Abstract
In peanut (Arachis hypogaea L.), the customization of fatty acid profile is an evolving area to fulfill the nutritional needs in the modern market. A total of 174 peanut genotypes, including 167 Indian cultivars, 6 advanced breeding lines and “SunOleic95R”—a double mutant line, were investigated using AS-PCRs, CAPS and gene sequencing for the ahFAD2 allele polymorphism, along with its fatty acid compositions. Of these, 80 genotypes were found having substitution (448G>A) mutation only in ahFAD2A gene, while none recorded 1-bp insertion (441_442insA) mutation in ahFAD2B gene. Moreover, 22 wild peanut accessions found lacking both the mutations. Among botanical types, the ahFAD2A mutation was more frequent in ssp. hypogaea (89%) than in ssp. fastigiata (17%). This single allele mutation, found affecting not only oleic to linoleic acid fluxes, but also the composition of other fatty acids in the genotypes studied. Repeated use of a few selected genotypes in the Indian varietal development programs were also eminently reflected in its ahFAD2 allele polymorphism. Absence of known mutations in the wild-relatives indicated the possible origin of these mutations, after the allotetraploidization of cultivated peanut. The SNP analysis of both ahFAD2A and ahFAD2B genes, revealed haplotype diversity of 1.05% and 0.95%, while Ka/Ks ratio of 0.36 and 0.39, respectively, indicating strong purifying selection pressure on these genes. Cluster analysis, using ahFAD2 gene SNPs, showed presence of both mutant and non-mutant genotypes in the same cluster, which might be due the presence of ahFAD2 gene families. This investigation provided insights into the large number of Indian peanut genotypes, covering various aspects related to O/L flux regulation and ahFAD2 gene polymorphism.
Keywords: FAD2 gene, allele-specific PCR, fatty-acids, groundnut, O/L ratio, single nucleotide polymorphism
Introduction
Cultivated peanut or groundnut (Arachis hypogaea L.) is an allotetraploid crop (2n = 4x = 40, AABB) having two subspecies, spp. hypogaea and spp. fastigiata (Krapovickas and Rigoni, 1960). It is cultivated in more than 100 countries, mostly by the small and marginal farmers, under low-input conditions (Bhauso et al., 2014; Sarkar et al., 2014). Peanut is one among the major oilseed crops, which contribute to the bulk of total oil production worldwide (Mishra et al., 2015). Indian vegetable oil economy is world's fourth largest after USA, China and Brazil. India ranks first in acreage, occupying 5.25 M ha under cultivation and second in production (9.47 M tons) in the world, after China (16.91 M tons) (FAOSTAT, 2014).
In peanut seed oil, two unsaturated fatty acids (UFA), oleic acid (C18:1, Δ9), a mono UFA (MUFA) and linoleic acid (C18:2, Δ9, Δ12), a poly UFA (PUFA) contribute around 80% of the total oil composition. Further, a saturated fatty acid (SFA), palmitic acid contributing to about 10%, whereas, rest 10% is constituted of up to 9 other fatty acids (Janila et al., 2016). Thus, the flavor, shelf-life, and nutritional quality of peanut seeds and its products are reliant on the proportion of three main fatty acids viz., oleic, linoleic and palmitic acid present in its oil (Derbyshire, 2014). Oils containing high percentage of linoleic acid are prone to oxidation, leading to rancidity, off-flavors, and short shelf-life (Mondal et al., 2010). Oleic acid has 10-fold higher auto-oxidative stability than linoleic acid; therefore, high O/L peanut has a longer shelf life (O'Keefe et al., 1993). The palmitic acid is reported to increase the risk for multiple life-threatening diseases such as cardio-vascular diseases (CVD) and atrial-fibrillation (Wang et al., 2015a).
High-oleic oil is an excellent solution for food manufacturers, looking for healthy alternatives to saturated or hydrogenated oils (Cao et al., 2013). Its neutral flavor and odor, make the oil highly suitable for a wide range of food related applications, including spray coating and frying (Pandey et al., 2014). A diet with a high oleic acid and low palmitic acid, is an exceptional way to reduce the risk of heart diseases, promotes a healthier ratio of high density lipoprotein (HDL) to low density lipoprotein (LDL), and reduces triacylglycerol and blood glucose levels (Vassiliou et al., 2009; Janila et al., 2013).
Normal peanut genotypes contain about 36–70% oleic while 15–43% linoleic acid (Knauft et al., 1993). However, in 1987, Norden and co-workers identified the first high-oleate mutant lines, F435 with about 80% oleic acid and 2% linoleic acid. So far, more than 50 high-oleate peanut cultivars were registered worldwide, which are derived through traditional breeding, mutagenesis, marker-assisted selection (MAS) and marker assisted backcross breeding (MABB) (Wang et al., 2015c; Janila et al., 2016). Following conventional breeding methods, the first high oleate peanut line, “SunOleic95R” was bred in USA (Gorbet and Knauft, 1997); whereas, “Tifguard High O/L” was developed using MAS (Chu et al., 2011).
In plants, oleate to linoleate conversion is catalyzed by oleoyl-PC (phosphatidyl choline) desaturase, which exists in endoplasmic reticula and chloroplast membrane, and incorporates a second double bond to oleic acid (Schwartzbeck et al., 2001). In peanuts, two homeologous genes, ahFAD2A and ahFAD2B having 99% sequence similarity, are reported to regulate the desaturase activity (Jung et al., 2000b; Lopez et al., 2000). A single base pair (bp) substitution (G:C/A:T) mutation at 448 bp position in ahFAD2A gene, results in a missense amino acid from aspartic acid to asparagine (D150N). While, 1-bp insertion (A:T) mutation in ahFAD2B gene, at 442 bp position results in frame-shift mutation, which generates a premature stop codon (Jung et al., 2000b; Lopez et al., 2000). Both these mutations lead to the altered ahFAD2 gene expression, resulting in reduced enzymatic activity, which leads to high O/L ratio in the mutant genotypes (Jung et al., 2000a; Chu et al., 2009). Recently, Wang et al. (2015b) has identified two natural mutant lines (PI342664 and PI342666) with 80% oleic acid, having substitutions of G448A in FAD2A (same as previously identified) and C301G in FAD2B (new mutation) for both mutant lines, resulting in a missense amino acid substitution of D150N, and H101D, respectively. However, many reports also indicate the epistatic regulation, presence of ahFAD2 gene families, and existence of other unidentified genetic factors, conditioning fatty-acid content in peanut (Fang et al., 2012; Wang et al., 2015b,d). The various QTLs for ahFAD2 genes were reported to be located on 6th and 9th linkage group of both A and B genomes of cultivated peanut (Pandey et al., 2014; Wang et al., 2015a).
To enhance the efficiency of peanut breeding program for high oleate, different molecular tools for accurate genotyping of ahFAD2 genes includes; cleaved amplified polymorphic sequences (CAPS) markers for ahFAD2A (Chu et al., 2007) and ahFAD2B alleles (Chu et al., 2009), real-time PCR (Barkley et al., 2010, 2011) and allele-specific PCR (AS-PCR) assays (Chen et al., 2010; Yu et al., 2013). These tools have been successfully utilized for the screening of peanut germplasm (Chu et al., 2007; Wang et al., 2011a,b, 2013b; Mukri et al., 2012). However, no such efforts have been reported for the Indian peanut cultivars and advanced breeding lines, which are being used in improving the O/L ratio of the Indian genotypes by the breeders.
Looking at the availability of various ahFAD2 gene mutant high oleic lines in peanut, identification of Indian peanut cultivars for ahFAD2 gene polymorphism and fatty acid composition seems essential for boosting the peanut breeding programme for oil quality traits. Therefore, the present study was aimed to find the relationship between ahFAD2 allele polymorphism and its fatty-acid composition, especially O/L fluxes in Indian peanut genotypes.
Materials and methods
Plant materials
A total of 196 peanut genotypes; including 167 Indian cultivars, 6 advance breeding lines, SunOleic 95R− a high oleic (~80%) genotype, and 22 wild-species, representing different section were used for this study (Tables 1, 2). The seeds of these genotypes were obtained from the Genetic Resources Section of the ICAR-Directorate of Groundnut Research (DGR), Gujarat (India).
