Abstract
Objective Reinventing data extraction from electronic health records (EHRs) to meet new analytical needs is slow and expensive. However, each new data research network that wishes to support its own analytics tends to develop its own data model. Joining these different networks without new data extraction, transform, and load (ETL) processes can reduce the time and expense needed to participate. The Informatics for Integrating Biology and the Bedside (i2b2) project supports data network interoperability through an ontology-driven approach. We use i2b2 as a hub, to rapidly reconfigure data to meet new analytical requirements without new ETL programming.
Materials and Methods Our 12-site National Patient-Centered Clinical Research Network (PCORnet) Clinical Data Research Network (CDRN) uses i2b2 to query data. We developed a process to generate a PCORnet Common Data Model (CDM) physical database directly from existing i2b2 systems, thereby supporting PCORnet analytic queries without new ETL programming. This involved: a formalized process for representing i2b2 information models (the specification of data types and formats); an information model that represents CDM Version 1.0; and a program that generates CDM tables, driven by this information model. This approach is generalizable to any logical information model.
Results Eight PCORnet CDRN sites have implemented this approach and generated a CDM database without a new ETL process from the EHR. This enables federated querying within the CDRN and compatibility with the national PCORnet Distributed Research Network.
Discussion We have established a way to adapt i2b2 to new information models without requiring changes to the underlying data. Eight Scalable Collaborative Infrastructure for a Learning Health System sites vetted this methodology, resulting in a network that, at present, supports research on 10 million patients’ data.
Conclusion New analytical requirements can be quickly and cost-effectively supported by i2b2 without creating new data extraction processes from the EHR.
Keywords: medical informatics, data integration, data models, ontology-driven data representation, patient centered outcomes research institute, PCORnet CDM, informatics for integrating biology and the bedside
BACKGROUND AND SIGNIFICANCE
The need for interoperable medical data and analytical tools to support clinical research has led to a variety of solutions. The Cancer Biomedical Informatics Grid (caBIG),1 the European Translational Information & Knowledge Management Services (eTRIKS) Innovative Medicines Initiative (IMI),2 the Shared Health Research Information Network (SHRINE),3 the Observational Medical Outcomes Partnership (OMOP),4 and the National Patient-Centered Clinical Research Network (PCORnet)5 are just a few of the initiatives designed to optimize enormous amounts of unstandardized data for clinical research. Each solution requires a participating institution to convert its data to target the syntactic and semantic standards for that solution, adding to the workload of already overburdened informatics teams at medical centers.
Data conversion is a complex, time intensive, and expensive endeavor, for several reasons:
Data must match the analytic standards of different networks, which requires a detailed analysis of the available transactional data.
New programs must be developed to extract, transform, and load (ETL) data at each network site.
Comprehensive tests need to be developed and executed to ensure data quality after potentially integrity-compromising data transformations.
Healthcare data governance is entangled with data conversion processes, often requiring multiple levels of regulatory approval before ETL development can begin.
Data must be collected in hospital use transactional data models for providing patient care and billing, which are significantly different from the analytical data models required for medical research and population analytics.
Data for individual patients from multiple systems frequently must be integrated with diverse access and security protocols and software platforms.
It would therefore be desirable to find a way to copy transactional data to new analytical models without having to duplicate ETL efforts for each new network and analysis.
Many data standards define both a distinct information model and a data model. An information model specifies the entirety of the data expected to be collected and presented to a network. Furthermore, information models define the coding systems (eg, the International Classification of Diseases 9 [ICD-9] diagnoses) used to express concepts from the data; modifiers (eg, a medication dose) used to express details about the concepts; and values (eg, numeric lab values) used to express the quantification of the concepts. Despite the extreme level of detail that may be expressed in the information model, it does not specify a physical way to represent data. In contrast, a data model is a concrete set of tables or data structures, which are often required by a data network to run specific computer programs across the network. Although both types of model are often defined in the same breath, they are distinct.
Informatics for Integrating Biology and the Bedside (i2b2) is a clinical data warehouse platform that uses a simple data model that is similar to the Entity-Attribute-Value (EAV) approach.6 Atomic “facts” in the data are placed in a narrow table, with each row representing a single observation about a patient. Ontology tables translate concepts, such as “diagnosis of diabetes mellitus” to local database codes. This makes i2b2’s model highly adaptable to new concepts and isolates i2b2 from site-specific coding – a feature that makes i2b2 easy to implement at a new site.
The i2b2 system’s adaptability also means that its data can be rearranged to make new tables for new analytical models, as long as the information exists in the i2b2 database. The key to this rearrangement is a method for linking the concepts in i2b2 to columns in the target analytical model’s data structure. By creating a new ontology that represents the target analytical data model, we can support the model’s logical structure directly in the i2b2 system. In this sense, each new ontology is an information model that represents the same i2b2 data.
We hypothesize that such information models could drive both i2b2 queries and transformations into other data models. This would represent a significant reduction of the effort required to support new data models, because a single ETL process of data from clinical systems into i2b2 could support multiple additional data models. A custom ETL code for these additional models would be replaced by a common process for mapping data to information models, which separates programming expertise from medical terminology expertise.
Scalable Collaborative Infrastructure for a Learning Health System
The Scalable Collaborative Infrastructure for a Learning Health System (SCILHS), pronounced “skills,” is a Clinical Data Research Network that participates in the Patient-Centered Outcomes Research Institute (PCORI) Network, PCORnet.4 SCILHS is comprised of 12 health centers across the United States that cover over 10 million patients.7 Each site uses i2b2 to store and analyze patient data for clinical research.
