Abstract
Objective The metadata reflecting the location of the infected host (LOIH) of virus sequences in GenBank often lacks specificity. This work seeks to enhance this metadata by extracting more specific geographic information from related full-text articles and mapping them to their latitude/longitudes using knowledge derived from external geographical databases.
Materials and Methods We developed a rule-based information extraction framework for linking GenBank records to the latitude/longitudes of the LOIH. Our system first extracts existing geospatial metadata from GenBank records and attempts to improve it by seeking additional, relevant geographic information from text and tables in related full-text PubMed Central articles. The final extracted locations of the records, based on data assimilated from these sources, are then disambiguated and mapped to their respective geo-coordinates. We evaluated our approach on a manually annotated dataset comprising of 5728 GenBank records for the influenza A virus.
Results We found the precision, recall, and f-measure of our system for linking GenBank records to the latitude/longitudes of their LOIH to be 0.832, 0.967, and 0.894, respectively.
Discussion Our system had a high level of accuracy for linking GenBank records to the geo-coordinates of the LOIH. However, it can be further improved by expanding our database of geospatial data, incorporating spell correction, and enhancing the rules used for extraction.
Conclusion Our system performs reasonably well for linking GenBank records for the influenza A virus to the geo-coordinates of their LOIH based on record metadata and information extracted from related full-text articles.
Keywords: phylogeography, information extraction, natural language processing
BACKGROUND AND SIGNIFICANCE
Information extraction (IE) involves the use of natural language processing techniques for automated extraction of structured information about entities, relations, or events from unstructured textual data. In recent years, the rapidly expanding field of IE has been applied to accelerate research in various biomedical domains. For instance, IE methods are currently being used to automatically extract relations between drugs, genes, and diseases from PubMed articles in order to populate the structured PharmGKB database,1 which in turn can be used for advancing personalized medicine.
Much less work in IE has explored supporting public health applications that heavily rely on detailed geospatial information about the sampling sites of genetic sequences. One example of such an application is phylogeography, which has recently grown into a popular means of tracking the spread of infectious pathogens and enhancing their epidemiological analysis.2–4 For instance, Hovmöller et al.5 used multiple phylogenetic trees to estimate the geographical transmission routes of a highly pathogenic strain of H5N1 virus and developed a web application for visualizing the estimated routes. Similarly, Janies et al.6 combined phylogenetic analyses with visualization techniques to study the global spread of H7 influenza A viruses. In addition to advancing the surveillance of infectious diseases, such forms of sequence-based analysis, incorporating the location of the infected host (LOIH) from which the pathogen sequence was isolated, can also assist the design and distribution of vaccines, and help clinical researchers better understand the etiology of various diseases.2,7,8
The geographic metadata required for studies involving the spatial modeling of sequences are often obtained from public databases such as GenBank,9 which is part of the International Nucleotide Sequence Database Collaboration and includes data deposited by researchers all over the world. Each GenBank record contains separate fields for holding various forms of sequence-related metadata such as strain name, date of collection, LOIH, and the type of host. The LOIH is typically present in the country field of the record. Despite its name, this field does not simply contain the name of the country in which the host was found; it may include locations with varying levels of specificity or may not contain any location at all. For instance, in GenBank record AY282759 (of note, this is the accession number of the record, not the GenBank identifier), this field does not contain any geographic metadata; in M63769 it contains a single ambiguous place name, “Cambridge” without indicating the specific country in which it resides; in GU332632 it contains the name of a state “USA:Iowa”; and in CY024354 it contains the name of a city “China:Shantou.” Since building precise spatial models often requires very specific information about the LOIH of the sequences being studied, the geospatial metadata in GenBank, even when available, may not be sufficient for the researcher. For instance, Raghwani et al.10 used district-level phylogeographic analysis to study dengue virus migration within Ho Chi Minh City in Vietnam. With only country-level or province-level geospatial metadata, such an analysis would not have been feasible.
Because of the absence or inadequacy of geospatial metadata in GenBank records, researchers might need to search full-text publications linked to the records for more specific information. If found, the more specific metadata from the paper is incorporated within their study. However, a manual survey of articles is a time-consuming and tedious process and presents a major bottleneck for data collection. Moreover, many studies may require the specific latitude/longitude coordinates of the sampling sites; thus, simply finding the name of the locations may not be enough. For instance, for continuous phylogeography studies11 and disease spread visualization tools,12 obtaining the specific geo-coordinates is crucial. In this case, researchers wishing to use the information would need to perform an additional step of mapping each location to its correct geospatial coordinates using a database such as GeoNames,13 which lists 10 million geospatial locations across the world. This is not a trivial process, since some locations can be highly ambiguous and possibly map to a large number of unique coordinates. Consider, for example, “Malang, Indonesia,” which is mapped in GeoNames to 23 distinct locations. An additional problem ancillary to the manual process is that, depending on how the ambiguities are resolved, it is possible that different coordinates would be derived for the same study by different researchers.