Table 1.
Fatty acid composition (%), O/L ratio and ahFAD2A substitution mutation in the 167 peanut varieties and six advance breeding lines.
| S. no. | Genotypes | Market type | Palmitic acid (C16:0) | Stearic acid (C18:0) | Oleic acid (C18:1) | Linoleic acid (C18:2) | Arachidic acid (C20:0) | Gadoleic acid (C20:1) | Behenic acid (C22:0) | Lignoceric acid (C24:0) | O/L ratio | Substitution mutation in ahFAD2A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SunOleic95R | VR | 6.93 | 2.28 | 80.18 | 7.34 | 0.78 | 1.26 | 1.00 | 0.23 | 10.92 | + |
| 2 | RG510 | VR | 10.07 | 3.98 | 63.45 | 19.04 | 1.10 | 0.83 | 1.21 | 0.31 | 3.33 | + |
| 3 | Kadiri71-1 | VR | 9.64 | 3.20 | 63.50 | 20.12 | 1.06 | 0.94 | 1.19 | 0.36 | 3.16 | + |
| 4 | GG16 | VR | 9.00 | 2.25 | 63.96 | 20.58 | 0.93 | 1.01 | 1.75 | 0.52 | 3.11 | + |
| 5 | UF70-103 | VR | 9.90 | 3.20 | 62.22 | 20.33 | 1.18 | 1.16 | 1.58 | 0.42 | 3.06 | + |
| 6 | GG14 | VR | 10.65 | 3.17 | 59.50 | 21.17 | 1.29 | 1.10 | 2.21 | 0.88 | 2.81 | + |
| 7 | GJG17 | VR | 9.77 | 2.30 | 61.35 | 22.05 | 1.03 | 1.08 | 1.87 | 0.54 | 2.78 | + |
| 8 | GG13 | VR | 10.25 | 2.60 | 59.63 | 23.01 | 1.15 | 1.24 | 1.78 | 0.31 | 2.59 | + |
| 9 | GG11 | VR | 10.46 | 2.52 | 58.86 | 22.73 | 1.21 | 1.23 | 2.28 | 0.54 | 2.59 | + |
| 10 | GG 15 | VR | 10.43 | 2.32 | 59.95 | 23.32 | 0.95 | 1.02 | 1.60 | 0.41 | 2.57 | + |
| 11 | Karad 4-11 | VR | 10.02 | 1.98 | 60.76 | 23.77 | 0.75 | 1.01 | 1.14 | 0.27 | 2.56 | + |
| 12 | RS1 | VR | 10.43 | 2.13 | 59.97 | 23.57 | 1.04 | 0.89 | 1.30 | 0.41 | 2.54 | + |
| 13 | M13 | VR | 11.51 | 2.86 | 57.81 | 24.10 | 1.04 | 1.04 | 1.22 | 0.22 | 2.40 | + |
| 14 | M548 | VR | 11.03 | 2.83 | 57.08 | 24.61 | 1.18 | 0.91 | 1.78 | 0.55 | 2.32 | + |
| 15 | S230 | VR | 10.19 | 3.04 | 58.33 | 25.18 | 0.99 | 0.66 | 1.12 | 0.23 | 2.32 | + |
| 16 | Punjab1 | VR | 10.95 | 2.45 | 58.09 | 25.11 | 0.75 | 0.80 | 1.24 | 0.42 | 2.31 | + |
| 17 | GAUG10 | VR | 11.15 | 2.63 | 57.47 | 24.94 | 1.12 | 0.92 | 1.38 | 0.39 | 2.30 | + |
| 18 | M37 | VR | 10.77 | 2.54 | 57.80 | 25.50 | 0.92 | 0.98 | 1.16 | 0.25 | 2.27 | + |
| 19 | Faizapur1-5 | VR | 10.68 | 2.56 | 57.21 | 25.36 | 1.00 | 1.06 | 1.66 | 0.49 | 2.26 | + |
| 20 | DSG1 | VR | 10.79 | 2.48 | 56.14 | 26.00 | 1.13 | 1.18 | 1.72 | 0.53 | 2.16 | + |
| 21 | TMV4 | VR | 10.70 | 2.17 | 55.86 | 25.98 | 1.09 | 1.24 | 2.13 | 0.82 | 2.15 | + |
| 22 | GG12 | VR | 11.09 | 2.34 | 55.03 | 26.14 | 0.97 | 1.09 | 1.80 | 0.54 | 2.11 | + |
| 23 | CSMG9510 | VR | 10.77 | 3.16 | 55.35 | 26.94 | 1.07 | 0.84 | 1.57 | 0.32 | 2.05 | + |
| 24 | M335 | VR | 11.90 | 2.94 | 54.42 | 26.64 | 1.16 | 1.04 | 1.61 | 0.29 | 2.04 | + |
| 25 | TMV1 | VR | 10.15 | 2.40 | 55.72 | 27.52 | 0.91 | 1.02 | 1.62 | 0.66 | 2.02 | + |
| 26 | Chandra | VR | 12.12 | 2.70 | 54.17 | 27.50 | 1.01 | 0.93 | 1.26 | 0.24 | 1.97 | + |
| 27 | CSMG84-1 | VR | 12.79 | 4.08 | 50.48 | 25.91 | 1.76 | 1.05 | 3.30 | 0.52 | 1.95 | + |
| 28 | TMV3 | VR | 10.50 | 2.13 | 54.71 | 28.58 | 0.95 | 1.12 | 1.54 | 0.48 | 1.91 | + |
| 29 | T28 | VR | 10.68 | 1.65 | 53.99 | 28.59 | 1.68 | 1.61 | 1.38 | 0.42 | 1.89 | + |
| 30 | Chitra / MA10 | VR | 12.94 | 3.63 | 50.55 | 28.67 | 1.31 | 1.00 | 1.53 | 0.28 | 1.76 | + |
| 31 | GJG-HPS-1 | VR | 10.99 | 2.19 | 51.53 | 30.09 | 0.99 | 1.26 | 2.21 | 0.74 | 1.71 | + |
| 32 | Somnath | VR | 10.94 | 2.25 | 49.54 | 32.04 | 1.50 | 0.89 | 2.00 | 0.51 | 1.55 | + |
| 33 | GJG22 | VB | 9.25 | 3.33 | 65.42 | 17.41 | 1.27 | 1.06 | 1.68 | 0.58 | 3.76 | + |
| 34 | GG21 | VB | 9.51 | 3.68 | 64.78 | 17.86 | 1.24 | 0.82 | 1.70 | 0.43 | 3.63 | + |
| 35 | GG20 | VB | 10.16 | 3.62 | 63.93 | 19.68 | 0.91 | 0.52 | 0.82 | 0.19 | 3.25 | + |
| 36 | TMV10 | VB | 7.92 | 0.56 | 66.57 | 21.89 | 0.82 | 0.70 | 1.55 | 0.00 | 3.04 | + |
| 37 | RG425 | VB | 10.00 | 3.54 | 61.52 | 21.11 | 0.95 | 0.87 | 1.65 | 0.36 | 2.91 | + |
| 38 | BAU13 | VB | 9.63 | 2.90 | 63.30 | 21.98 | 0.69 | 0.30 | 1.00 | 0.20 | 2.88 | + |
| 39 | ICGS76 | VB | 11.73 | 2.60 | 58.42 | 24.10 | 0.75 | 0.69 | 0.84 | 0.34 | 2.42 | + |
| 40 | MA16 | VB | 11.02 | 2.58 | 57.65 | 24.33 | 1.08 | 0.86 | 1.81 | 0.60 | 2.37 | + |
| 41 | HNG10 | VB | 8.97 | 1.98 | 59.81 | 25.53 | 0.96 | 0.82 | 1.41 | 0.53 | 2.34 | + |
| 42 | ICGV86325 | VB | 9.93 | 2.04 | 59.37 | 25.52 | 0.87 | 0.87 | 0.67 | 0.58 | 2.33 | + |
| 43 | Kadiri 7 Bold | VB | 11.51 | 2.86 | 57.51 | 24.74 | 0.97 | 0.84 | 1.16 | 0.41 | 2.32 | + |
| 44 | T64 | VB | 12.06 | 1.73 | 57.65 | 25.07 | 0.70 | 0.77 | 1.36 | 0.35 | 2.30 | + |
| 45 | ALR1 | VB | 10.88 | 3.76 | 55.84 | 24.39 | 1.65 | 0.99 | 2.13 | 0.36 | 2.29 | + |
| 46 | BG2 | VB | 10.98 | 1.89 | 58.47 | 26.00 | 0.47 | 0.98 | 0.98 | 0.23 | 2.25 | + |
| 47 | M145 | VB | 10.75 | 2.26 | 57.63 | 25.81 | 0.92 | 0.88 | 1.27 | 0.49 | 2.23 | + |
| 48 | ICGS5 | VB | 10.89 | 1.73 | 58.03 | 26.15 | 0.80 | 0.80 | 1.03 | 0.43 | 2.22 | + |
| 49 | Kaushal | VB | 11.07 | 2.69 | 56.63 | 26.25 | 1.02 | 1.03 | 1.00 | 0.23 | 2.16 | + |
| 50 | CSMG884 | VB | 11.87 | 2.28 | 55.51 | 26.31 | 1.14 | 0.86 | 1.74 | 0.31 | 2.11 | + |
| 51 | Kadiri2 | VB | 11.54 | 2.20 | 55.93 | 27.03 | 0.80 | 1.14 | 1.08 | 0.28 | 2.07 | + |
| 52 | AK303 | VB | 12.29 | 1.68 | 55.86 | 27.56 | 0.70 | 0.88 | 0.75 | 0.28 | 2.03 | + |
| 53 | BG3 | VB | 11.27 | 2.31 | 54.39 | 27.52 | 1.03 | 1.07 | 1.62 | 0.54 | 1.98 | + |
| 54 | AK265 | VB | 11.19 | 3.15 | 54.38 | 27.68 | 1.09 | 0.68 | 1.37 | 0.46 | 1.96 | + |
| 55 | RSB87 | VB | 11.79 | 1.87 | 54.16 | 27.87 | 0.88 | 1.19 | 1.61 | 0.21 | 1.94 | + |
| 56 | TG1 | VB | 11.88 | 2.88 | 53.26 | 28.70 | 1.04 | 0.77 | 1.14 | 0.33 | 1.86 | + |
| 57 | TGLPS3 | VB | 11.60 | 2.21 | 53.99 | 29.38 | 0.68 | 0.63 | 1.14 | 0.37 | 1.84 | + |
| 58 | B95 | VB | 12.26 | 2.30 | 53.08 | 29.29 | 0.95 | 0.68 | 1.17 | 0.27 | 1.81 | + |
| 59 | RS138 | VB | 11.41 | 2.38 | 52.38 | 29.22 | 1.14 | 0.87 | 2.17 | 0.43 | 1.79 | + |
| 60 | M197 | VB | 12.80 | 3.50 | 51.20 | 29.06 | 1.15 | 0.80 | 1.31 | 0.19 | 1.76 | + |
| 61 | HNG69 | VB | 11.41 | 2.11 | 51.90 | 29.56 | 1.04 | 1.18 | 2.13 | 0.67 | 1.76 | + |
| 62 | Kadiri3 | VB | 11.78 | 1.70 | 49.52 | 28.97 | 1.21 | 0.92 | 1.60 | 4.30 | 1.71 | − |
| 63 | DRG17 | VB | 12.53 | 3.10 | 49.24 | 30.94 | 1.19 | 0.90 | 1.61 | 0.48 | 1.59 | + |
| 64 | Malika | VB | 9.99 | 2.52 | 51.57 | 32.92 | 0.73 | 0.70 | 1.26 | 0.30 | 1.57 | + |
| 65 | BG1 | VB | 13.32 | 2.46 | 48.47 | 32.23 | 0.89 | 0.71 | 1.34 | 0.35 | 1.50 | − |
| 66 | Kadiri8 Bold | VB | 11.38 | 2.67 | 47.25 | 34.21 | 1.07 | 1.03 | 1.85 | 0.55 | 1.38 | + |
| 67 | Konkan Gaurao | VB | 13.23 | 2.52 | 46.16 | 33.82 | 1.07 | 0.80 | 1.73 | 0.68 | 1.36 | − |
| 68 | Girnar2 | VB | 13.23 | 3.40 | 44.94 | 34.63 | 1.12 | 0.69 | 1.52 | 0.47 | 1.30 | − |
| 69 | M522 | VB | 14.05 | 3.06 | 44.99 | 34.98 | 0.91 | 0.54 | 1.00 | 0.20 | 1.29 | − |
| 70 | BAU19* | VB | 11.50 | 2.54 | 42.30 | 39.28 | 1.07 | 1.00 | 1.69 | 0.62 | 1.08 | − |
| 71 | ICGV87846 | VB | 12.67 | 2.05 | 41.25 | 39.44 | 1.06 | 0.97 | 2.16 | 0.40 | 1.05 | − |
| 72 | LGN2 | VB | 13.62 | 2.42 | 39.84 | 38.35 | 1.32 | 1.03 | 2.46 | 0.92 | 1.04 | − |
| 73 | MH2 | VAL | 12.92 | 3.44 | 38.24 | 39.90 | 1.42 | 0.81 | 2.50 | 0.78 | 0.96 | − |
| 74 | MH4 | VAL | 13.19 | 3.08 | 38.24 | 40.91 | 0.98 | 0.80 | 2.12 | 0.67 | 0.93 | − |
| 75 | Gangapuri | VAL | 12.32 | 2.83 | 41.60 | 39.22 | 1.13 | 0.74 | 1.73 | 0.45 | 1.06 | − |
| 76 | Kopargaon3 | VAL | 11.77 | 3.26 | 44.79 | 35.97 | 1.22 | 1.10 | 1.53 | 0.36 | 1.25 | − |
| 77 | TMV11 | VAL | 12.65 | 4.03 | 44.77 | 34.28 | 1.36 | 0.69 | 1.80 | 0.42 | 1.31 | − |
| 78 | VRI5 | SB | 9.37 | 2.83 | 62.69 | 21.64 | 0.75 | 0.69 | 1.59 | 0.44 | 2.90 | + |
| 79 | Tirupati3 | SB | 10.20 | 3.28 | 60.21 | 21.53 | 1.41 | 0.97 | 1.81 | 0.47 | 2.80 | + |
| 80 | TPG41 | SB | 10.88 | 3.33 | 58.27 | 24.10 | 0.98 | 0.65 | 1.25 | 0.54 | 2.42 | + |
| 81 | TAG24 | SB | 10.03 | 2.83 | 58.67 | 25.55 | 0.89 | 0.67 | 1.13 | 0.23 | 2.30 | − |
| 82 | JL24 | SB | 11.75 | 1.93 | 57.88 | 25.26 | 0.74 | 0.78 | 1.22 | 0.42 | 2.29 | − |
| 83 | GPBD4 | SB | 10.73 | 2.67 | 55.12 | 26.09 | 1.34 | 1.08 | 2.44 | 0.54 | 2.11 | + |
| 84 | DH4-3* | SB | 10.83 | 2.77 | 55.83 | 26.53 | 1.05 | 1.16 | 1.18 | 0.46 | 2.10 | + |
| 85 | Chintamani1* | SB | 10.51 | 1.79 | 56.24 | 27.03 | 0.94 | 1.06 | 1.67 | 0.76 | 2.08 | + |
| 86 | Konkan Tapora* | SB | 11.43 | 2.82 | 54.17 | 27.77 | 1.08 | 0.87 | 1.47 | 0.39 | 1.95 | + |
