes critical in the management of several maladies including cancer and HIV infection, confounding the treatment process17. Additionally, microbial dysbiosis is also correlated with gut barrier compromise and bacterial translocation, leading to increased inflammation and endotoxemia23,39,40. One of the major physiological consequences of opioid use is severe constipation, which is speculatively implicated for morphine induced gut pathologies including barrier compromise and bacterial translocation. In our studies, we have seen that constipation resulting from non-opioid inducers, e.g. low fiber diet, does not result in gut barrier disruption/bacterial translocation (Fig S6). In various disease models describing morphine-mediated co-morbidities, gut barrier compromise, bacterial translocation and uncontrolled inflammation play a dominant role4,19,20. Here, we have shown a distinct gram-positive skew in the microbial composition following morphine treatment, which strongly correlates to the clinical presentations attributed to relative increase of gram-positive phyla within the microbiota23,41. While gut commensal flora constitutes a complex ecosystem, stress and disease in the host allows for certain simplified, yet strongly indicative changes in the microbial composition. The Firmicutes/Bacroidetes ratio is one of those “markers” of pro-inflammatory changes in the microbiome, so far studies mostly in the context of aging, obesity and diabetes9,16. In this study, we demonstrate a skew of this ratio towards a pro-inflammatory phenotype, well corroborated with the host immune status involving innate responses. The role of TLRs is well established in mucosal pathogenic complications. Both TLR2 and 4 play a major role in morphine mediated gut barrier compromise and inflammation4,24,40,42–44. Hence, as shown here, morphine induced microbial dysbiosis and inflammation is affected through gut barrier compromise and commensal bacterial translocation through gut mucosa, in a TLR2 and μ-opioid receptor dependent manner3,40. We have recently shown3,23 that morphine treatment results in gut barrier compromise and translocation of predominantly, gram-positive bacteria across gut mucosa. We have also shown3 that TLR stimulation results in myosin light chain kinase (MLCK) mediated withdrawal of tight-junction proteins from the gut epithelial membrane and resultant barrier compromise. In this manuscript, we show the essential role of host immune system in morphine’s effect on microbial dysbiosis (no effect of morphine on microbiome in immune-compromised NSG animals; Figure 2C), implying that morphine’s effects on the microbiome is routed via immune modulation. Finally, we show here, that TLR2KO animals, like NSG, do not show microbial dysbiosis, implicating the role of TLR2 in morphine mediated immune changes, resulting in microbial dysbiosis. One of the functional consequences of morphine-induced microbial dysbiosis is the reprogramming of the host immune status17,24,39–41,45,46. In this study, we clearly show that morphine-induced dysbiotic microbiome alone, can recapitulate diseased gut pathology and immune responses and it is possible to reverse microbial dysbiosis and restore gut immune homeostasis with fecal transplant, which has immense therapeutic potential, as shown previously for treating C. difficile infection25–28. The second and more direct physiological effect of morphine-mediated microbial dysbiosis is its consequence on hepatoenteric circulation of bile acids. Recent reports indicate that bile acid metabolism, its pool in the host and release into the gut, plays a significant role in the manifestation of gut barrier pathology and resultant inflammation. Recently, modulation of cholesterol-7α-hydroxylase (CYP7A1) in the liver and Farnesoid-x-Receptor (FxR) in liver and intestine have been implicated in bile acid dysbiosis and gut barrier compromise13,14,47,48. In this study, morphine induced accumulation of cholesterol in the liver and its excessive conversion to coprostanol in the intestine. Primary and secondary bile acids, however, decreased in the intestine, indicating morphine induced altered cholesterol metabolism in the liver and intestine. At the same time, altered bile release in the gut has adverse consequences in the expansion and maintenance of specific bacterial communities, where bile acids, and their conjugation status influence both sporulation and germination process12,33,49,50.
Our results clearly show a linear correlation between morphine-mediated microbial dysbiosis, disruption of cholesterol/bile acid metabolism and barrier disruption, promoting sustained inflammation in the host, although, the sequence of events are still not clear. We have also demonstrated that microbial reconstitution and timely blockade of TLR2/MOR signaling can restore gut homeostasis in morphine-implanted animals. Additional studies are required to understand the temporal relationship between morphine-treatment, bile acid imbalance, microbial dysbiosis and role and status of bile regulatory receptors e.g. FxR51, bile transporters and the feedback loop including CYP7A135 in the host liver to effectively exploit microbial and bile acid modulation as secondary therapeutic strategy on patients maintained on morphine for pain management.