Therefore, an automated system for the extraction of geospatial metadata from GenBank records and related full-text articles can help make this process faster and more systematic, positively impacting public health research. To the best of our knowledge, no such system currently exists. The Bacteria Biotope Task14 of BioNLP Shared Task 2013 included the extraction of localization relations between bacterial species and geographical entities, but participants were not required to map the geo-entities to their latitude/longitude coordinates and the task corpus consisted of web documents rather than full-text scientific articles. Additionally, Tamames and Lorenzo15 performed toponym (location name) resolution (detection and disambiguation) in full-text articles mentioning the sampling sites of bacterial sequences, and achieved a precision and recall of 0.92 and 0.86, respectively, for this task. However, their work did not focus on linking the sequences to their collection sites in the articles. Other studies have analyzed or used GenBank metadata, often in combination with other resources, for various applications16–20 but none specifically attempted the enhancement of geospatial metadata in GenBank using information extracted from full-text articles.
OBJECTIVE
The objective of the present study was to provide an IE framework for automatically linking GenBank record sequences to the latitude and longitude coordinates of their LOIH in order to help advance public health research. Our system attempted to make this location as specific as possible by extracting geospatial metadata from GenBank record fields and related full-text PubMed Central (PMC) articles. As our primary case study, we present a detailed evaluation of our system on a set of manually annotated GenBank records for the influenza A virus. We chose this virus because of the large sample of influenza A sequences in GenBank as well as its significance in public health research. In addition, to test the generalizability of our system, we also report its accuracy on a smaller sample of GenBank records for St. Louis Encephalitis (SLE), Eastern equine encephalitis (EEE), Western equine encephalitis (WEE), West Nile virus (WNV), rabies, and hantavirus, which are some of the most widely studied zoonotic viruses (viruses transmittable between animals and humans) by public health, agricultural, and wildlife state departments in United States.21
MATERIALS AND METHODS
Our methodology for conducting this study can be divided into three broad stages: selection and download of GenBank records and related PMC articles, development of the IE system, and evaluation of the IE system. Each of these stages is described in detail below.
Selection and Download of GenBank Records and Related PMC Articles
For the influenza case study, we used stratified random sampling to select ∼10% of all PMC articles linked to GenBank records for the influenza A virus (stratification was performed based on the number of records linked to each article; further details given in Appendix A). This produced a corpus of 60 PMC articles corresponding to 5728 GenBank records. We manually downloaded the PDF versions of these papers and used Xpdf22 to convert them into text files (of note, we chose to use the PDF version of each article instead of parsing its HTML version or searching for its XML version in PMC Open Access because not all articles have an HTML version and only a limited subset of PMC articles are available through PMC Open Access). In addition, we used the National Center for Biotechnology Information (NCBI) Entrez Utilities application programming interface (API)23 to download relevant metadata fields from the selected GenBank records, including: country, strain, organism, isolate, date, and host.
For our secondary study, we first gathered a list of PMC articles linked to at least 10 GenBank records associated with the remaining six viruses (SLE, EEE, WEE, WNV, rabies, and hantavirus) and randomly selected two PMC articles for each virus. For each selected article, we randomly selected 10 records for inclusion within our evaluation sample. This resulted in a total of 120 records from 12 articles, equally distributed among the 6 viruses.
IE System Development
Our geospatial IE system is largely dependent on the GeoNames13 database, a collection of over 10 million geospatial locations across the world that has been effectively used in several existing systems for toponym resolution.15,24,25 In addition to location names, GeoNames also contains several useful features about each entry in the database such as population data, country code, and the latitude and longitude coordinates of the location’s centroid. For the purpose of this project, we downloaded the GeoNames data available online and imported it into a local database. In addition, we also imported data from the Socrata dataset,26 which contains geospatial data for 243 countries, since several country names were too ambiguous in GeoNames. For instance, the results from the query “Italy” in GeoNames does not include the country “Italy”—it contains populated places in other countries. One needs to query for “Repubblica Italiana” to retrieve this country. France, on the other hand, is listed as “Republic of France” and not “Republique Francaise.” If we include alternate names for these countries we obtain names such as “Farani” for France, which are not as likely to be referring to the country in a scientific article written in the English language and may generate more false positives (FP). Therefore, we opted to include the Socrata dataset, which focuses solely on countries.