| 87 | GG5 | SB | 12.79 | 4.08 | 50.48 | 25.91 | 1.76 | 1.05 | 3.30 | 0.52 | 1.95 | − |
| 88 | R2001-3 | SB | 11.64 | 3.19 | 52.63 | 28.67 | 1.06 | 0.73 | 1.63 | 0.46 | 1.84 | + |
| 89 | Kadiri9 | SB | 11.82 | 2.71 | 51.53 | 28.81 | 1.26 | 1.02 | 2.32 | 0.53 | 1.79 | + |
| 90 | S206 | SB | 11.67 | 2.47 | 52.47 | 29.40 | 1.09 | 0.75 | 1.83 | 0.32 | 1.78 | − |
| 91 | VG9816 | SB | 11.11 | 3.42 | 52.44 | 29.60 | 1.13 | 0.61 | 1.38 | 0.31 | 1.77 | + |
| 92 | ALR2 | SB | 11.84 | 2.94 | 51.84 | 29.66 | 1.13 | 0.86 | 1.49 | 0.25 | 1.75 | − |
| 93 | R2001-2 | SB | 12.23 | 2.91 | 51.69 | 29.73 | 1.00 | 0.67 | 1.39 | 0.39 | 1.74 | + |
| 94 | TG17 | SB | 13.11 | 2.15 | 52.08 | 30.15 | 0.44 | 0.60 | 0.56 | 0.56 | 1.73 | + |
| 95 | GJG9 | SB | 12.31 | 3.86 | 49.87 | 29.80 | 1.39 | 0.65 | 1.60 | 0.51 | 1.67 | − |
| 96 | Girnar3 | SB | 12.55 | 2.75 | 49.72 | 32.27 | 0.73 | 0.53 | 0.85 | 0.61 | 1.54 | − |
| 97 | TLG45 | SB | 11.84 | 2.91 | 48.47 | 33.02 | 1.06 | 0.70 | 1.56 | 0.45 | 1.47 | + |
| 98 | Kadiri Harithandra | SB | 11.99 | 3.05 | 48.33 | 33.06 | 1.03 | 0.73 | 1.39 | 0.42 | 1.46 | − |
| 99 | TG26 | SB | 12.16 | 3.19 | 47.95 | 32.83 | 1.18 | 0.79 | 1.47 | 0.43 | 1.46 | − |
| 100 | Jyothi | SB | 13.22 | 4.88 | 45.05 | 31.30 | 1.75 | 0.73 | 2.69 | 0.38 | 1.44 | − |
| 101 | TKG19 A | SB | 10.96 | 2.01 | 49.40 | 34.43 | 1.13 | 0.70 | 1.00 | 0.38 | 1.43 | + |
| 102 | TMV12 | SB | 13.91 | 3.54 | 46.15 | 32.54 | 1.74 | 0.38 | 0.65 | 1.09 | 1.42 | − |
| 103 | VRI4 | SB | 13.64 | 2.53 | 46.93 | 33.11 | 1.05 | 0.81 | 1.49 | 0.45 | 1.42 | − |
| 104 | GG4 | SB | 13.40 | 3.87 | 45.59 | 32.36 | 1.43 | 0.70 | 2.10 | 0.55 | 1.41 | − |
| 105 | GG7 | SB | 13.40 | 3.87 | 45.59 | 32.36 | 1.43 | 0.70 | 2.01 | 0.55 | 1.41 | − |
| 106 | J11 | SB | 12.42 | 4.70 | 44.11 | 31.78 | 1.33 | 0.81 | 3.71 | 1.15 | 1.39 | − |
| 107 | ICG (FDRS 10) | SB | 13.69 | 1.84 | 46.87 | 34.02 | 0.48 | 0.38 | 0.62 | 0.25 | 1.38 | − |
| 108 | DRG12 | SB | 13.30 | 2.26 | 46.31 | 34.02 | 0.95 | 1.08 | 1.50 | 0.35 | 1.36 | − |
| 109 | SG99 | SB | 12.76 | 2.46 | 46.52 | 34.32 | 1.01 | 0.79 | 1.66 | 0.48 | 1.36 | − |
| 110 | DH40* | SB | 12.68 | 3.86 | 45.35 | 33.49 | 1.46 | 0.69 | 1.98 | 0.49 | 1.35 | − |
| 111 | TMV2 | SB | 13.84 | 4.60 | 43.66 | 32.69 | 1.65 | 0.61 | 2.14 | 0.48 | 1.34 | − |
| 112 | OG52-1 | SB | 13.14 | 4.21 | 44.43 | 33.31 | 1.45 | 0.65 | 2.31 | 0.51 | 1.33 | − |
| 113 | Tirupati1 | SB | 12.01 | 2.06 | 47.50 | 35.70 | 0.50 | 0.40 | 0.61 | 0.75 | 1.33 | − |
| 114 | KRG1 | SB | 14.87 | 3.79 | 44.19 | 33.30 | 1.15 | 0.56 | 1.60 | 0.27 | 1.33 | − |
| 115 | AK12-24 | SB | 13.79 | 4.58 | 43.73 | 33.20 | 1.64 | 0.65 | 1.97 | 0.41 | 1.32 | − |
| 116 | LGN1 | SB | 12.89 | 3.68 | 44.57 | 33.96 | 1.34 | 0.71 | 2.36 | 0.50 | 1.31 | − |
| 117 | Girnar1 | SB | 15.47 | 2.73 | 45.33 | 34.59 | 0.40 | 0.22 | 0.26 | 0.32 | 1.31 | − |
| 118 | GJG31 | SB | 13.54 | 2.98 | 45.03 | 35.08 | 1.09 | 0.62 | 1.25 | 0.42 | 1.28 | − |
| 119 | GG6 | SB | 13.95 | 4.28 | 43.16 | 33.82 | 1.39 | 0.65 | 2.23 | 0.53 | 1.28 | − |
| 120 | SB XI | SB | 13.76 | 4.44 | 44.02 | 34.53 | 1.16 | 0.46 | 1.30 | 0.13 | 1.27 | − |
| 121 | Pratap Mungphali2 | SB | 13.45 | 3.43 | 44.48 | 35.10 | 1.15 | 0.51 | 1.40 | 0.48 | 1.27 | − |
| 122 | Kadiri5 | SB | 13.32 | 3.44 | 43.19 | 34.32 | 1.48 | 0.77 | 2.58 | 0.91 | 1.26 | − |
| 123 | GG8 | SB | 14.41 | 3.95 | 42.63 | 34.57 | 1.43 | 0.56 | 1.94 | 0.52 | 1.23 | − |
| 124 | TMV9 | SB | 13.61 | 4.27 | 42.93 | 34.85 | 1.51 | 0.50 | 1.88 | 0.46 | 1.23 | − |
| 125 | Kisan | SB | 14.29 | 3.42 | 43.22 | 35.15 | 0.92 | 1.07 | 0.95 | 0.16 | 1.23 | − |
| 126 | VRI3 | SB | 13.93 | 4.29 | 42.85 | 34.91 | 1.36 | 0.43 | 1.61 | 0.40 | 1.23 | − |
| 127 | R8808 | SB | 13.73 | 2.16 | 43.56 | 35.72 | 1.17 | 1.01 | 1.95 | 0.41 | 1.22 | − |
| 128 | Tirupati4 | SB | 13.38 | 5.20 | 41.50 | 34.18 | 1.87 | 0.66 | 2.63 | 0.56 | 1.21 | − |
| 129 | Sp. Improved | SB | 15.28 | 4.95 | 40.52 | 33.55 | 1.78 | 0.69 | 2.60 | 0.46 | 1.21 | − |
| 130 | Tirupati2 | SB | 15.28 | 4.95 | 40.52 | 33.55 | 1.78 | 0.69 | 2.60 | 0.46 | 1.21 | − |
| 131 | TMV (GN)13 | SB | 13.75 | 3.80 | 42.72 | 35.38 | 1.23 | 0.56 | 2.00 | 0.58 | 1.21 | − |
| 132 | ALR3 | SB | 10.73 | 2.85 | 45.97 | 38.25 | 0.58 | 0.24 | 1.10 | 0.29 | 1.20 | − |
| 133 | DH86 | SB | 13.35 | 3.82 | 42.78 | 35.64 | 1.36 | 0.73 | 1.82 | 0.51 | 1.20 | − |
| 134 | ICGS44 | SB | 13.48 | 1.59 | 44.46 | 37.16 | 0.54 | 0.77 | 0.54 | 0.19 | 1.20 | − |
| 135 | DH2000-1* | SB | 13.17 | 4.66 | 42.12 | 35.31 | 1.49 | 0.58 | 1.91 | 0.77 | 1.19 | − |
| 136 | Kadiri4 | SB | 13.98 | 4.07 | 41.54 | 35.08 | 1.39 | 0.77 | 2.27 | 0.91 | 1.18 | − |
| 137 | CO1 | SB | 13.76 | 3.75 | 43.19 | 36.58 | 0.86 | 0.49 | 0.73 | 0.20 | 1.18 | − |
| 138 | Co2 | SB | 13.82 | 2.95 | 44.04 | 37.32 | 0.65 | 0.36 | 0.37 | 0.13 | 1.18 | − |
| 139 | DH101 | SB | 13.76 | 3.40 | 42.56 | 36.15 | 1.19 | 0.69 | 1.76 | 0.50 | 1.18 | − |
| 140 | AK159 | SB | 13.14 | 3.63 | 42.38 | 36.01 | 1.40 | 0.74 | 2.15 | 0.53 | 1.18 | − |
| 141 | JGN23 | SB | 13.34 | 4.01 | 41.76 | 35.56 | 1.49 | 0.63 | 2.70 | 0.52 | 1.17 | − |
| 142 | BSR1 | SB | 13.70 | 2.40 | 41.51 | 35.36 | 1.29 | 1.14 | 3.16 | 1.45 | 1.17 | − |
| 143 | JL220 | SB | 13.95 | 3.84 | 41.78 | 35.93 | 1.54 | 0.71 | 1.82 | 0.44 | 1.16 | − |
| 144 | JCG88 | SB | 11.12 | 2.24 | 44.31 | 38.44 | 0.79 | 0.95 | 1.70 | 0.45 | 1.15 | − |
| 145 | ICGS37 | SB | 14.66 | 2.41 | 42.06 | 36.52 | 1.19 | 0.95 | 1.48 | 0.54 | 1.15 | − |
| 146 | JGN3 | SB | 13.43 | 3.78 | 41.42 | 36.28 | 1.47 | 0.75 | 2.42 | 0.46 | 1.14 | − |
| 147 | GG3 | SB | 13.64 | 3.71 | 41.55 | 36.46 | 1.28 | 0.76 | 2.04 | 0.46 | 1.14 | − |
| 148 | TMV7 | SB | 14.55 | 3.10 | 41.68 | 36.75 | 1.08 | 0.73 | 1.72 | 0.39 | 1.13 | − |
| 149 | SG84 | SB | 14.59 | 1.93 | 42.81 | 37.96 | 0.25 | 0.33 | 0.30 | 0.11 | 1.13 | − |
| 150 | ICGV00350 | SB | 13.37 | 1.66 | 42.61 | 37.87 | 0.92 | 1.01 | 1.99 | 0.57 | 1.13 | − |
| 151 | ICGS11 | SB | 12.87 | 1.97 | 43.55 | 38.72 | 0.30 | 0.41 | 0.38 | 0.19 | 1.12 | − |
| 152 | CO(GN)4 | SB | 14.20 | 3.08 | 41.44 | 36.94 | 1.17 | 0.84 | 1.92 | 0.41 | 1.12 | − |
| 153 | Vemana/K134 | SB | 13.91 | 2.17 | 43.07 | 38.57 | 0.84 | 0.56 | 0.67 | 0.22 | 1.12 | − |
| 154 | RG141 | SB | 14.87 | 2.68 | 41.42 | 37.50 | 0.65 | 0.40 | 0.63 | 1.00 | 1.10 | − |
| 155 | Jawan | SB | 14.29 | 3.30 | 42.18 | 38.67 | 0.36 | 0.28 | 0.30 | 0.17 | 1.09 | − |
| 156 | GAUG1 | SB | 13.45 | 3.52 | 41.33 | 37.93 | 1.09 | 0.50 | 1.36 | 0.83 | 1.09 | − |
| 157 | VRI2 | SB | 13.86 | 1.94 | 42.88 | 39.64 | 0.30 | 0.19 | 0.19 | 0.38 | 1.08 | − |
| 158 | Dh8 | SB | 14.68 | 2.93 | 40.61 | 37.84 | 0.81 | 0.35 | 0.41 | 0.27 | 1.07 | − |
| 159 | GG2 | SB | 14.81 | 2.02 | 42.08 | 39.51 | 0.36 | 0.29 | 0.28 | 0.30 | 1.07 | − |
| 160 | JL286 | SB | 13.96 | 3.61 | 40.41 | 37.95 | 1.29 | 0.60 | 1.84 | 0.35 | 1.06 | − |
| 161 | JL501 | SB | 12.44 | 2.81 | 41.73 | 39.36 | 1.04 | 0.62 | 1.64 | 0.36 | 1.06 | − |
| 162 | TG3 | SB | 15.38 | 2.75 | 41.21 | 38.94 | 0.66 | 0.25 | 0.24 | 0.22 | 1.06 | − |
| 163 | ICGV91114 | SB | 13.37 | 3.97 | 39.57 | 37.47 | 1.63 | 0.78 | 2.66 | 0.55 | 1.06 | − |
| 164 | CO3 | SB | 14.42 | 2.93 | 38.69 | 36.90 | 1.41 | 1.04 | 3.25 | 1.33 | 1.05 | − |
| 165 | ICGS1 | SB | 13.09 | 1.14 | 42.90 | 40.92 | 0.31 | 0.47 | 0.26 | 0.20 | 1.05 | − |
| 166 | TG51 | SB | 14.21 | 3.53 | 40.36 | 38.74 | 1.07 | 0.48 | 1.25 | 0.37 | 1.04 | − |
| 167 | Dh3-30 | SB | 13.70 | 2.47 | 39.94 | 38.35 | 1.26 | 1.03 | 2.46 | 0.79 | 1.04 | + |
| 168 | TG37A | SB | 13.75 | 2.81 | 40.62 | 39.05 | 0.93 | 0.62 | 1.61 | 0.62 | 1.04 | − |
| 169 | ICGV86590 | SB | 11.60 | 2.30 | 41.52 | 40.14 | 1.09 | 0.89 | 1.91 | 0.44 | 1.03 | − |
| 170 | TG22 | SB | 11.60 | 2.30 | 41.52 | 40.14 | 1.09 | 0.89 | 1.91 | 0.44 | 1.03 | + |
| 171 | Pratap Mungphali1 | SB | 11.35 | 2.17 | 41.16 | 40.08 | 0.99 | 1.19 | 2.26 | 0.80 | 1.03 | + |
| 172 | Kadiri6 | SB | 12.69 | 3.36 | 39.91 | 39.34 | 1.37 | 0.78 | 2.03 | 0.53 | 1.01 | − |
| 173 | ICGV86031 | SB | 13.78 | 3.15 | 39.29 | 39.48 | 1.21 | 0.81 | 1.92 | 0.37 | 1.00 | − |
| 174 | MH1 | SB | 13.58 | 3.02 | 37.87 | 40.65 | 1.23 | 0.80 | 2.24 | 0.61 | 0.93 | − |
| Mean** | 12.26 | 2.93 | 49.43 | 31.29 | 1.08 | 0.79 | 1.60 | 0.48 | – | − |
Where; (+) Indicates ahFAD2A mutant allele; (−) Indicates ahFAD2A non-mutant allele; ND, Not Detected;
Advance breeding lines;
In the calculation of mean the data of “SunOleic95R” was not considered.