METHODS
Materials and reagents
Antibodies for flow-cytometry were purchased from BD biosciences (San Jose, CA). Cytokine levels were determined using 13-plex Cytometric bead arrays (CBA) from Biolegend (San Diego, CA). Mass-spectrometry reagents were sourced from various vendors as follows: J.T. Baker Ultra LC/MS-grade acetonitrile (ACN), methanol (MeOH), and water were purchased from VWR International (Radnor, PA). LC/MS Ultra-grade formic acid (Fluka) was purchased from Sigma-Aldrich (Saint Louis, MO, USA). Internal standards 2,2,4,4-D4 cholic acid, 2,2,4,4-D4 deoxycholic acid, 2,2,4,4-D4 chenodeoxycholic acid, 2,2,4,4-D4 ursodeoxycholic acid, and 2,2,4,4-D4 lithocholic acid were purchased from Cambridge Isotope Laboratories (Andover, MA USA). 2,2,4,4-D4 taurocholic acid was purchased from AlsaChim (France). Millipore Amicon Ultrafree PTFE membrane centrifugal filters (0.2 μm), and Millipore Amicon Ultra 0.5 mL 3,000 MWCO centrifugal filters were purchased from Thermo Fisher Scientific (Waltham, MA USA)
Mice
C57BL/6 and NSG mice were purchased from Jackson Laboratories (Bar Harbor, Maine). TLR2KO and MORKO mice were bred in-house. All animals were maintained in pathogen-free facilities and all procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee. Typically, 8–10 week old animals were used for our studies.
Placebo/Morphine/Naltrexone pellet Implantation
Slow release morphine pellets (25mg; ~1μM serum levels of morphine for 5–6 days) and corresponding placebo or naltrexone pellets, as appropriate, were kindly provided by National Institute of Drug Abuse (NIDA, National Institutes of Health, Rockville, MD). The implantation procedure involved 3% isoflurane induced anesthesia, followed by making a small incision at the dorsal torso of the mice. The appropriate pellet was inserted into the small pocket created during incision and the wound was closed using stainless steel wound-clips. The whole process was carried out under aseptic conditions.
Fecal transplant
Two batches of C57Bl6/j (WT) animals (10 each) were implanted with placebo or morphine pellets as mentioned above, for 24 or 48 hours and their fecal contents collected and pooled. The fecal content was processed according to fecal microbial transplant (FMT) procedure described for human patients52 with modifications. Briefly, the fecal contents were suspended in PBS (10mg/ml; w/v), filtered through a 40μ mesh and centrifuged at 6,000Xg for 20 minutes. The resultant microbial pellet was resuspended in half volume of chilled PBS and aliquoted into volumes for single thaw and use. Aliquotes destined for later use were reconstituted to 10% sterile glycerol (Sigma) and stored at −80°. A total of 32 WT mice were used for the transplant experiment. Animals were implanted with placebo or morphine pellets (12 each) and the stored microbiota was administered (106 CFUs/dose) via oral gavage every 24 hours thrice according to the following scheme: [a] Placebo pellet implanted animals getting placebo microbiome (PP; n=8), [b] Placebo pellet implanted animals getting morphine microbiome (PM; n=8), [c] Morphine pellet implanted animals getting placebo microbiome (MP; n=8) and [d] Morphine pellet implanted animals getting morphine microbiome (MM; n=8). Animals were sacrificed 24hours after third transplant and fecal contents and tissues harvested for various downstream analyses as described.