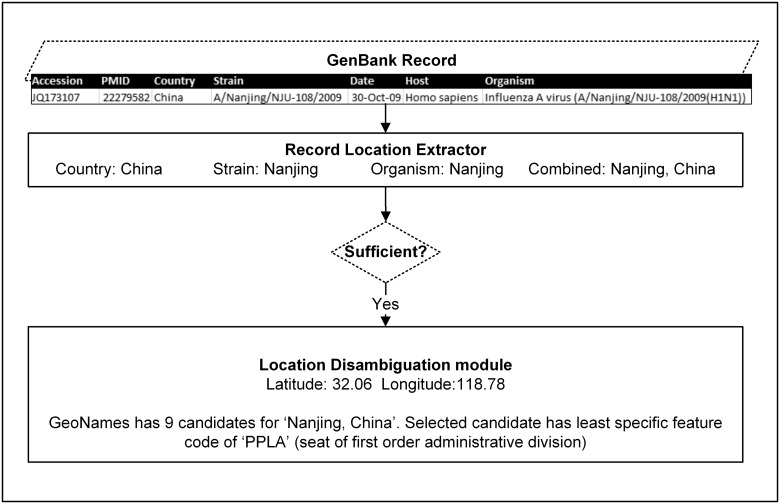
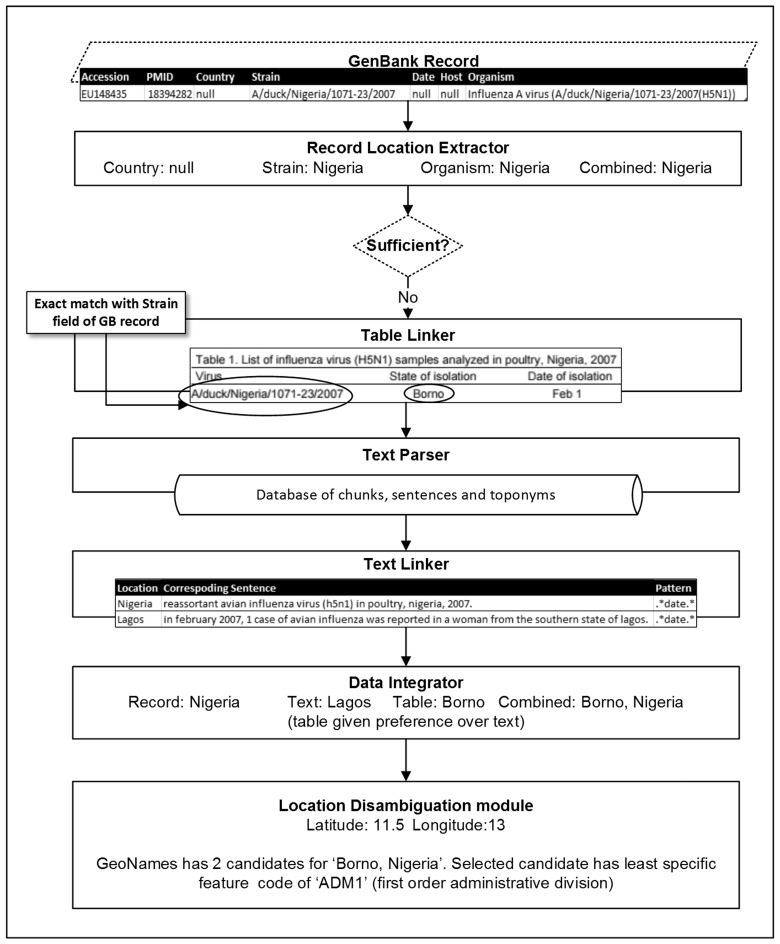
In order to introduce a stopping criterion for our system, we defined any location more specific than first order administrative division (ADM1) level as “sufficient” based on our prior study.27 This criterion includes counties, districts, cities, towns, or any other form of populated place within a country that is less specific than states and provinces. As illustrated in Figure 1, if a record already contains sufficient geographic metadata within the record, our system directly proceeds to assigning geo-coordinates to the locations present in the record metadata instead of processing the related paper. For records with insufficient geographic metadata, the system attempts to extract more specific information from the related article, if available, until it finds a LOIH that is considered sufficient (see Figure 2). Therefore, our system is capable of finding locations more specific than ADM1 (such as districts and cities) but does not necessarily search for more specific geospatial information once it finds such a location. To ensure retrieval of even more specific locations, when available, the stopping criterion would need to be altered.
Figure 1.
System pipeline for a GenBank record with sufficient geospatial metadata (location of infected host more specific than state or province level).
Figure 2.
System pipeline for a GenBank record with insufficient geospatial metadata (location of infected host not more specific than state or province level).
For our current study, we integrated data from the strain, isolate, and organism fields of GenBank records, in addition to the country field, for extracting existing geospatial metadata from the records. This is because virus nomenclature often includes the unique practice of incorporating the LOIH of sequences within their taxon names. For instance, the strain field of GenBank record JF340084 contains “A/St.Petersburg/14/2010” while the country field contains “Russia,” thereby implying that the sequence was isolated from “St. Petersburg, Russia.”
The individual steps in the system pipeline are described below.