Table 2.
| S. no. | Accession no. | Species | Section | Genome | Ploidy (x = 10) |
|---|---|---|---|---|---|
| 1 | NRCG11802 | A. duranensis Krapov and WC Gregory | Arachis | AA | 2x |
| 2 | NRCG11837 | A. glabrata Benth | Rhizomatosae | RR | 2x |
| 3 | NRCG11786 | A. appressipila Krapov and WC Gregory | Procumbentes | EE | 2x |
| 4 | NRCG11793 | A. paraguariensis Chodat and Hassl | Erectoides | EE | 2x |
| 5 | NRCG12017 | Arachis correntina (Burkart) Krapov and WC Gregory | Arachis | AA | 2x |
| 6 | NRCG11781 | Arachis diogoi Hoehne | Arachis | AA | 2x |
| 7 | NRCG12989 | A. hermanii Krapov and WC Gregory | Erectoides | EE | 2x |
| 8 | NRCG12018 | A. batizocoi Krapov and WC Gregory | Arachis | BB | 2x |
| 9 | NRCG12046 | A. cardenasii Krapov and WC Gregory | Arachis | AA | 2x |
| 10 | NRCG12984 | A. cruziana Krapov, WC Gregory and CE Simpson | Arachis | BB | 2x |
| 11 | NRCG12031 | A. batizogaea Krapov And A.Fernández | Arachis | BB | 2x |
| 12 | NRCG12032 | A. rigonii Krapov and WC Gregory | Procumbentes | EE | 2x |
| 13 | NRCG11800 | A. monticola Krapov and Rigoni | Arachis | AABB | 4x |
| 14 | NRCG12029 | A. kretschmeri Krapov. and WC Gregory | Procumbentes | EE | 2x |
| 15 | NRCG17206 | A. marginata Gardner | Rhizomatosae | EX | 2x |
| 16 | NRCG17205 | A. prostrate Benth | Rhizomatosae | EX | 2x |
| 17 | NRCG12990 | A. pintoi Krapov and WC Gregory | Caulorhizae | CC | 2x |
| 18 | NRCG14871 | A. valida Krapov and WC Gregory | Arachis | BB | 2x |
| 19 | NRCG14855 | A. matiensis Krappov | Procumbentes | PP | 2x |
| 20 | NRCG14862 | A. stenosperma Krapov and WC Gregory | Arachis | AA | 2x |
| 21 | NRCG14868 | A. sylvestris Chev | Heteranthae | Am | 2x |
| 22 | NRCG468363 | Arachis sp. | Rhizomatosae | – | 2x |
DNA extraction
Two seeds of each genotype were grown in plastic pots filled with sand, under controlled conditions and leaf samples from the 10–15 days old seedlings were drawn for DNA isolation, using the cetyltrimethyl ammonium bromide (CTAB) extraction method (Cuc et al., 2008). However, for wild-species, leaf samples were taken from the peanut field gene-bank of DGR, Junagadh (India). The DNA quality was checked on agarose gel (0.8%, w/v) and quantification was done using NanoDropND-1000 (NanoDrop products, DE, USA) and working concentration was adjusted to 20 ng μL−1.
Allele-specific PCR (AS-PCR) and cleaved amplified polymorphic sequences (CAPS) assays
For the identification of known mutations in the ahFAD2 genes, all the genotypes were screened using known AS-PCR markers (Chen et al., 2010; Yu et al., 2013). Further, CAPS assays were also deployed to identify the zygosity of genotypes for ahFAD2A (448G>A as given by Chu et al., 2007) and ahFAD2B (441_442insA as given by Chu et al., 2009) mutations (Table S1).
For both, AS-PCR and CAPS analysis, the PCR mixtures (10 μl) contains, template DNA (1 μl, 20 ng), 5X PCR buffer (2 μl, Promega, USA), 25 mM MgCl2 (0.8 μl, Promega, USA), 2 mM dNTP (0.7 μl, Thermo Fisher Scientific, USA), primers (0.5 μl, 25 p moles), 5U Taq polymerase (0.2 μl, Promega, USA) and sterile double distilled water (4.3 μl). Amplification was performed in a thermal cycler (Eppendorf, USA) using thin walled 96 wells PCR plates (Sorenson™ Bioscience, USA).
The touchdown PCR was done with initial denaturation at 94.0°C for 3 min and then 5 cycles of the following: 94.0°C for 30 s (−1.0°C reduction per cycle), 65–60°C for 30 s and 72.0°C for 1 min. This was followed by another 35 cycles of 94.0°C for 30 s, 60.0°C for 30 s and 72.0°C for 1 min of denaturation, annealing and extension, respectively. Final extension was 72.0°C for 10 min. Amplification was performed twice and amplified products were analyzed using 2% agarose gel in 1 × TBE buffer at 225 volts for about 2.5–3.0 h and stained with ethidium bromide. The gels were documented in automated gel documentation system (Fujifilm FLA-5000) and scored.
For CAPS analysis of ahFAD2A and ahFAD2B mutations, the amplified products (10 μl) of 826 and 1200 bp were digested using 0.4 μl each, of Hpy99I and Hpy188I (0.4 U; New England Biolabs, MA) restriction enzymes, respectively, in 2.0 μl digestion buffer, 2.0 μl BSA (0.1%, Takara, Japan) and 5.6 μl distilled water. The restriction digestion of ahFAD2A and ahFAD2B gene specific amplicon was done at 37°C for 4 h, and 37°C for overnight with Hpy99I and Hpy188I enzymes, respectively.
Fatty acid profiling
The fatty acids were analyzed using gas chromatography system (Thermo fisher, Trace GC 1100) equipped with flame ionization detector (FID). The fatty acid methyl esters were passed through capillary column (TR-wax), and the esters of fatty acids were estimated (Misra and Mathur, 1998). The inlet, FID detector were set to 240°C and oven at 190°C whereas carrier gas (nitrogen) and fuel gas (hydrogen) were maintained at 30 mL per min. Total run time for each sample was 12 min and the peaks were identified by comparison to a FAME standard mix RM-3 (Sigma-Aldrich, St. Louis, MO).
Pearson's coefficient analysis was performed using SPSS 15.0 software to determine correlations among different fatty acids. An analysis of variance was performed on mean values to test the significant differences in major fatty acids among the mutant and non-mutant genotypes using Tukey's multiple comparison test (Brown, 1979).
Single nucleotide polymorphism (SNP) analysis of ahFAD2A gene
A set of 30 peanut genotypes were selected based on ahFAD2A gene mutation and O/L ratio so as to find the allelic variations as SNPs (Table S2). The functional domain flanking sequence information of desaturase enzyme was used to design FAD2 gene specific primers, using Primer3plus software (http://primer3plus.com), with expected amplicon size of 1148 bp (Table S1). PCR reaction conditions were same as of AS-PCR and CAPS assays; except, it is carried out in a final volume of 20 μl. The PCR products from each sample were visualized by agarose gel electrophoresis.
Cloning of full length ahFAD2 gene
The ahFAD2 gene specific bands were excised and purified using QIAquick gel extraction kits (QIAGEN), and used for direct sequencing as-well-as cloning, using pGEM®-T Easy cloning kit (Promega). The amplified products were ligated to pGEM®-T easy vector, using T4 DNA ligase (4°C), and kept overnight. Transformations were carried out via conventional heat shock method into competent E. coli (Dh5-Alpha) cells, and selection of transformed cells was done on LB/ampicillin/IPTG/X-Gal plates. Positive colonies were identified using vector specific (SP6 and T7 primers) and gene specific primers. Positive colonies were grown overnight and plasmid DNA was isolated using QIAgen kit and utilized for DNA sequencing. The sequencing was performed using SciGenome's (Kerala, India) Sanger sequencing services using ABI 3730Xl platform.
Sequence analysis
The raw sequences were edited and aligned using CAP3 programme (Huang and Madan, 1999), to generate consensus sequences. Further, the chromatograms were also checked manually, so as to find any miscalled bases in the identified SNPs. The ahFAD2A and ahFAD2B gene sequences were distinguished based on the previously reported SNPs. Subsequently, both ahFAD2A and ahFAD2B sequences were aligned using ClustalW programme. The phylogenetic tree was constructed using Neighbor-Joining method of MEGA 6 program (Tamura et al., 2013), and visualized using online tool iTOL (Letunic and Bork, 2011). The haplotype numbers, haplotype diversity, nucleotide diversity, non-synonymous (Ka) to synonymous (Ks) ratio were generated using DnaSP 5.10 software (Librado and Rozas, 2009).
Results
Detection of mutations in ahFAD2 genes using AS-PCR and CAPS assays
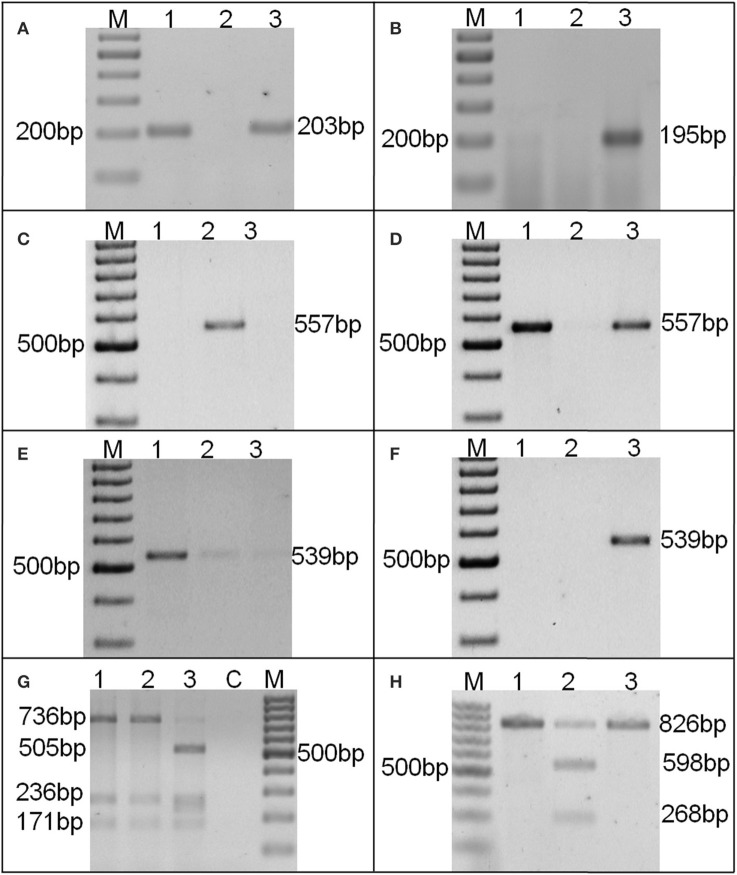
The AS-PCR assay (Chen et al., 2010) for substitution allele-specific primer combination, amplified 203 bp amplicon in 80 genotypes, confirming a single bp substitution (G:C/A:T) at 448 position in ahFAD2A allele (Figure 1A). While, insertion (A:T at 442 bp position) allele-specific primer combination for ahFAD2B allele, did not amplify in any genotypes. Only a known mutant genotype, SunOleic95R showed a 195 bp amplicon (Figure 1B). In addition, the primer combination F435-F and F435IC-R was used as internal control and primer combination F435-F and F435WT-R amplified a 193 bp amplicon in all the genotypes.
Figure 1.