Sequencing and 16S DNA analysis
Fecal content was collected from gut region encompassing distal cecum and approximately one inch of the colon and frozen on dry ice. The fecal matter was lysed using glass beads in MagnaLyser tissue disruptor (Roche) and total DNA isolated using Power-soil/fecal DNA isolation kit (Mo-Bio) as per manufacturer’s specifications. All samples was quantified via the Qubit® Quant-iT dsDNA Broad-Range Kit (Invitrogen, Life Technologies, Grand Island, NY) to ensure that they met minimum concentration and mass of DNA and submitted to either Second genome Inc. or University of Minnesota Genomic Center for microbiome analysis as follows: To enrich the sample for the bacterial 16S V4 rDNA region, DNA was amplified utilizing fusion primers designed against the surrounding conserved regions which are tailed with sequences to incorporate Illumina (San Diego, CA) flow cell adapters and indexing barcodes. Each sample was PCR amplified with two differently bar coded V4 fusion primers and were advanced for pooling and sequencing. For each sample, amplified products were concentrated using a solid-phase reversible immobilization method for the purification of PCR products and quantified by electrophoresis using an Agilent 2100 Bioanalyzer®. The pooled 16S V4 enriched, amplified, barcoded samples were loaded into the MiSeq® reagent cartridge, and then onto the instrument along with the flow cell. After cluster formation on the MiSeq instrument, the amplicons were sequenced for 250 cycles with custom primers designed for paired-end sequencing. Using QIIME, sequences were quality filtered and demultiplexed using exact matches to the supplied DNA barcodes. Resulting sequences were then searched against the Greengenes reference database of 16S sequences, clustered at 97% by uclust (closed-reference OTU picking). Analysis for alpha- and beta-diversity was done with standardized qiime workflow at the Minnesota Supercomputing Institute (University of Minnesota). To compute the “global changes with morphine” histogram (Fig 1D), relative abundance for all OTUs constituting a single phylum in morphine treated animals were normalized with the same OTUs in the placebo animals, including mismatched OTUs. The resulting ratio was analyzed using Prism (GraphPad) software to understand morphine-mediated perturbations in the 5 major phyla within the microbiome. The raw data files for 16s rDNA sequencing have been deposited with ArrayExpress with the accession numbers E-MTAB-3722 (Native effects of morphine) and E-MTAB-3723 (microbial transplant).
Mass Spectrometry for gut and liver metabolites
Fecal metabolites were analysed by Metabolon Inc. (Research Triangle Park, NC) using their proprietary 310 named biochemicals screen. Based on the results, independent analysis of liver bile acids was performed at the University of Minnesota Mass-spectrometric facility as follows: Samples for analysis by UPLC-MS/MS were spiked with a fixed volume of the isotopically-labeled bile acid internal standards described above. To each sample was added an equal volume of LC/MS-grade methanol. The samples were then centrifuged at 12,000 rpm for 5 min and the supernatants removed from the proteinaceous pellet. These supernatants were centrifuged through an Amicon Ultrafree low-binding hydrophilic PTFE membrane (0.2 μm) at 12,000 rpm and the filtrates collected. These filtrates were then centrifuged at 14,000 × g through an Amicon Ultra 3kDa MW cutoff filtration column (Millipore) for 30 min, and the flow-through collected for UPLC/MS/MS analysis. A Waters Acquity UPLC coupled to a Waters triple quadrupole mass spectrometer (Acquity TQD) was used for separation and detection of bile acids. A Waters CORTECS C18 2.1 mm × 100 mm column (2.7 μm particles) at 40 ° C was used during the following 19 min gradient separation with A: water containing 0.1% formic acid and B: ACN containing 0.1% formic acid, at a flow rate of 0.6 mL/min: 35% B to 40% B, 0 min to 1.5 min; 40% B to 50% B, 1.5 min to 6.0 min; 50% B 6.0 min to 7.0 min; 50% B to 97% B, 7.0 min to 14.0 min; 97% B, 14.0 min to 16.0 min; 97% B to 35% B, 16.0 min to 17.0 min; and 35% B, 17.0 min to 19.0 min. By directly infusing each of the bile acids and corresponding internal standards, cone voltages and collision energies for each selected reaction monitoring (SRM) transition were optimized. The transitions that produced the highest sensitivity for the determination of each analyte were selected for quantification. Note that MS1 selected ion monitoring of the precursor ions specified lacked the sensitivity and selectivity of MS2 measurement of the following precursor-to-precursor MS/MS transitions. Note also that method development and validation included identity verification via unique precursor-fragment MS/MS transitions for each analyte. The following transitions were selected for quantitative analysis: lithocholic acid: 375.4 to 375.4; 2,2,4,4-D4 lithocholic acid: 379.4 to 379.4; cheno-urso- and deoxycholic acids: 391.4 to 391.4; 2,2,4,4-D4 cheno-urso- and deoxycholic acids: 395.4 to 395.4; cholic acid: 407.3 to 407.3; 2,2,4,4-D4 cholic acid: 411.3 to 411.3; taurocholic acid: 514.1 to 514.1; 2,2,4,4-D4 taurocholic acid: 518.1 to 518.1. Dwell time for each transition was 0.05 s. For electrospray ionization tandem mass spectrometry (ESI-MS/MS) in negative ionization mode, parameters were as follows: capillary, 3.2 kV; cone, 70 V; extractor, 3 V; rf lens, 0.3 V; source temperature, 150 °C; desolvation temperature, 500 °C; desolvation flow, 800 L/h; cone gas flow, 20 L/h; low-mass resolution (Q1), 15 V; high-mass resolution (Q1), 15 V; ion energy (Q1), 0.2 V; entrance −5 V; exit, 1 V; collision energy 5 V; low-mass resolution (Q2), 15 V; high-mass resolution (Q2), 15 V; ion energy (Q2) 3.5 V. For standardization, 6 levels of calibration mixtures for each bile acid ranging from 0 ng/mL to 250 μg/mL were prepared to achieve 6 different response ratios in the mixtures. These solutions were then analyzed by UPLC-MS/MS, and the data were subjected to a linear least squares analysis with the Waters Targetlynx™ software program. The peak area ratios of analyte:internal standard were then used in conjunction with the calibration curves to determine the concentrations of bile acids in the samples.