Record Location Extractor
The first step in our system pipeline involves the integration of existing geospatial metadata present within the country, strain, isolate, and organism fields of the GenBank record in order to identify the most specific LOIH available within the record itself. For instance, if the country field contains the location “China” while the strain field contains the location “Guangdong,” then the most specific LOIH for this record based on record data will be “Guangdong, China.” The record location extractor module uses our integrated database of geospatial locations for detecting location names mentioned within the record, determining the administrative level of each location detected and pairing extracted location names with any mention of their parent ADM1-level location and/or parent country in the record.
Sufficiency Analyzer
Depending on the final output produced by the record location extractor, a GenBank record may be classified as either sufficient or insufficient. To be considered sufficient, the GenBank record must either contain a location name more specific than ADM1-level along with the name of the country in which it resides; e.g., “San Diego, USA” or a location name more specific than ADM1-level that can only be present in a single country; e.g., “Beijing,” which can only be present in “China.” A record classified as insufficient may contain no location information, only country-level information; e.g., “China,” ADM1-level information along with associated country name; e.g., “Guangdong, China” or ambiguous location information for which no matching country name could be found within the record; e.g., “Osaka,” which is a place that can be present in Japan, USA, South Africa, or the Solomon Islands. For all records that are classified as insufficient, our system searches the related full-text PMC article for more specific geospatial information using our table linker, text parser, and text linker modules.
Table Linker
The purpose of the table linker module is to identify possible LOIHs using table data (if available) in referenced PMC articles. Since the conversion from PDF to text does not allow the tables in the document to retain their defined structure, we directly parsed the HTML content of the articles in PMC to extract table information. For every table in each article related to at least one insufficient record, the table linker analyzes the table headers to determine whether or not it possesses relevant information that could be used to link a GenBank record to a geographic location. A table is considered relevant if one of its headers contains the word location or any of its synonyms (location column) and another contains the words accession, strain, date, host, or one of their synonyms (GenBank metadata column). We manually compiled a list of synonyms for each of these words. If the data from the GenBank metadata column of one of the rows of a relevant table matches the metadata of the record, the table linker links data from the location column of the row to that record (see Figure 2). Aside from date metadata, we used exact match as our means of establishing equivalency between table data and GenBank record data. For dates, we first normalized the data before comparing them. Here, we used Stanford SUTime28 along with a separate rule-based program that we wrote for normalizing date metadata in GenBank records, presented in formats such as “12-May” and “12-Jun-2007,” which Stanford SUTime was unable to parse, into TIMEX expressions.
Text Parser
To allow effective IE from the textual content of the article, we first used the text parser module to extract all sentences, tokens, and toponyms and stored them in a local database. For toponym detection, we used our system presented by Weissenbacher et al.,29 which was found to have a precision, recall, and f-score of 0.599, 0.904, and 0.72, respectively, for this task. For sentence segmentation, we used the ANNIE module from the GATE platform,30 while for word segmentation, parts-of-speech tagging, and chunking, we used the Genia tagger.31
Text Linker
The text linker module links the geospatial entities identified by our toponym detection system within an article to relevant GenBank records using a rule-based approach. A geospatial location extracted from an article is linked to a record referencing the article if the sentence containing the location also mentions other record metadata, such as strain name and accession number, or fits a few simple patterns that we developed. In Appendix B, we list the patterns used by our system. For every geospatial entity identified by our toponym detection system in an article related to a specific insufficient record, our text linker first determines whether the entity is present in a relevant section of the article (of note, the Author Affiliation, Acknowledgment, and Reference sections are considered to be the only “irrelevant” sections of an article) and for those that are, it proceeds to analyze the sentence containing the entity to check if it fits any of the utilized patterns; if it does, the entity is considered to be a possible candidate for the location of the virus.
Data Integrator
The data integrator module assimilates information extracted by the record location extractor, table linker, and text linker modules for a given GenBank record to produce a coherent set of geographical locations that are possible LOIHs for the record. This set of locations is referred to as final locations in the remainder of the present article. The first step in this process involves the elimination of all locations extracted by the text linker and table linker that are inconsistent with the output produced by the record location extractor. A location is said to be inconsistent if one of the following is true: 1) It does not belong to the same country as the record location; or 2) it does not belong to the same ADM1-level location as the record location (e.g., “Philadelphia” is inconsistent with “Arizona, USA”). Once all the inconsistent locations have been removed, the data integrator uses output from the table linker and text linker to increase the specificity of the record location until a sufficient location is found; if no sufficient location is found, it outputs the most specific location retrieved. In case of locations with the same level of specificity, preference is given to table-derived locations over text-derived locations. This is because tables tend to link each individual record to its precise LOIH (one-to-one mapping) whereas paragraphs in the article typically provide a list of locations related to all records referenced in them (see Figure 2). Therefore, adding information from the text linker, when sufficient table data is present, may reduce system precision. The final output from this module consists of distinct, non-overlapping locations considered by the system to be the set of most specific LOIHs available for the record based on the sources analyzed. This may include more than one location if the heuristics fail to narrow it down to a single LOIH. For every location, the parent country name and ADM1-level location is also included in the output, if found by the system.