A representative picture of AS-PCR (A–F) and CAPS assays (G,H) AS-PCR assay (Chen et al., 2010) showing (A) ahFAD2A mutant allele specific 203 bp amplification; (B) ahFAD2B mutant allele specific 195 bp amplification; AS-PCR assay (Yu et al., 2013) showing (C) ahFAD2A non-mutant allele specific 557 bp amplification; (D) ahFAD2A mutant allele specific 557 bp amplification; (E) ahFAD2B non-mutant allele specific 539 bp amplification and (F) ahFAD2B mutant allele specific 539 bp amplification; Amplicons generated by CAPS assay for (G) ahFAD2B and (H) ahFAD2A alleles. Lanes: 1: GG20, 2: Kadiri3, 3: SunOleic95R, M: 100 bp DNA ladder.
Since, the AS-PCR assay of Chen et al. (2010) was unable to identify the zygosity of both ahFAD2A and ahFAD2B genes; therefore, AS-PCR assay of Yu et al. (2013) was used (Table S1). All the 80 genotypes were found having ahFAD2A mutant allele in homozygous condition (ol1ol1/Ol2Ol2) (Figures 1C,D). The result of ahFAD2B gene revealed non-mutant gene specific amplification in all the genotypes (Figures 1E,F).
Further, CAPS assay was also used for the identification of 448G>A mutation in ahFAD2A gene and 441_442insA mutation in ahFAD2B gene (Table S1). Full-length amplification of ahFAD2A gene generated an 826 bp product, which was digested into 598 and 228 bp fragments (with Hpy991 enzyme) in 93 genotypes, predicting their wild-type nature. However, PCR products of 80 genotypes remain undigested, showed mutation in the ahFAD2A gene. Moreover, a 1200 bp band of ahFAD2B gene, digested into 736, 263, 171 bp bands with enzyme Hpy1881, indicating its non-mutant nature. Only SunOleic95R produced banding pattern akin to ahFAD2B mutant allele (Figures 1G,H). All the 22 wild-species (Table 2), when tested using AS-PCR assay (Chen et al., 2010), showed the presence of non-mutant alleles.
Pedigree analysis of indian peanut cultivars for ahFAD2A gene
Since, 46% of the Indian peanut cultivars recorded ahFAD2A mutation; therefore, the pedigree was also analyzed so as to find the association of ahFAD2A mutant allele with its parents. Out of 26 varieties having ahFAD2A mutant allele, both the parents of 3 varieties and one parent of 23 varieties found containing mutant ahFAD2A gene. Moreover, in 34 non-mutant varieties, 12 had either non-mutant ahFAD2 gene in both the parents or were derived from the selection. The details of all the genotypes are given in Table S3.
Cultivated peanut is known to have an extremely narrow genetic base due to the inbreeding among a few select parental lines in commercial breeding programs (Nigam, 2000; Simpson et al., 2001). This was found true, when pedigree of 167 Indian cultivars were critically analyzed. It is found that 08 genotypes were most frequently (4 times or more) used as one of the parent, leading to the release of 59 cultivars (Table 3). Thus, there is need of better utilization of existing variability in Indian peanut improvement programs.
Table 3.
List of varieties frequently used in the Indian varietal development programme.
| S. no. | Genotypes/varieties | No of times used for variety development | Varieties developed |
|---|---|---|---|
| 1 | JL24 | 14 | VRI2, GG3, K134, Tirupati4, GG5, JGN39, CO3, AK159, Kadiri5, Kadiri6, AK303, LGN1, JGN23 and GG8 |
| 2 | M13 | 6 | Somnath, B95, M335, GG11, BAU13 and Girnar2 |
| 3 | ICGS11 | 5 | R8808, AK265, Girnar3, R2001-2 and R2001-3 |
| 4 | GAUG10 | 4 | GG20, GG11, GG13 and GG12 |
| 5 | J11 | 4 | GG2, Jawan, VRI3 and JCG88 |
| 6 | Spanish Improved | 4 | TG1, DH330, Kisan and TG3 |
| 7 | Robut 33-1 | 18 | ICGS11, Kadiri3, ICGS44, ICGS1, GG20, ICGS5, TG22, VRI3, RG141, ICGS37, DRG17, DRG12, BSR1, LGN2, ALR3, GG14, HNG10 and Girnar2 |
| 8 | Chico | 4 | ICGS76, GG4, R8808 and TG51 |
| Total | 59 | – |
Fatty acid profiling
In the 174 peanut genotypes studied, oleic (C18:1), linoleic (C18:2) and palmitic (C16:0) acids were the major fatty acids, of which, the first two constitute about 80% of total fatty acids. Behenic (C22:0), stearic (C18:0), arachidic (C20:0), gadoleic acid (C20:1), lignoceric (C24:0), were also present in smaller amounts in all the genotypes (Table 1 and Figure S1). Myristic acid was found in 19 genotypes; interestingly all of which belongs to Spanish bunch group (Table S4).
Fatty acid profiling revealed highest and lowest contents of oleic acid in TMV10 (66.57%) and MH1 (37.87%); while, linoleic acid in ICGS1 (40.92%) and GJG22 (17.41%), respectively (Table 1). Similar trends were obtained for ICRISAT mini-core collections; where highest, 71% of oleic acid and lowest 15.81% linoleic acid were reported (Mukri et al., 2012). The average oleic and linoleic acid contents were recorded as 49.61% and 31.15%, respectively; while, Bishi et al. (2015) reported an average of 46.47% oleic acid and 34.51% linoleic acid in 41 Indian peanut cultivars.
The mean of oleic acid content was found significantly higher for 80 ahFAD2A mutant lines (55.91%), than the 93 non-mutant lines (43.85%). Fourteen mutant genotypes for ahFAD2A gene recorded more than 60% oleic acid content (Table 1); thus, for increasing the O/L ratio, these genotypes should be the first choice. The high oleate trait can be combined with other traits like disease resistance, for which foliar disease resistance genotypes viz. GPBD4 and AK265 as well as peanut bud necrosis resistant genotype R2001-3, could be taken into breeding program (Table 1). Since, these genotypes already have one mutant allele (ahFAD2A) in homozygous condition; therefore, another mutant gene (ahFAD2B) can be easily transferred using marker assisted backcross breeding (MABB) in allotetraploid peanut.
Among the genotypes studied, SunOleic95R, TMV10, HNG10 and GG16 were found to have less than 9% pamitic acid, which along with high oleic and low linoleic acid can make the peanut oil healthier for consumers. The genotypes, GG13, BAU13, ICGS76 had low oil (<45%), high sucrose (>6%), high oleic acid (>58%), and high O/L ratio (>2%) which is highly desired combination for confectionery peanut industry.
Classification of mutant varieties on the basis of botanical types
The 173 cultivated peanut genotypes studied, belong to two subspecies viz. ssp. fastigiata (102) and ssp. hypogaea (71); which recorded 17 and 62 mutant, while 85 and 8 non-mutant genotypes, respectively. In general, mutant lines showed higher mean O/L ratio than the non-mutant botanical types. Interestingly, no genotype of Valencia group recorded mutation in ahFAD2A gene, while all the genotypes of Virginia runner group showed substitution mutation (Table 4). Mutant Virginia runner genotypes, recorded mean O/L ratio of 2.34 with highest oleic (57.23%), lowest palmatic (10.75%) and linoleic acid contents (25%). Highest range of O/L ratio was observed in Virginia bunch mutant type (1.38–3.76); whereas, ssp. hypogaea recorded high oleic, low linoleic and palmitic acid than the ssp. fastigiata (Table 4). These results are in accordance with Wang et al. (2013b), Mukri et al. (2012), and Wang et al. (2009).
Table 4.
Classification of peanut genotypes on the basis of botanical types and major fatty acid composition.
| Botanical type | Habit group | ahFAD2A status | No. of genotypes | Palmitic acid (%) | Oleic acid (%) | Linoleic acid (%) | Mean O/L ratio | Range of O/L ratio |
|---|---|---|---|---|---|---|---|---|
| ssp. fastigiata | Spanish Bunch (vars. vulgaris) | Mutant | 17 | 11.37 | 51.96 | 29.86 | 1.84 | 1.00–2.90 |
| Non-mutant | 80 | 13.45 | 43.90 | 35.29 | 1.26 | 0.93–2.30 | ||
| Valencia (vars. fastigiata) | Mutant | Nil | – | – | – | – | – | |
| Non-mutant | 5 | 12.57 | 41.53 | 38.05 | 1.10 | 0.93–1.31 | ||
| ssp. hypogaea | Virginia Runner (vars. hypogaea) | Mutant | 31 | 10.75 | 57.23 | 25.0 | 2.34 | 1.55–3.33 |
| Non-mutant | Nil | – | – | – | – | – | ||
| Virginia Bunch (vars. hypogaea) | Mutant | 32 | 10.98 | 56.77 | 27.08 | 2.25 | 1.38–3.76 | |
| Non-mutant | 08 | 12.92 | 44.68 | 35.21 | 1.29 | 1.04–1.71 | ||
| Total/mean | 173 | 12.26 | 49.43 | 31.29 | 1.69 | 0.93–3.76 |
Correlations studies and effect of ahFAD2A mutation on fatty acid composition
In correlation studies, oleic acid was found negatively correlated with all the SFAs including palmitic acid (r = −0.87), which most likely represents an increased rate of palmitic acid elongation to stearic acid, with rapid desaturation to oleic acid via Delta-9 desaturase (Wang et al., 2015a). A significant positive correlations are observed between linoleic acid and palmitic acid (r = 0.78), stearic acid and arachidic acid (r = 0.68) and also between arachidic acid and behenic acid (r = 0.78) (Table 5).
Table 5.
Correlation coefficients among fatty acid components of different peanut genotypes.
| Palmitic acid (C16:0) | Stearic acid (C18:0) | Oleic acid (C18:1) | Linoleic acid (C18:2) | Arachidic acid (C20:0) | Gadoleic acid (C20:1) | Behenic acid (C22:0) | Lignoceric acid (C24:0) | |
|---|---|---|---|---|---|---|---|---|
| Palmitic acid | 1 | |||||||
| Stearic acid | 0.38** | 1 | ||||||
| Oleic acid | −0.87** | −0.33** | 1 | |||||
| Linoleic acid | 0.78** | 0.13 | −0.96** | 1 | ||||
| Arachidic acid | 0.15* | 0.68** | −0.22** | 0.02 | 1 | |||
| Gadoleic acid | −0.50** | −0.26** | 0.40** | −0.44** | 0.28** | 1 | ||
| Behenic acid | 0.05 | 0.44** | −0.22** | 0.05 | 0.78** | 0.44** | 1 | |
| Lignoceric acid | 0.55 | −0.09 | −0.15* | 0.07 | 0.25** | 0.16* | 0.33** | 1 |
Where,
denotes significance level at 0.05%, 0.01%, respectively.
The effect of ahFAD2A gene mutation, on both O/L ratio, and other fatty acids composition, revealed significant reduction in the palmitic acid, stearic acid and linolenic acid contents in mutant genotypes. However, significant increase in the oleic acid, gadoleic acid and O/L ratio was observed. No significant difference was recorded for the fatty acids like arachidic acid and behenic acid in both, mutant and non-mutant genotypes (Figure 2).
Figure 2.
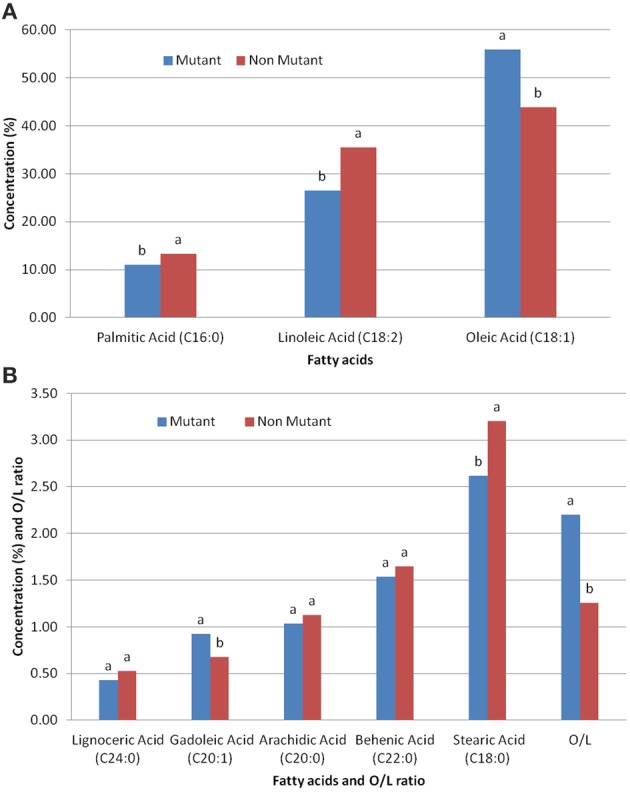
Effect of ahFAD2A mutant allele on (A) various fatty acids; (B) various fatty acids and O/L ratio.