Bile Salt Hydrolase Assay
For microbial bile salt hydrolase assay, method described in Kumar et. al.37 was used with modifications. Briefly, weight-matched fecal content from placebo and morphine-treated animals were resuspended in chilled PBS and filtered sequentially through 100, 40 and 20μ mesh (BD). The filtered suspension was centrifuged at 500×g (supernatant collected), 1000×g (supernatant collected) and finally, 10,000×g (pellet collected). The microbial pellets were resuspended in 100μl of 0.5M citrate buffer and bacterial cells were disrupted using an ultrasonic homogenizer. Bacterial cytoplasmic content was separated from debris by centrifugation (20,000×g) and the supernatant was used for Bsh assay. Bacterial cytoplasmic fraction was incubated with or without 0.5M tauro-deoxycholate (Fisher) at 37° for 30 minutes and release of free taurine was measured using 1% ninhydrin at 570nm. Bacterial cytoplasmic fraction, without tauro-deoxycholate incubation was used to determine free amino acids and used as a background for normalization.
Histology
Tissues were harvested and preserved in 10% formaldehyde. H&E staining was performed by the Comparative Pathology Shared Resource (CPSR and Bionet) at the University of Minnesota and slides were imaged using a Leica DM5500 B microscope. Representative images are shown.
Statistical Analysis
Microbiome analysis OTU tables were rarefied to the sample containing the lowest number of sequences in each analysis. Qiime 1.8 was used to calculate alpha diversity (alpha_rarefaction.py) and to summarize taxa (summarize_taxa_through_plots.py). Principal Coordinate Analysis was done within this program using observation ID level. Heatmaps were generated using family level (L5) taxonomic data using R based Phyloseq or using Explicet as described. The Adonis test was utilized for finding significant whole microbiome differences among discrete categorical or continuous variables. In this randomization/Monte Carlo permutation test, the samples were randomly reassigned to the various sample categories, and the mean normalized cross-category differences from each permutation are compared to the true cross-category differences. The fraction of permutations with greater distinction among categories (larger cross-category differences) than that observed with the non-permuted data reported as the p-value for the Adonis test. Cytokine concentrations and bile acid changes from plasma and liver is expressed as ± SEM. Significance is defined as p<0.05 between groups in an unpaired student’s t test. Bacterial counts were reported as means of CFU and were analyzed by the Mann-Whitney U test (GraphPad Prism). For metabolite analysis by mass-spectrometry, Welch’s two-sample t-test was used to identify biochemicals that differed significantly between experimental groups. An estimate of the false discovery rate (q-value<0.10) was calculated to take into account the multiple comparisons that normally occur in metabolomic-based studies. Biochemical importance plot was obtained using random forest analysis (a statistical tool for biomarker selection utilizing a supervised classification technique based on an ensemble of decision trees). In this study, the metabolic profiles of fecal samples from 5 groups (WT+Placebo, WT+Morphine, WT+Morphine+NTX, TLR2KO+Placebo and TLR2KO+Morphine) were compared amongst each other. Figure S4-A lists the top 30 candidates based on importance to separating genotype/treatment. A predictive accuracy of 97% was observed based on key differences in lipid and bile metabolism.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH grants RO1 DA 12104, RO1 DA 022935, RO1 DA031202, K05DA033881, P50 DA 011806, 1R01DA034582 and 1R01DA037843 to SR and T32DA007097 (SR and others), and 1R21HL125021(SB).