Location Disambiguation Module
The location disambiguation module links each final location to its specific latitude and longitude coordinates. First, the system queries our geospatial database to retrieve all possible latitude/longitudes for the location. Next, it sorts them based on their feature codes (code in GeoNames denoting the type of the location, e.g., country, state, city, etc.) and chooses the group of coordinates belonging to the least specific feature codes. For instance, in GeoNames, “Arizona” can be both a state in United States with feature code of ADM1 and a populated place in the state of Texas, United States with feature code of PPL but our system will only select the former since it has a less specific feature code. This heuristic is based on the assumption that in the majority of cases, when an author mentions a location name that can refer to multiple places on earth with varying levels of specificity, he/she is referring to the one that is more widely known across the world and the less specific a place is, the more widely known it tends to be. Lastly, the module sorts the group of coordinates selected in the previous step based on their population, and outputs the set with the highest population. This is a popular heuristic32 within the field of toponym disambiguation since it is generally assumed that places that have higher populations are better known among people and are more likely to be mentioned. When querying the database for the latitude/longitudes of a given location, we included the country code and ADM1 code of the location, if known.
System Evaluation
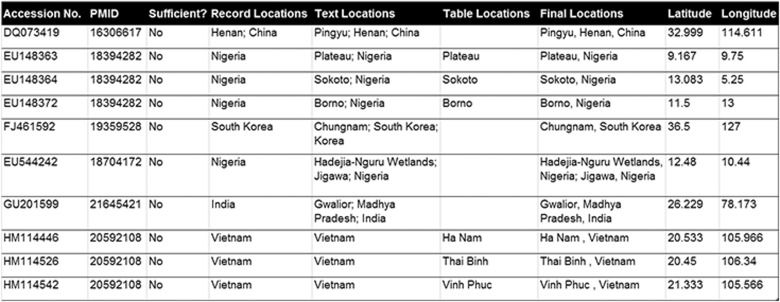
In order to evaluate our system for the influenza case study, three annotators manually annotated the 5728 GenBank records linked to the 60 related papers. The annotators followed a set of guidelines created prior to development of the corpus and documented relevant data for evaluating each individual module within the system pipeline (see Appendix C for annotation details and Figure 3 for example). We calculated the inter-rater agreement (IRR) for “sufficiency” annotation of 2017 records related to six randomly selected PMC articles and the final location (defined in the Data Integrator section) annotation of 1477 records related to 36 randomly selected PMC articles. We used the traditional IRR metric of Cohen’s kappa statistic as our measure of IRR for “sufficiency” annotation. However, for the other annotations, we used f-score as our measure of IRR, holding one annotator as the gold standard each time. This is because Cohen’s kappa calculation requires well-defined negative cases, which we lack for these annotations, and f-score has been shown to be a reliable IRR measure for information retrieval tasks.33 After calculation of the IRR, the annotators performed multiple rounds of annotation for all records to ensure that the guidelines were followed and any mistakes corrected before creation of the gold-standard corpus (included as supplementary file in Appendix D).
Figure 3.
Example of annotated GenBank records for the influenza case study.
We evaluated the different components of our system for the influenza case study using Exact Match per record-location linkage criteria. In this case, a true positive indicates that a given record-location linkage extracted by our system is equivalent to one annotated in the gold standard. For the final locations, we normalized the country codes to allow for fairer comparison but partial matches were not allowed. For instance if the annotated final location for a record is “New York City, New York, USA” and our program simply outputs “New York, USA,” then we count this linkage as both a FP and a false negative. When evaluating the disambiguation module, we considered two geo-coordinates of non-country locations to be equivalent if they were within 10 miles of each other; geo-coordinates of country-level locations were considered equivalent if they were within 200 miles of each other. We used a larger margin for country mentions since our annotators used the GeoNames website online to annotate these locations while our system used the Socrata dataset and there were slight discrepancies between the two sources.
For our secondary study, we annotated only the final location for each record and manually verified whether the final location extracted by our program matched our annotated location for each record. If any part of the correct location was missing or additional FPs were present within the final location for a record, then we counted it as a single error. This allowed us to estimate the accuracy of our system for other viruses.
RESULTS
Corpus Statistics
According to the results from our gold standard annotation, 75% of the 5728 GenBank records selected for our influenza case study were found to be insufficient using data from all four fields in the GenBank records. The specificity of the LOIH of 38% of insufficient records was increased using information from the PMC articles. For 90% of these records, it was necessary to read the full-text content of the articles, rather than the abstract only, to make the LOIH more specific.
The percentage of insufficient records in our secondary study was 61%. The specificity of 70% of the insufficient records was increased using information from the full-text content of the PMC articles, primarily tables.