SNP analysis of ahFAD2 gene in selected peanut genotypes
Since a gradient in the O/L ratio was observed in the genotypes studied; therefore, a set of 30 genotypes, having 17 mutant and 13 non-mutant lines for ahFAD2A gene were selected for further sequencing of the ahFAD gene (Table S2). To amplify the full functional domain of ahFAD2A and ahFAD2B genes, the primers were designed using GenBank accession >gi|307697073|gb|HM359250.1| (Table S1), which amplified an 1148bp amplicon. Both, direct sequencing of PCR products, and clone based sequencing was performed for both ahFAD2A and ahFAD2B genes.
The chromatogram analysis of PCR products revealed double peaks at 19 positions, since it consisted of amplicons from both ahFAD2A and ahFAD2B genes (Figure S2). Previously, overlapped peak were reported to identify the heterozygous state of allele (Wang et al., 2011b; Yu et al., 2013). To distinguish the sequences of A and B genomes in the genotypes studied, 1148 bp gene specific amplicons were cloned and sequenced. Further, the sequence information pertaining to ahFAD2A (GenBank accession no. HM359250.1 and GQ412349) and ahFAD2B (GenBank accession No. HM359251 and HM359252.1) genes were used to identify the sequences of ahFAD2A and ahFAD2B genes in the genotypes studied (Tables S5, S6).
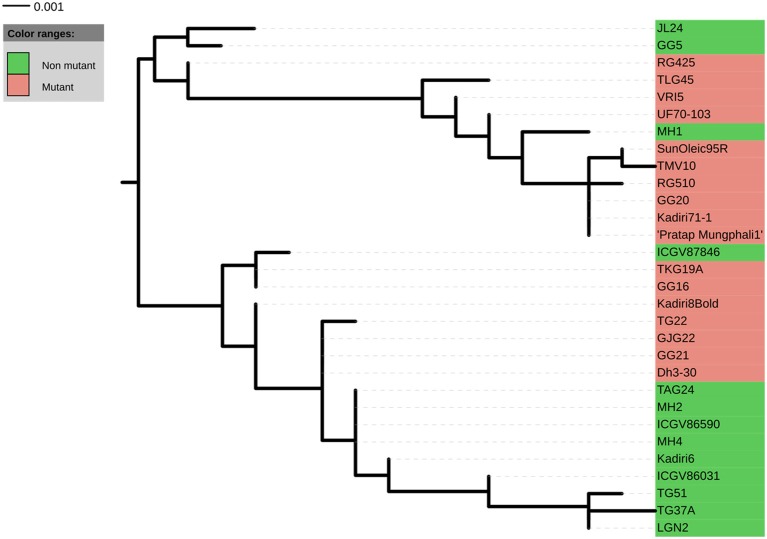
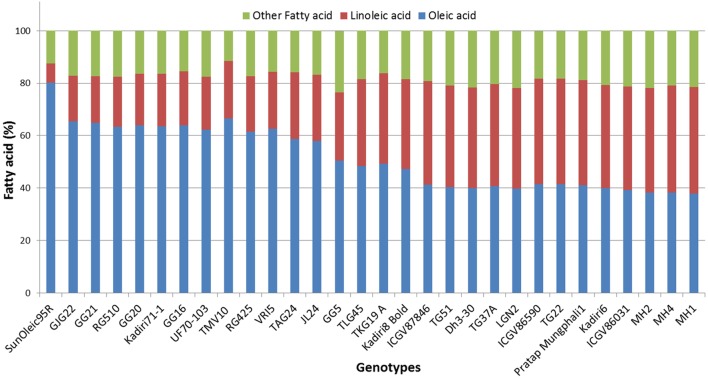
A total of 22 and 21 haplotypes numbers (H) with haplotype diversity (Hd) of 0.970 and 0.952 were detected in ahFAD2A and ahFAD2B genes, respectively. The nucleotide diversity (Pi) for ahFAD2A and ahFAD2B were 1.05% and 0.95%, respectively. In present study, the ratio of non-synonymous substitution (Ka) to synonymous substitution rates (Ks) or Ka/Ks ratio of 0.36 and 0.39 were obtained for ahFAD2A and ahFAD2B gene sequences, respectively (Table 6). Combined phylogenetic analysis of SNPs present in the ahFAD2A and ahFAD2B genes, among 30 genotypes revealed two major clusters. Cluster 1 comprised of mainly mutant genotypes except MH1 which is a non-mutant genotype. However, cluster 2 consisted of both mutant and non-mutant genotypes. Overall, no clear clustering pattern has been observed based on the SNP sequences of desaturase gene (Figure 3). Similar pattern was obtained when clustering was done using ahFAD2A and ahFAD2B genes separately (Figures S4, S5). Fatty acid profiles of selected genotypes representing high, medium and low, oleic, linoleic and other fatty-acids are presented in Figure 4.
Table 6.
Summary of nucleotide polymorphism analysis of ahFAD2A gene among selected peanut genotypes.
| Gene | Genome | Haplotypes | Pi | Synonymous SNPs (Ks) | Non-synonymous SNPs (Ka) | Total number of SNPs | Ka/Ks ratio | InDel | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | Hd | A/G | T/C | A/T | G/C | A/C | G/T | ||||||
| ahFAD2 | A | 22 | 0.970 ± 0.018 | 0.0105 | 4 | 10 | 1 | 1 | 2 | 1 | 19 | 0.36 | 0 |
| B | 21 | 0.952 ± 0.027 | 0.0095 | 3 | 10 | 1 | 1 | 2 | 1 | 19 | 0.39 | 1 | |
Where, H, Haplotype numbers; Hd, Haplotype diversity; Pi, nucleotide diversity.
Figure 3.
Cladogram generated from cluster analysis using ahFAD2A nucleotide sequence data of 30 genotypes.
Figure 4.
Fatty acid profile of selected peanut genotypes representing high, medium and low oleic acid to linoleic acid ratio.
Discussion
Out of 167 varieties and 6 advanced breeding lines studied; 77 (46%) and 3 (50%) genotypes, respectively, showed the presence of ahFAD2A muation, while none had ahFAD2B mutant allele. Similarly, in US mini-core collections, 44% (Wang et al., 2011b), 31.6% (Chu et al., 2007) and 41% (Wang et al., 2013b); while in ICRISAT mini-core collections, 49.5% (Mukri et al., 2012) ahFAD2A mutant allele was reported. But, none found having ahFAD2B mutant allele. However, all the 22 wild-species, showed absence of mutation in the both AhFAD2A and ahFAD2B alleles. Similar results were also observed in 27 Arachis duranensis accessions (Chu et al., 2007), and in 39 wild-species including the two putative progenitors of the cultivated peanut (Wang et al., 2010). Therefore, it seems that the substitution mutation in ahFAD2 genes might have occurred after the polyploidization of cultivated peanut (Chu et al., 2007; Wang et al., 2010); and ahFAD2A mutant allele appears to have arisen before the global distribution of peanut from its center of origin (South America). This could be the reason that, only ahFAD2A mutant allele has been reported in a range of genotypes across the world (Chu et al., 2007; Wang et al., 2011b, 2013b; Mukri et al., 2012); and the absence of ahFAD2B mutation, might be due to its recent origin (Norden et al., 1987).
Since, the AS-PCR assay of Chen et al. (2010) is unable to detect the zygosity of the genes; and even in CAPS assay for ahFAD2A mutation, problem of incomplete digestion of PCR product was observed (Jung et al., 2000b; Chu et al., 2007). Therefore, the zygosity of ahFAD2A gene was studied using AS-PCR assay of Yu et al. (2013); which identified all the 80 mutant genotypes as homozygous. The results are expected, since all these lines were highly stable in nature. Moreover, for ahFAD2B gene, both AS-PCR (Yu et al., 2013) and CAPS (Chu et al., 2009) assays were found efficient for the zygosity detection.
Pedigree analysis has clearly revealed that, the mutant ahFAD2A gene has moved from its parents to the varieties. However, in the variety Kadari9, although both the parents were found non-mutant; but, it still had ahFAD2A mutation. The possible explanations could be the origin of a spontaneous mutation, although it seems a rare possibility in a varietal cross. On the contrary, a non-mutant variety, MH1 was developed through selection of Faizpur1-5, having ahFAD2A mutant gene (Table S3). Thus, considering the polyploid nature of peanut, it might take several generations of inbreeding to have a plant without a mutant allele. Another possibility could be that, during the development of these varieties, the lines might have accidently crossed with some other mutant and non-mutant lines, respectively. This could have resulted in fixation of mutant and non-mutant allele in Kadiri9 and MH1, respectively, during the course of selection. This needs further confirmation, using in-depth molecular diversity studies. Overall, it is clearly revealed that, large number of Indian peanut cultivars share common parent, which was also shown by the presence of ahFAD2A mutant allele in 46% of the varieties.
In this study, the analysis of peanut botanical types, reported more frequent ahFAD2A mutation in ssp. hypogaea (89%) compared to ssp. fastigiata (17%). Similarly, Mukri et al. (2012) reported 84.52% of the ICRISAT mini-core accessions belonging to ssp. hypogaea and 19.39% of ssp. fastigiata types carried the ahFAD2A mutant allele. Wang et al. (2013b) also reported 33.82, 6.37, 1.96, and 0% ahFAD2A mutation in vars. hypogaea, vulgaris, fastigiata and peruviana, respectively. This is also in agreement with Chu et al. (2007) and Wang et al. (2009), where in the U.S. peanut mini-core collections more frequent ahFAD2A mutation was reported in ssp. hypogaea than in ssp. fastigiata. The pedigree analysis of mutant genotypes of ssp. fastigiata revealed that, only five genotypes were derived from TG1, and one from TMV10, as a parent having ahFAD2A mutation (Table S3). Therefore, the occurrence of very low frequency of ahFAD2A mutation in ssp. fastigiata can be explained by the hypothesis of origin of ssp. hypogaea from ssp. fastigiata (Singh, 1988).
The mutant genotypes showed significantly higher average O/L ratio (2.20) than the non-mutants genotypes (1.26), and average O/L ratio of all the genotypes was 1.69 with a range of 0.93 (MH1) to 3.76 (GJG22). Similarly, in ahFAD2A mutant germplasm lines, the range of mean O/L ratio recorded was 1.11–6.93 with an average of 2.43, whereas, in the non-mutant accessions it was 1.26 with a range of 0.77–2.55 (Mukri et al., 2012). Majority of the ahFAD2A mutant genotypes were found having O/L ratio above 1.5, while non-mutant lines had less than that, with some exceptions (Table 1). Nearly similar results were reported by Wang et al. (2013b). This means, genotyping for ahFAD2A mutant allele using molecular makers is quite reliable and robust method to discriminate or identify the genotypes with high oleic acid. Overall, three genotypes viz. GG20, ICGV86325 and BG2 were found having high oil (>48%), low behenic acid (<1%), high oleic acid (>58%) (Table 1), which are more desirable traits for the oil industry (Grosso et al., 2000).
Highly negative correlation was observed between oleic acid and linoleic acid (r = −0.96), as also observed by Wang et al. (2013b, 2015a). The accumulation of oleic acid might have led to the increase in gadoleic acid content, which is formed from it. On the other hand, since more palmitic acid (C16:0) is mobilized toward the oleic acid (C18:1), it may have led to the decrease in the contents of palmitic acid, stearic acid, arachidic acid, behenic acid and lignoceric acid. This could be the reason for the negative correlation of oleic acid with all these SFAs (Wang et al., 2015a).
As reported in this study, other results also showed two mutant fatty acid desaturase (ahFAD) alleles, controlling three major fatty acids viz. oleic, linoleic and palmitic acid (Wang et al., 2013b, 2015a; Pandey et al., 2014; Janila et al., 2016). Some reduction in the production of linoleic, palmitic, stearic and arachidic acid, while no influence of these mutant alleles on the behenic and lignoceric acid was observed as also reported by Wang et al. (2013b, 2015a). Since, gadoleic acid is formed from oleic acid; therefore, an increase in the oleic acid content results in an increase in gadoleic acid content. On the other hand, since more palmitic acid is mobilized toward the oleic acid formation, the oleic acid acts as a signal to trigger a negative feedback loop to deal with an excess of SFAs, which might have resulted in decrease in the contents of palmitic acid, stearic acid, arachidic acid, behenic and lignoceric acid as also reported by Lim et al. (2013) and Harvey et al. (2010).
Interestingly, the ahFAD2B gene of all the peanut genotypes, found transcribing full 379 amino acid protein functional domains; whereas, the ahFAD2A gene of 19 genotypes showed stop codons (Figure S3). Similarly, five partial truncated sequences were also identified by Wang et al. (2015e), which might have translated a truncated fatty-acid desaturase protein. The changes in coding sequences might have caused varying efficiency of metabolic processes of fatty acid desaturase enzyme formation. Recently, Wang et al. (2015e) identified six distinct novel members in the ahFAD2 gene family involved in the regulation of linoleic acid synthesis. Among these, ahFAD2-2, ahFAD2-3 and ahFAD2-4 variants, displayed a high degree of sequence similarity with the known peanut ahFAD2 sequences. As we have used single primer pair, targeting coding regions of ahFAD2 gene, the resulting sequences may be representing different members of ahFAD2 gene. This might be one of the possible reasons for the mixing of both mutant and non-mutant genotypes in the cladogram, which needs further confirmations.