Footnotes
Additional Information
The author(s) declare no competing financial or conflict of interests.
Author Contributions
SR conceptualized the project. SB, TJ and SR designed experiments. SB, GS, FW, JJ, US, LZ, PD, CC and JD did experiments. SB and SR wrote the manuscript.
SB, GS and FW contributed equally to this work.
References
- 1.Ninković J, Roy S. Morphine decreases bacterial phagocytosis by inhibiting actin polymerization through cAMP-, Rac-1-, and p38 MAPK-dependent mechanisms. Am J Pathol. 2012;180:1068–79. doi: 10.1016/j.ajpath.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy S, et al. Opioid drug abuse and modulation of immune function: consequences in the susceptibility to opportunistic infections. J neuroimmune Pharmacol. 2011;6:442–65. doi: 10.1007/s11481-011-9292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng J, et al. Morphine induces bacterial translocation in mice by compromising intestinal barrier function in a TLR-dependent manner. PLoS One. 2013;8:e54040. doi: 10.1371/journal.pone.0054040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee S, et al. Morphine induced exacerbation of sepsis is mediated by tempering endotoxin tolerance through modulation of miR-146a. Sci Rep. 2013;3:1977. doi: 10.1038/srep01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilburger ME, et al. Morphine induces sepsis in mice. J Infect Dis. 1997;176:183–8. doi: 10.1086/514021. [DOI] [PubMed] [Google Scholar]
- 6.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura K, Slusher N. Role of the gut microbiota in defining human health. Expert Rev anti- …. 2010;8:435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J Physiol. 2012;590:441–6. doi: 10.1113/jphysiol.2011.222133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr. 2014;111:387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- 10.Bourzac K. The bacterial tightrope. Nature. 2014;516:S14–S16. doi: 10.1038/516S14a. [DOI] [PubMed] [Google Scholar]
- 11.Dawson PA, Karpen SJ. Intestinal Transport and Metabolism of Bile Acids. J Lipid Res. 2014 doi: 10.1194/jlr.R054114. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Chiang JYL. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenman LK, Holma R, Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J Gastroenterol. 2012;18:923–929. doi: 10.3748/wjg.v18.i9.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:G227–34. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 15.Sindberg GM, et al. An Infectious Murine Model for Studying the Systemic Effects of Opioids on Early HIV Pathogenesis in the Gut. J Neuroimmune Pharmacol. 2014 doi: 10.1007/s11481-014-9574-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs JP, Braun J. Immune and genetic gardening of the intestinal microbiome. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breslow JM, et al. Potentiating effect of morphine on oral Salmonella enterica serovar Typhimurium infection is μ-opioid receptor-dependent. Microb Pathog. 2010;49:330–5. doi: 10.1016/j.micpath.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Barke Ra, Charboneau R, Roy S. Morphine impairs host innate immune response and increases susceptibility to Streptococcus pneumoniae lung infection. J Immunol. 2005;174:426–34. doi: 10.4049/jimmunol.174.1.426. [DOI] [PubMed] [Google Scholar]
- 20.El-Hage N, Dever SM, Fitting S, Ahmed T, Hauser KF. HIV-1 coinfection and morphine coexposure severely dysregulate hepatitis C virus-induced hepatic proinflammatory cytokine release and free radical production: increased pathogenesis coincides with uncoordinated host defenses. J Virol. 2011;85:11601–14. doi: 10.1128/JVI.05239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babrowski T, et al. Pseudomonas aeruginosa virulence expression is directly activated by morphine and is capable of causing lethal gut-derived sepsis in mice during chronic morphine administration. Ann Surg. 2012;255:386–93. doi: 10.1097/SLA.0b013e3182331870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ocasio FM, Jiang Y, House SD, Chang SL. Chronic morphine accelerates the progression of lipopolysaccharide-induced sepsis to septic shock. J Neuroimmunol. 2004;149:90–100. doi: 10.1016/j.jneuroim.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Meng J, et al. Opioid Exacerbation of Gram-positive sepsis, induced by Gut Microbial Modulation, is Rescued by IL-17A Neutralization. Sci Rep. 2015;5:10918. doi: 10.1038/srep10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asselin C, Gendron F-P. Shuttling of information between the mucosal and luminal environment drives intestinal homeostasis. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.02.049. [DOI] [PubMed] [Google Scholar]