Inter-rater Agreement
The IRR for the sufficiency annotation of 2017 GenBank records from the influenza case study was found to be 0.984 on average, using Cohen’s kappa statistics as a measure of agreement. Table 1 presents the IRR between each pair of annotators for this task.
Table 1:
Inter-rater agreement for sufficiency annotation based on kappa statistic
| Measurements | A;B | A;C | B;C |
|---|---|---|---|
| Sufficient;Sufficient | 1329 | 1312 | 1316 |
| Insufficient;Insufficient | 683 | 684 | 683 |
| Sufficient;Insufficient | 0 | 17 | 18 |
| Insufficient;Sufficient | 5 | 4 | 0 |
| Kappa value | 0.994 | 0.977 | 0.980 |
A, B, and C represent the three annotators, respectively.
For the final location annotation of 1477 GenBank records from the influenza case study, the IRR was found to be 0.755 on average, using f-score for exact match per record-location linkage as a measure of agreement between each pair of annotators (the individual f-scores were 0.677, 0.699, and 0.888, respectively).
Performance Statistics of the IE System
For determination of the sufficiency of records, our system had a Cohen’s kappa value of 0.988 when compared to the gold standard annotation.
Of the 5728 GenBank records used for the influenza case study, our system was able to correctly link 5011 records to the correct final location. For two of the records, neither our system nor our annotators were able to find any geospatial information about their LOIH. For the remaining 715 records, the final location extracted by our system did not exactly match the final location identified by our annotators. However, for 604 of these records, our system output for final location included true positive matches in addition to FPs. For instance, for record GQ463225, our system output for final location was “Guangdong China; Fujian China” while the annotated final location was “Guangdong, China.”
The precision, recall, and f-score of individual tasks performed by the system based on the evaluation criteria described in The Materials and Methods section for the influenza case study is given in Table 2.
Table 2:
Recall, precision, and f-score for individual tasks performed by the system
| Task | Recall | Precision | f-score |
|---|---|---|---|
| Extraction of the location of infected host from GenBank record metadata | 0.996 | 0.953 | 0.974 |
| Linkage of consistent, text-derived locations of infected host (locations extracted from textual content of related article and consistent with record metadata) to GenBank records | 0.800 | 0.847 | 0.823 |
| Linkage of consistent, table-derived locations of infected host (locations extracted from tabular content of related article and consistent with record metadata) to GenBank records | 0.838 | 1.0 | 0.912 |
| Linkage of final locations of infected host (locations produced after integrating geospatial data from record, text, and tables) to GenBank records | 0.980 | 0.876 | 0.925 |
| Mapping of correctly identified final locations of infected host to their correct latitude and longitude coordinates (disambiguation) | 0.984 | 0.948 | 0.965 |
| Linkage and disambiguation of final locations | 0.967 | 0.832 | 0.894 |
The accuracy of our system for linking the 120 records used in our secondary study to their correct final location was found to be 75% (90 records had correct final location).
DISCUSSION
The results indicate that our system is capable of linking GenBank records to the correct location of sequence collection with a high level of recall and precision. However, since our system evaluation was primarily performed on GenBank records related to the influenza A virus, the results may not be a true reflection of its performance level for GenBank records related to other pathogens. Using the remaining 120 records allowed us to obtain a rough estimate of its accuracy for other viruses but the generalizability of this secondary study is limited by its small sample size. Moreover, at its current state our system is only capable of analyzing records related to PMC articles and therefore, we were limited in our selection of the GenBank records for this study.
Our IRR for sufficiency annotation was very high and matched by the performance of our automated module for this task. However, for final location, we had a relatively low IRR due to misinterpretation of annotation guidelines and missed locations in linked articles, illustrating the difficulty of this task. For instance, one of our annotators listed “New York City, New York, USA” as the final location while the second annotator simply listed “New York, USA” due to missed information in the article. Through repeated annotations, we minimized these mistakes in our gold standard.
For the majority of records whose specificity was increased using information from a related article, it was necessary to retrieve data from the full-text content of the article, including tables. This supports our decision to parse full-text PMC articles rather than PubMed abstracts for this study.