Mutations in general and SNPs in particular are the major driving forces causing genetic variation in peanut ahFAD genes. An understanding of the mutations in these genes and proteins in peanut is of prime importance due to its fundamental role in regulating the O/L ratio. The values of Ka/Ks give a clear picture into the evolution of ahFAD gene (Hurst, 2002) and the observed Ka/Ks ratio of <1 for both ahFAD2A and ahFAD2B gene sequences of the Indian peanut genotypes, implies that most amino acid substitutions have been eliminated by the strong purifying or stabilizing selection. It is also reported that the proteins with rigorous functional or structural requirements are subject to such negative selection pressure, resulting in smaller numbers of amino acid changes and tend to evolve gradually (Kondrashov et al., 2002). Thus, our analysis showed that the Ka/Ks ratio can certainly be a practical and robust measure to infer the direction and magnitude of natural selection acting on any gene in peanut.
The O/L ratio among the ahFAD2A mutant genotypes ranged from 1.03 to 3.76 (Table 1). Similarly, Mukri et al. (2012) and Wang et al. (2010) have recorded O/L ratio in the range of 1.11–6.93 and 1.86–3.42, respectively. Since, the genetic backgrounds for each genotype in our study are different; this might have affected the levels of oleic acid and linoleic acid. Moreover, reports on recombinant inbred lines (RILs) also revealed considerable phenotypic variations for different fatty acids (Sarvamangala et al., 2011; Pandey et al., 2014; Wang et al., 2015a). Even for the introgression lines (ILs) with homozygous ahFAD2 mutant alleles, considerable variations in the oleic and linoleic acid content from 62 to 82% and 02 to 20%, respectively, was recorded (Janila et al., 2016).
The double mutant line SunOleic95R is reported to have 79–81% oleic acid and 2.5 to 4.7% linoleic acid under US conditions (Gorbet and Knauft, 1997; Barkley et al., 2010, 2011). However, the seeds of same line which were grown under Junagadh, India (21.31° N, 70.36° E, 200 m above mean sea level, AMSL) conditions, recorded 80.18% oleic acid and 7.34% linoleic acid. Similarly, Janila et al. (2016) also reported 78.3% oleic acid and 6% linoleic acid for this genotype at ICRISAT-Patancheru, India (17.50° N, 78.27° E, 545 m AMSL). The oleic acid content seems nearly the same under both Indian and US environmental conditions, while linoleic acid content was relatively higher under Indian conditions. This clearly implies the involvement of other factors, influencing the production of linoleic acid.
The O/L ratio in two non-mutant genotypes, JL24 and TAG24 in the present investigation was recorded as >2.0; however, Bishi et al. (2015) and Mukri et al. (2012) recorded, <1.5 for these genotypes. On the other hand, Mondal et al. (2010) for the genotype TAG24 recorded O/L ratio as 1.85. On the similar note, normal oleic seeds were also identified in seed-lots of the high-oleic peanut cultivar “Brantley” (Chamberlin et al., 2011). The peanut fatty-acid compositions were found influenced by the genetic factors and its interaction with environment (Andersen and Gorbet, 2002; Singkham et al., 2010) and also by various environmental factors like temperature (Sun et al., 2014), growing season (Singkham et al., 2010), and maturity (Hinds, 1995).
Wang et al. (2013a) reported that 60% of the variation in oleic or linoleic acid content can be explained by the genotypic effect of ahFAD2A and ahFAD2B genes. Pandey et al. (2014) found two marker intervals with phenotypic variance (PVE) i.e., ahFAD2B gene (26.54, 25.59, and 41.02% PVE) and ahFAD2A gene (8.08, 6.86, and 3.78% PVE) for oleic acid (C18:1), linoleic acid (C18:2), and oleic/linoleic acid ratio (O/L ratio), respectively. All these reports indicate the presence of other factors, regulating the O/L flux. Recently, some novel members of ahFAD2 genes with varying expressing levels were identified in peanut. Besides, presence of other candidate genes controlling oleate levels in developing seeds and/or presence of complex gene networks controlling the fluxes between the endoplasmic reticulum and the chloroplast within the peanut cells cannot be ruled out (Wang et al., 2015e).
Conclusions
As per the US Food and Drug Administration determination, various health risks are associated with the consumption of trans-fat. As oleic acid is less prone to oxidation, the food products made from high oleic peanuts will have more shelf-life, which is most preferred trait of both oil and food processing industry. Such oil also has high impact on the markets due to the consumer's preference for several health benefits. Therefore, to make the oil healthier, breeding of improved peanut lines with high oleic and low linoleic acid becomes one of the most important breeding goals of recent times.
Though, India is one of the major oilseeds producing country, till now, there is no report of high oleate cultivar in any crop including peanut. Therefore, it is required to breed high oleate peanut cultivars in India, so as to enhance the livelihoods of small and marginal farmers, along with benefiting all the stakeholders of the value chain. Significant variation for oleic acid content was found in the genotypes that were identified having mutant ahFAD2 gene; which confirms the complex mechanisms regulating O/L ratio, and involvement of more genes and other factors in increasing the linoleic acid. In this backdrop, phenotypic confirmation is essential to advance the selected lines for further evaluations.
The peanut genotypes identified having ahFAD2A mutant allele in homozygous condition, should be intensively used for the development of double mutant lines, using MAS. It is therefore proposed that the combined approach of both, genotypic- as well as phenotypic- based selection should be used for the effective identification of lines having mutant ahFAD alleles. Additional studies on the expression of other FAD2 genes in various tissues may identify varying expression patterns of these genes including partitioning of expression. Moreover, further search for the novel mutant alleles or SNPs in ahFAD gene family are also underway in different laboratories of the world, which may help in significantly increasing increase the O/L ratio of different cultivars, in time to come. Currently, we are in the process of utilizing these assays using mutant lines for ahFAD alleles, in our peanut breeding programme to facilitate the development of lines with desired oil profile in a very precise and efficient manner.
Author contributions
BN: Performed the experiments, analyzed the data and wrote the paper; TB: Analyzed the data; RT: Designed the experiments, wrote the paper; AR: Designed the experiments; AK: Performed the analysis; JD: Performed the experiments; RK: Designed the experiments; GM: Designed the experiment, performed analysis, wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support received from Science and Engineering Board of Department of Science and Technology, New Delhi, India for the project SERB/LS-50/2013 is gratefully acknowledged.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01271
Fatty acid profile of peanut genotypes.
Chromatograms of PCR sequencing showing heterozygous peaks at different locations.
Comparison of the deduced amino acid sequences of ahFAD2A gene in 30 peanut genotypes.
Cladogram generated from cluster analysis using ahFAD2B nucleotide sequence data of 30 genotypes.
Cladogram generated from cluster analysis using ahFAD2A and ahFAD2B combined nucleotide sequence data of 30 genotypes.
Details of markers used for the identification of ahFAD2A and ahFAD2B mutant alleles in the peanut genotypes.
List of peanut varieties selected for ahFAD2 gene sequencing.
Pedigree analysis of Indian cultivars for ahFAD2A mutant allele.
Pedigree and fatty acid (Myristic, Linoleic and Erucic acid) composition (%), in the 167 peanut varieties and six advance breeding lines.
Full length FAD2A gene sequence among selected peanut genotypes.
Full length FAD2B gene sequence among selected peanut genotypes.
References
- Andersen P. C., Gorbet D. W. (2002). Influence of year and planting date on fatty acid chemistry of high oleic acid and normal peanut genotypes. J. Agric. Food Chem. 50, 1298–1305. 10.1021/jf0113171 [DOI] [PubMed] [Google Scholar]
- Barkley N. A., Chamberlin K. D., Wang M. L., Pittman R. N. (2010). Development of a real-time PCR genotyping assay to identify high oleic acid peanuts (Arachis hypogaea L.) Mol. Breed. 25, 541–548. 10.1007/s11032-009-9338-z [DOI] [Google Scholar]
- Barkley N. A., Wang M. L., Pittman R. (2011). A real-time PCR genotyping assay to detect FAD2A SNPs in peanuts (Arachis hypogaea L.). Electron. J. Biotechnol. 14, 1–12. 10.2225/vol14-issue1-fulltext-12 [DOI] [Google Scholar]
- Bertioli D. J., Seijo G., Freitas F. O., Valls J. F. M., Bertioli S. C. M., Moretzsohn M. C. (2011). An overview of peanut and its wild relatives. Plant Genet. Resour. 9, 134–149. 10.1017/S1479262110000444 [DOI] [Google Scholar]
- Bhauso T. D., Radhakrishnan T., Kumar A., Mishra G. P., Dobaria J. R., Rajam M. V. (2014). Over-expression of bacterial mtlD gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic peanut (Arachis hypogaea) through accumulation of mannitol. Aust. J. Crop Sci. 8, 413–421. 10.1155/2014/125967 [DOI] [Google Scholar]
- Bishi S. K., Lokesh K., Mahatma M. K., Khatediya N., Chauhan S. M., Misra J. B. (2015). Quality traits of Indian peanut cultivars and their utility as nutritional and functional food. Food Chem. 167, 107–114. 10.1016/j.foodchem.2014.06.076 [DOI] [PubMed] [Google Scholar]
- Brown L. D. (1979). A Proof that the Tukey-Kramer Multiple Comparison Procedure for Differences between Treatment Means is Level for 3, 4, or 5 Treatments. Technical Report, Department of Mathematics, Cornell University. Available online at: https://statistics.wharton.upenn.edu/files/?whdmsaction=public:main.file&fileID=1120
- Cao S., Zhu Q. H., Shen W., Jiao X., Zhao X., Liu L., et al. (2013). Comparative profiling of miRNA expression in developing seeds of high linoleic and high oleic safflower (Carthamus tinctorius L.) plants. Front. Plant Sci. 4:489. 10.3389/fpls.2013.00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin K. D., Melouk H. A., Madden R., Dillwith J. W., Bannore Y., Rassi Z. E., et al. (2011). Determining the oleic/linoleic acid ratio in a single peanut seed: a comparison of two methods. Peanut Sci. 38, 78–84. 10.3146/PS11-3.1 [DOI] [Google Scholar]
- Chen Z. B., Wang M. L., Barkley N. A., Pittman R. N. (2010). A simple allele-specific PCR assay for detecting FAD2 alleles in both A and B genomes of the cultivated peanut for high-oleate trait selection. Plant Mol. Biol. Rep. 28, 542–548. 10.1007/s11105-010-0181-5 [DOI] [Google Scholar]
- Chu Y., Holbrook C. C., Ozias-Akins P. (2009). Two alleles of ahFAD2B control the high oleic acid trait in cultivated peanut. Crop Sci. 49, 2029–2036. 10.2135/cropsci2009.01.0021 [DOI] [Google Scholar]
- Chu Y., Ramos L., Holbrook C. C., Ozias-Akins P. (2007). Frequency of a loss-of-function mutation in oleoyl-PC desaturase (ahFAD2A) in the mini-core of the US peanut germplasm collection. Crop Sci. 47, 2372–2378. 10.2135/cropsci2007.02.0117 [DOI] [Google Scholar]
- Chu Y., Wu C. L., Holbrook C. C., Tillman B. L., Person G., Ozias-Akins P. (2011). Marker assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 4, 110–117. 10.3835/plantgenome2011.01.0001 [DOI] [Google Scholar]
- Cuc L. M., Mace E. S., Crouch J. H., Quang V. D., Long T. D., Varshney R. K. (2008). Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea L.). BMC Plant Biol. 8:55. 10.1186/1471-2229-8-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire E. J. (2014). A review of the nutritional composition, organoleptic characteristics and biological effects of the high oleic peanut. Int. J. Food Sci. Nutr. 65, 781–790. 10.3109/09637486.2014.937799 [DOI] [PubMed] [Google Scholar]
- Fang C. Q., Wang C. T., Wang P. W., Tang Y. Y., Wang X. Z., Cui F. G., et al. (2012). Identification of novel mutation in FAD2B from a peanut EMS mutant with elevated oleate content. J. Oleo Sci. 61, 143–148. 10.5650/jos.61.143 [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2014). Available online at: http://faostat.fao.org/ (Accessed January 15, 2016).