- 25.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the Composition of the Human Fecal Microbiome After Bacteriotherapy for Recurrent. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 26.Khoruts a, Sadowsky MJ. Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol. 2011;4:4–7. doi: 10.1038/mi.2010.79. [DOI] [PubMed] [Google Scholar]
- 27.Kelly CR, et al. Fecal Microbiota Transplant for Treatment of Clostridium difficile Infection in Immunocompromised Patients. Am J Gastroenterol. 2014;109:1065–71. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoruts A, Sadowsky MJ, Hamilton MJ. Development of Fecal Microbiota Transplantation Suitable for Mainstream Medicine. Clin Gastroenterol Hepatol. 2014 doi: 10.1016/j.cgh.2014.11.014. in press. [DOI] [PubMed] [Google Scholar]
- 29.O’Connor W, Zenewicz La, Flavell Ra. The dual nature of T(H)17 cells: shifting the focus to function. Nat Immunol. 2010;11:471–476. doi: 10.1038/ni.1882. [DOI] [PubMed] [Google Scholar]
- 30.Meng J, Li D, Roy S, Wang F. Morphine exacerbates experimental sepsis by modulating IL17/IL22 response (IRC8P.488) J Immunol. 2014;192:190.16–190.16. [Google Scholar]
- 31.Takahashi N, et al. IL-17 produced by Paneth cells drives TNF-induced shock. J Exp Med. 2008;205:1755–61. doi: 10.1084/jem.20080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappu R, Rutz S, Ouyang W. Regulation of epithelial immunity by IL-17 family cytokines. Trends Immunol. 2012;33:343–9. doi: 10.1016/j.it.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–8. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloch K, Berg B, Rittenberg D. The Biological Conversion of Cholesterol to cholic acid. J Biol Chem. 1943;149:511–517. [Google Scholar]
- 35.Jones ML, Tomaro-Duchesneau C, Prakash S. The gut microbiome, probiotics, bile acids axis, and human health. Trends Microbiol. 2014;22:306–8. doi: 10.1016/j.tim.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Nat Acad Sci. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar R, Grover S, Batish VK. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase-producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br J Nutr. 2011;105:561–73. doi: 10.1017/S0007114510003740. [DOI] [PubMed] [Google Scholar]
- 38.Cardona M, Vanay V, Midtveldt T, Norin KE. Probiotics in gnotobiotic mice. Microb Ecol Health Dis. 2000;12:219–224. [Google Scholar]
- 39.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–84. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–73. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez-Hurtado I, et al. Gut microbiota dysbiosis is associated with inflammation and bacterial translocation in mice with CCl4-induced fibrosis. PLoS One. 2011;6:e23037. doi: 10.1371/journal.pone.0023037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Ding J, Chang TL. TLR2 activation enhances HIV nuclear import and infection through T cell activation-independent and -dependent pathways. J Immunol. 2012;188:992–1001. doi: 10.4049/jimmunol.1102098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutta R, et al. Morphine Modulation of Toll-Like Receptors in Microglial Cells Potentiates Neuropathogenesis in a HIV-1 Model of Coinfection with Pneumococcal Pneumoniae. J Neurosci. 2012;32:9917–30. doi: 10.1523/JNEUROSCI.0870-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garsin Da, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, prebiotics and immunomodulation of gut mucosal defences: homeostasis and immunopathology. Nutrients. 2013;5 doi: 10.3390/nu5061869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renga B, et al. The Bile Acid Sensor FXR Is Required for Immune-Regulatory Activities of TLR-9 in Intestinal Inflammation. PLoS One. 2013:8. doi: 10.1371/journal.pone.0054472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakiyama G, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2014;306:929–937. doi: 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weingarden AR, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–9. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weingarden AR, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–9. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaik FB, Prasad DVR, Narala VR. Role of farnesoid X receptor in inflammation and resolution. Inflamm Res. 2015;64:9–20. doi: 10.1007/s00011-014-0780-y. [DOI] [PubMed] [Google Scholar]
- 52.Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:761–7. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.