Although we presented evaluation results for the various tasks performed by our system for the influenza case study, we will only present a detailed discussion of its performance in the extraction and disambiguation of final locations, which is the principal objective of the system. Upon conducting a thorough error analysis for the task of final location extraction, we found that 613 of the 715 records with incorrect final locations were caused by the system’s failure to correctly identify the parent ADM1 code of the locations. A total of 605 of these errors were a direct result of the ambiguity of the location “Philadelphia, USA” in GeoNames. Philadelphia was one of the several locations mentioned in a paper linked to over 600 records and our system produced locations such as “Philadelphia, Virginia, USA” and “Philadelphia, New York, USA,” respectively, for records containing “Virginia” or “New York” in the GenBank metadata fields. Since we are evaluating the system on a per record-location linkage basis, a single paper associated with a large number of records can have a substantial effect on the system performance. However, by collecting a stratified random sample of papers from the list of all PMC articles related to GenBank records for the influenza A virus, based on the number of records linked to them, we attempted to prevent this from skewing the results significantly (see Appendix E for table showing the distribution of GenBank records and final location errors across the articles)
For 72 records (linked to 8 papers), our system failed to correctly identify the final location due to errors produced by the text linker. Forty-one of these errors (representing 4 papers) were due to the text linker missing relevant relations in the related paper since they did not fit any of the utilized patterns while the remaining resulted from its lack of precision. Although the system’s failure to correctly identify the parent ADM1 code of the locations accounted for a significantly greater number of errors in our current evaluation set, the limitations of the text linker, by leading to errors in a larger number of articles, has the potential to be a greater problem in the future depending on the number of records linked to the affected articles.
Spelling errors in the GenBank metadata (e.g., “Jilangsu” recorded in JN804364 instead of “Jiangsu”) and missing location names in GeoNames (e.g., “Pasteur Institute, France”) accounted for incorrect final locations in 17 records, none of which had more specific information in the paper. The remaining errors were primarily caused by locations missed by the table linker due to the presence of split cells in the relevant table of a single paper and the failure of the disambiguation module to select the correct candidate for a single ambiguous toponym mention (e.g., “Cambridge”).
The coordinates chosen by our disambiguation module were incorrect for a total of seven correctly identified final locations (corresponding to 90 GenBank records) because of the rule-based nature of our program. For five of these locations, the population recorded in GeoNames was 0 and hence our population heuristic had no effect.
The errors in our secondary study primarily resulted from two papers. One of the papers contained more specific information within a table but our program was unable to parse this table since an HTML version of the paper was not available. The second paper utilized two-letter codes to describe the city and state in Brazil from which the virus sample was isolated and our annotator was able to use this data to infer the LOIH of the related sequences. For example, based on the isolate field “CA_SP_P1/0” for record EU170195, our annotator was able to deduce that the LOIH for the sequence was Cássia dos Coqueiros, Sao Paolo, Brazil. Our program was unable to make such inferences. The majority of remaining errors were a result of missing locations in GeoNames.
CONCLUSION
Our system is capable of linking genetic sequences in GenBank records to the coordinates of their LOIH, using data from the record itself or related PMC articles, with reasonably high accuracy. However, as our error analysis showed, even a single error type can lead to a significant reduction in system performance if a large number of records happen to be affected by this error type. Therefore, as future work, we will attempt to address the different limitations of our system by incorporating additional databases for geographic data such as the Wikipedia dictionary, adding a spell check component to the record location extractor module, and modifying the table linker so that it is capable of parsing more complex tables. In addition, since the rule-based nature of the text linker was a major cause of errors produced by the system, we will test the use of machine learning approaches in this module. Finally, to determine the extensibility of the system, we will evaluate it on other corpora including different species of viruses and other pathogens.
CONTRIBUTORS
This study was performed under the supervision of G.G., M.S., and G.W. and they all provided significant contributions toward its design and implementation. In addition, G.G. and M.S. helped considerably with drafting and reviewing the content of the manuscript. T.T. helped with system design and development, evaluated the system based on gold standard data and drafted and revised the manuscript. D.W. helped with system design and development and made significant edits to the manuscript. R.R. performed the stratified random sampling of PMC articles, calculated IRR and made significant edits to the manuscript. R.R., R.B., and M.F. devised the annotation guidelines and annotated the GenBank records. All authors helped review the manuscript.
FUNDING
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R56AI102559 to G.G. and M.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
COMPETING INTERESTS
The authors have no competing interests to declare.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Whirl-Carrillo M, McDonagh EM., Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. [DOI] [PubMed] [Google Scholar]
- 3.Magee D, Beard R, Suchard MA, Lemey P, Scotch M. Combining phylogeography and spatial epidemiology to uncover predictors of H5N1 influenza A virus diffusion. Arch Virol. 2015;160:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray RR, Salemi M. Integrative molecular phylogeography in the context of infectious diseases on the human-animal interface. Parasitology. 2012;139:1939–1951. [DOI] [PubMed] [Google Scholar]
- 5.Hovmöller R, Alexandrov B, Hardman J, et al. Tracking the geographical spread of avian influenza (H5N1) with multiple phylogenetic trees. Cladistics. 2010;26:1–13. [DOI] [PubMed] [Google Scholar]
- 6.Janies DA, Pomeroy LW, Krueger C, et al. Phylogenetic visualization of the spread of H7 influenza A viruses. Cladistics. 2015;31:679–691. [DOI] [PubMed] [Google Scholar]
- 7.Chan J, Holmes A, Rabadan R. Network analysis of global influenza spread. PLoS Comput Biol. 2010;6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott P, Wartenberg D. Spatial epidemiology: current approaches and future challenges. Environ Health Perspect. 2004;112:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghwani J, Rambaut A, Holmes EC, et al. Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmission. PLoS Pathog. 2011;7:e1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faria NR, Suchard MA, Rambaut A, et al. Toward a quantitative understanding of viral phylogeography. Curr Opin Virol. 2011;1:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janies D, Hill AW, Guralnick R, et al. Genomic analysis and geographic visualization of the spread of avian influenza (H5N1). Syst Biol. 2007;56:321–329. [DOI] [PubMed] [Google Scholar]
- 13.GeoNames. http://www.geonames.org/. Accessed September 5, 2013.