- Gajjar K. N., Mishra G. P., Radhakrishnan T., Dodia S. M., Rathnakumar A. L., Kumar N., et al. (2014). Validation of SSR markers linked to the rust and late leaf spot diseases resistance in diverse peanut genotypes. Aust. J. Crop Sci. 8, 927–936. Available online at: http://www.cropj.com/thankapan_8_6_2014_927_936.pdf [Google Scholar]
- Gorbet D. W., Knauft D. A. (1997). Registration of ‘SunOleic95R’ peanut. Crop Sci. 37, 1392 10.2135/cropsci2000.0032rcv [DOI] [Google Scholar]
- Grosso N. R., Nepote V., Guzma C. A. (2000). Chemical composition of some wild peanut species (Arachis L.) J. Agric. Food Chem. 48, 806–809. 10.1021/jf9901744 [DOI] [PubMed] [Google Scholar]
- Harvey K. A., Walker C. L., Xu Z., Whitley P., Pavlina T. M., Hise M., et al. (2010). Oleic acid inhibits stearic acid-induced inhibition of cell growth and pro-inflammatory responses in human aortic endothelial cells. J. Lipid Res. 51, 3470–3480. 10.1194/jlr.M010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds M. J. (1995). Fatty acid composition of Caribbean-grown peanut (Arachis hypogaea L.) at three maturity stages. Food Chem. 53, 7–14. 10.1016/0308-8146(95)95779-6 [DOI] [Google Scholar]
- Huang X., Madan A. (1999). CAP3: a DNA sequence assembly program. Genome Res. 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18, 486–487. 10.1016/S0168-9525(02)02722-1 [DOI] [PubMed] [Google Scholar]
- Janila P., Nigam S. N., Pandey M. K., Nagesh P., Varshney R. K. (2013). Groundnut improvement: use of genetic and genomic tools. Front. Plant Sci. 4:23. 10.3389/fpls.2013.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janila P., Pandey M. K., Shasidhar Y., Variath M. T., Sriswathi M., Khera P., et al. (2016). Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 242, 203–213. 10.1016/j.plantsci.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Jung S., Powell G., Moore K., Abbott A. (2000a). The high oleate trait in the cultivated peanut (Arachis hypogaea L.) II. Molecular basis and genetics of the trait. Mol. Gen. Genet. 263, 806–811. 10.1007/s004380000243 [DOI] [PubMed] [Google Scholar]
- Jung S., Swift D., Sengoku E., Patel M., Teule F., Powell G., et al. (2000b). The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol. Gen. Genet. 263, 796–805. 10.1007/s004380000244 [DOI] [PubMed] [Google Scholar]
- Knauft D. A., Moore K. M., Gorbet D. W. (1993). Further studies on the inheritance of fatty acid composition in peanut. Peanut Sci. 20, 74–76. 10.3146/i0095-3679-20-2-2 [DOI] [Google Scholar]
- Kondrashov A. S., Sunyaev S., Kondrashov F. A. (2002). Dobzhansky–Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. U.S.A. 99, 14878–14883. 10.1073/pnas.232565499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapovickas A., Rigoni V. A. (1960). La nomenclatura de las subspecies y variedades de Arachis hypogaea L. Rev. Invest. Agric. 14, 198–228. [Google Scholar]
- Letunic I., Bork P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lim H. J., Hines Z. G., Dominy J. E., Lee Y., Kim S., Tabata M., et al. (2013). Oleic acid stimulates complete oxidation of fatty acids through PKA-dependent activation of SIRT1/PGC1 complex. J. Biol. Chem. 288, 7117–7126. 10.1074/jbc.M112.415729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Y., Nadaf H. L., Smith O. D., Connell J. P. (2000). Isolations and characterization of the Δ12 fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphism for the high oleate trait in Spanish market-type lines. Theor. Appl. Genet. 101, 1131–1138. 10.1007/s001220051589 [DOI] [Google Scholar]
- Mishra G. P., Radhakrishnan T., Kumar A., Thirumalaisamy P. P., Kumar N., Bosamia T. C., et al. (2015). Advancements in molecular marker development and their applications in the management of biotic stresses in peanuts. Crop Prot. 77, 74–86. 10.1016/j.cropro.2015.07.019 [DOI] [Google Scholar]
- Misra J. B., Mathur R. S. (1998). A simple and economic procedure for transmethylation of fatty acids of groundnut oil for analysis by GLC. Int. Arachis Newslett. 18, 40–42. [Google Scholar]
- Mondal S., Badigannavar A. M., Dsouza S. F. (2010). Induced variability for fatty acid profile and molecular characterization of high oleate mutant in cultivated groundnut (Arachis hypogaea L.). Mol. Breed. 130, 242–247. 10.1111/j.1439-0523.2010.01787.x [DOI] [Google Scholar]
- Mukri G., Nadaf H. L., Bhat R. S., Gowda M. V. C., Upadhyaya H. D., Sujay V. (2012). Phenotypic and molecular dissection of ICRISAT mini core collection of peanut (Arachis hypogaea L.) for high oleic acid. Plant Breed. 131, 418–422. 10.1111/j.1439-0523.2012.01970.x [DOI] [Google Scholar]
- Nigam S. N. (2000). Some strategic issues in breeding for high and stable yield in groundnut in India. J. Oilseeds Res. 17, 1–10. Available online at: http://oar.icrisat.org/1910/1/JOilseedsRes17_1_1-10_2000.pdf [Google Scholar]
- Norden A. J., Gorbet D. W., Knauft D. A., Young C. T. (1987). Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci. 14, 7–11. 10.3146/i0095-3679-14-1-3 [DOI] [Google Scholar]
- O'Keefe S. F., Wiley V. A., Knauft D. A. (1993). Comparison of oxidative stability of high and normal oleic peanut oils. J. Am. Oil Chem. Soc. 70, 489–492. 10.1007/BF02542581 [DOI] [Google Scholar]
- Pandey M. K., Wang M. L., Qiao L., Feng S., Khera P., Wang H., et al. (2014). Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 15:133. 10.1186/s12863-014-0133-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T., Thankappan R, Kumar A., Mishra G. P., Dobaria J. R. (2014). Heterologous expression of the AtDREB1A gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS ONE 9:e110507. 10.1371/journal.pone.0110507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvamangala C., Gowda M. V. C., Varshney R. K. (2011). Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.) Field Crop Res. 122, 49–59. 10.1016/j.fcr.2011.02.010 [DOI] [Google Scholar]
- Schwartzbeck J. L., Jung S., Abbott A. G., Mosley E., Lewis S., Pries G. L., et al. (2001). Endoplasmic oleoyl-PC desaturase references the second double bond. Phytochemistry 57, 643–652. 10.1016/S0031-9422(01)00081-4 [DOI] [PubMed] [Google Scholar]
- Simpson C. E., Krapovickas A., Valls J. F. M. (2001). History of Arachis including evidence of A. hypogaea L. progenitors. Peanut Sci. 28, 78–80. 10.3146/i0095-3679-28-2-7 [DOI] [Google Scholar]
- Singh A. K. (1988). Putative genome donors of Arachis hypogaea (Fabaceae), evidence from crosses with synthetic amphidiploids. Plant Syst. Evol. 160, 143–151. 10.1007/BF00936041 [DOI] [Google Scholar]
- Singkham N., Jogloy S., Kesmala T., Swatsitang P., Jaisil P., Puppala N. (2010). Genotypic variability and genotype by environment interactions in oil and fatty acids in high, intermediate and low oleic acid peanut genotypes. J. Agric. Food Chem. 58, 6257–6263. 10.1021/jf903728e [DOI] [PubMed] [Google Scholar]
- Sun M., Spears J. F., Isleib T. G., Jordan D. L., Penny B., Johnson D., et al. (2014). Effect of production environment on seed quality of normal and high- oleate large seeded Virginia-type peanut (Arachis hypogaea L.). Peanut Sci. 41, 90–99. 10.3146/PS12-16.1 [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassiliou E. K., Gonzalez A., Garcia C., Tadros J. H., Chakraborty G., Toney J. H. (2009). Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-α both in vitro and in vivo system. Lipids Health Dis. 8:25. 10.1186/1476-511X-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. T., Tang Y. Y., Wang X. Z., Chen D. X., Cui F. G., Chi Y. C., et al. (2011a). Evaluation of groundnut genotypes from China for quality traits. J. SAT Agric. Res. 9, 1–15. Available online at: http://ejournal.icrisat.org/Volume9/Groundnut/Evaluation.pdf [Google Scholar]
- Wang C. T., Tang Y. Y., Wang X. Z., Wu Q., Yang Z., Jiao K., et al. (2013a). Effect of FAD2A/FAD2B genes on fatty acid profiles in peanut seeds. Res. Crops 14, 1110–1113. Available online at: http://cropresearch.org/volume-14-number-4-december-2013/ [Google Scholar]
- Wang M. L., Barkley N. A., Chinnan M., Stalker H. T., Pittman R. N. (2010). Oil content and fatty acid composition variability in wild peanut species. Plant Genet. Resour. 8, 232–234. 10.1017/S1479262110000274 [DOI] [Google Scholar]
- Wang M. L., Chen C. Y., Davis J., Guo B., Stalker H. T., Pittman R. N. (2009). Assessment of oil content and fatty acid composition variability in different peanut subspecies and botanical varieties. Plant Genet. Resour. 1, 71–73. 10.1017/S1479262109990177 [DOI] [Google Scholar]
- Wang M. L., Chen C. Y., Tonnis B., Barkley N. A., Pinnow D. L., Pittman R. N., et al. (2013b). Oil, fatty acid, flavonoid, and resveratrol content variability and FAD2A functional SNP genotypes in the U.S. peanut mini-core collection. J. Agric. Food Chem. 61, 2875–2882. 10.1021/jf305208e [DOI] [PubMed] [Google Scholar]
- Wang M. L., Khera P., Pandey M. K., Wang H., Qiao L., Feng S., et al. (2015a). Genetic mapping of QTLs controlling fatty acids provided insights into the genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.) PLoS ONE 10:e0119454. 10.1371/journal.pone.0119454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. L., Pinnow D., Tonnis B., Tishchenko V., An Y. Q. C., Pederson G. A. (2015b). Newly identified natural high-oleate mutant from Arachis hypogaea L. subsp. hypogaea. Mol. Breed. 35, 186 10.1007/s11032-015-0377-3 [DOI] [Google Scholar]
- Wang M. L., Sukumaran S., Barkley N. A., Chen Z., Chen C. Y., Pittman R. N., et al. (2011b). Population structure and marker–trait association analysis of the US peanut (Arachis hypogaea L.) mini-core collection. Theor. Appl. Genet. 123, 1307–1317. 10.1007/s00122-011-1668-7 [DOI] [PubMed] [Google Scholar]
- Wang X. Z., Tang Y. Y., Wu Q., Sun Q. X., Wang Y. Y., Hu D. Q., et al. (2015c). Characterization of high-oleic peanut natural mutants derived from an intersectional cross. Grasas Aceites 66:e091 10.3989/gya.1070142 [DOI] [Google Scholar]
- Wang X. Z., Wu Q., Tang Y. Y., Sun Q. X., Wang C. T. (2015d). FAD2B from a peanut mutant with high oleic acid content was not completely dysfunctional, in Advances in Applied Biotechnology, Lecture Notes in Electrical Engineering, Vol. 332, eds Zhang T.-C., Nakajima M. (Berlin; Heidelberg: Springer-Verlag; ), 265–271. [Google Scholar]
- Wang Y., Zhang X., Zhao Y., Prakash C. S., He G., Yin D. (2015e). Insights into the novel members of the FAD2 gene family involved in high-oleate fluxes in peanut. Genome 58, 375–383. 10.1139/gen-2015-0008 [DOI] [PubMed] [Google Scholar]
- Yu H. T., Yang W. Q., Tang Y. Y., Wang X. Z., Wu Q., Hu D. Q., et al. (2013). An AS-PCR assay for accurate genotyping of FAD2A/FAD2B genes in peanuts (Arachis hypogaea L.) Grasas Aceites 64, 395–399. 10.3989/gya.118712 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fatty acid profile of peanut genotypes.
Chromatograms of PCR sequencing showing heterozygous peaks at different locations.
Comparison of the deduced amino acid sequences of ahFAD2A gene in 30 peanut genotypes.
Cladogram generated from cluster analysis using ahFAD2B nucleotide sequence data of 30 genotypes.
Cladogram generated from cluster analysis using ahFAD2A and ahFAD2B combined nucleotide sequence data of 30 genotypes.
Details of markers used for the identification of ahFAD2A and ahFAD2B mutant alleles in the peanut genotypes.
List of peanut varieties selected for ahFAD2 gene sequencing.
Pedigree analysis of Indian cultivars for ahFAD2A mutant allele.
Pedigree and fatty acid (Myristic, Linoleic and Erucic acid) composition (%), in the 167 peanut varieties and six advance breeding lines.
Full length FAD2A gene sequence among selected peanut genotypes.
Full length FAD2B gene sequence among selected peanut genotypes.