- 14.Bossy R, Golik W, Ratkovic Z, et al. BioNLP shared Task 2013–An Overview of the Bacteria Biotope Task. In: Proceedings of the BioNLP Shared Task Workshop, ACL. Sofia, Bulgaria: Omnipress, Inc; 2013:161–169. [Google Scholar]
- 15.Tamames J, de Lorenzo V. EnvMine: a text-mining system for the automatic extraction of contextual information. BMC Bioinformatics. 2010;11:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkar IN. Leveraging biomedical ontologies and annotation services to organize microbiome data from Mammalian hosts. AMIA Annu Symp Proc. 2010;2010:717–721. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ES, Sarkar IN. Towards structuring unstructured genbank metadata for enhancing comparative biological studies. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci. 2011;2011:6–10. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ES, Sarkar IN. MeSHing molecular sequences and clinical trials: a feasibility study. J Biomed Inform. 2010;43:442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller H, Norton CN, Sarkar IN. GenBank and PubMed: how connected are they? BMC Res Notes. 2009;2:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selama O, James P, Nateche F, et al. The world bacterial biogeography and biodiversity through databases: a case study of NCBI Nucleotide Database and GBIF Database. Biomed Res Int. 2013;2013:240175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tahsin T, Beard R, Rivera R, et al. Natural language processing methods for enhancing geographic metadata for phylogeography of zoonotic viruses. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci. 2014;2014:102–111. [PMC free article] [PubMed] [Google Scholar]
- 22.Xpdf: Download. http://www.foolabs.com/xpdf/download.html. Accessed March 4, 2014.
- 23.Sayers E. E-utilities Quick Start. 2013. http://www.ncbi.nlm.nih.gov/books/NBK25500/. Accessed May 13, 2015. [Google Scholar]
- 24.Lieberman MD, Samet H, Sankaranarayanan J. Geotagging with local lexicons to build indexes for textually-specified spatial data. In 2010 IEEE 26th International Conference on Data Engineering (ICDE 2010). Long Beach, CA, USA: IEEE, 2010: 201–212. [Google Scholar]
- 25.Ladra S, Luaces MR, Pedreira O, et al. A Toponym Resolution Service Following the OGC WPS Standard. In: Proceedings of the 8th International Symposium on Web and Wireless Geographical Information Systems. Shanghai, China: Springer-Verlag: 2008; 75–85. [Google Scholar]
- 26.Country List ISO 3166 Codes Latitude Longitude | Socrata. https://opendata.socrata.com/dataset/Country-List-ISO-3166-Codes-Latitude-Longitude/mnkm-8ram. Accessed June 18, 2014.
- 27.Scotch M, Sarkar IN, Mei C, et al. Enhancing phylogeography by improving geographical information from GenBank. J Biomed Inform. 2011;44 (Suppl 1):S44–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang AX, Manning C. SUTime: A library for recognizing and normalizing time expressions. In: Proceedings of the Eight International Conference on Language Resources and Evaluation (LREC'12). Istanbul, Turkey: European Language Resources Association (ELRA) 2012. [Google Scholar]
- 29.Weissenbacher D, Tahsin T, Beard R, et al. Knowledge-driven geospatial location resolution for phylogeographic models of virus migration. Bioinformatics. 2015;31:i348–i356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham H, Maynard D, Bontcheva K, et al. GATE: a framework and graphical development environment for robust NLP tools and applications. In: Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics. Philadelphia, PA, USA: Association for Computational Linguistics; 2002:168–175. [Google Scholar]
- 31.Tsuruoka Y, Tsujii J. Bidirectional inference with the easiest-first strategy for tagging sequence data. In: Proceedings of the Conference on Human Language Technology and Empirical Methods in Natural Language Processing - HLT ’05. Morristown, NJ, USA: Association for Computational Linguistics; 2005. 467–74. doi:10.3115/1220575.1220634. [Google Scholar]
- 32.Leidner JL. Toponym Resolution in Text: Annotation, Evaluation and Applications of Spatial Grounding. SIGIR Forum 2007;41:124–126. [Google Scholar]
- 33.Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005;12:296–298. [DOI] [PMC free article] [PubMed] [Google Scholar]