PCORnet-affiliated networks are required to adopt the Common Data Model (CDM) for at least 1 million patients.5 The PCORnet Distributed Research Network (DRN) will use these CDM databases to support nationwide distributed analytics on retrospective data, to quickly generate and refine hypotheses and find potential cohorts for clinical research. The CDM is based on the Mini-Sentinel project,8 but it is being significantly expanded by the PCORnet Coordinating Center, with some guidance from the PCORnet networks and the Office of the National Coordinator for Health Information Technology.
SCILHS has adopted the PCORnet-CDM as the foundation for interoperable data exchange in our network.
OBJECTIVE
SCILHS uses i2b2 to store patient data for clinical research, and PCORnet uses the PCORnet-CDM for inter-network analytics of that data. Therefore, a process that creates a PCORnet-CDM data model without a new ETL process represents a significant cost savings and demonstrates that new analytical models can be generated from i2b2 on demand. Such an approach can support the needs of a traditional i2b2 data network while simultaneously meeting the new analytical needs of PCORnet.
We formalized an approach to implement new information models (ontologies) in i2b2 without the need to develop new methods of ETL processing or even to change the underlying i2b2 data. We implemented this approach by developing an information model for PCORnet-CDM Version 1.0. Then, as a large-scale validation of our approach, we engaged eight SCILHS sites to “map” their data repositories to this i2b2-PCORnet-CDM Information Model. These sites implemented the i2b2 networking system (SHRINE)3 to perform intranetwork queries using the SCILHS Information Model. We then developed and validated a tool to transform the conformant i2b2 instances into the schema structure that PCORnet requires for cross-network queries.
METHODS
Our approach to data interchange between i2b2 and the PCORnet-CDM involves the design and development of an i2b2 logical information model and a process for mapping local data to this information model and managing its implementation. We further describe how the mapped data can be utilized to join an i2b2 network or to generate a CDM database.
Logical Information Model for PCORnet-CDM
First, we developed a logical model to represent PCORnet-CDM Version 1.0 in i2b2. An i2b2 ontology (ie, a particular “view” of the data in i2b2) can represent a logical model. Therefore, the ontology system can be used to develop a “mapping” from the data in i2b2 to a specific information model. i2b2 ontologies define their hierarchy by assigning each element a “path name,” which is distinct from its code. Developing the PCORnet ontology, which represents the PCORnet Information Model in i2b2, involved creating path names for each possible element in the PCORnet-CDM. We also included a “PCORnet code” column in the ontology, which contains the standardized PCORnet code and is used for materializing the data model.
Local sites can then take this standard PCORnet Information Model and modify the codes to match their local data, while keeping the paths consistent. Then, the information model-specific path names (eg, “\\PCORNET\Diagnosis\09…\250”) are matched to site-specific codes (eg, “ODA:DIABM”). Doing so creates a functional mapping of a site’s local codes to a new ontology, without changing the underlying local codes.9 This mapped information model is the underlying ontology used in i2b2 queries, and it serves as the basis for the materialization of the model into a physical schema.
A draft ontology was generated from code written by various collaborators to automatically create ontologies from the PCORnet-CDM specification.10 We edited this draft ontology extensively, based on site implementation experience, and we added terminology trees to each domain, as follows:
Demographics: We added a granular age tree, including pediatric ages, to support age queries.
Diagnoses: We added ICD-10 2014AA and ICD-9 2014AA codes, generated using existing i2b2 tools to extract ontologies from BioPortal.11
Procedures: We added ICD-9 2014AA codes from BioPortal, a Centers for Medicare & Medicaid Services Diagnosis-Related Group (CMS-DRG) tree provided by Partners Healthcare, a Medicare Severity-Diagnosis-Related Group (MS-DRG) tree provided by Beth Israel Deaconness Medical Center, and the Unified Medical Language System’s (UMLS) Healthcare Common Procedure Coding System (HCPCS) tree that a collaborator converted to i2b2 format.
Encounters: We added a tree of 3-digit zip codes to support querying the CDM’s “location code” through the ontology structure (rather than through manual entry).
Enrollment: We edited this table, with the intention of allowing sites to specify which patients have “complete longitudinal data” for selected date ranges, to automatically return all patients with clinical encounters in the specified date range.
Vitals: We added normal ranges to this simple vitals implementation, including height, weight, and blood pressure.
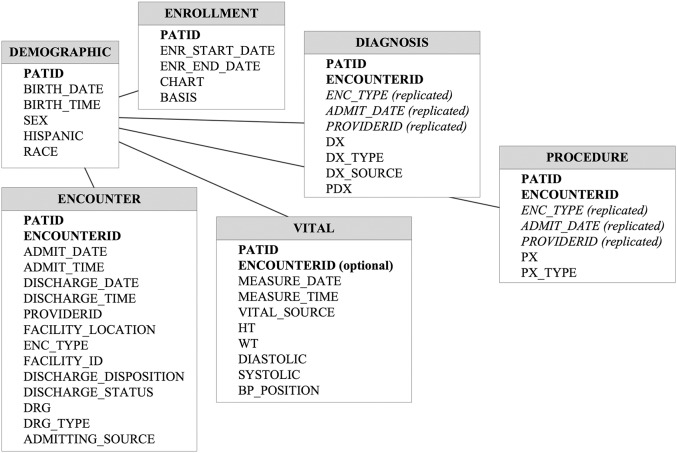
In production, the PCORnet-CDM is presently Version 1.0, which includes the six domains above. The database schema is shown in Figure 1. A new version of the CDM has been developed and will go into production at the end of 2015. This version will add specifications for medications, labs, death data, and patient-reported outcomes. Our PCORnet i2b2 ontology, currently Version 1.5.2, is available for download from the i2b2 community wiki.12 A new version that reflects the revised CDM is still under development.
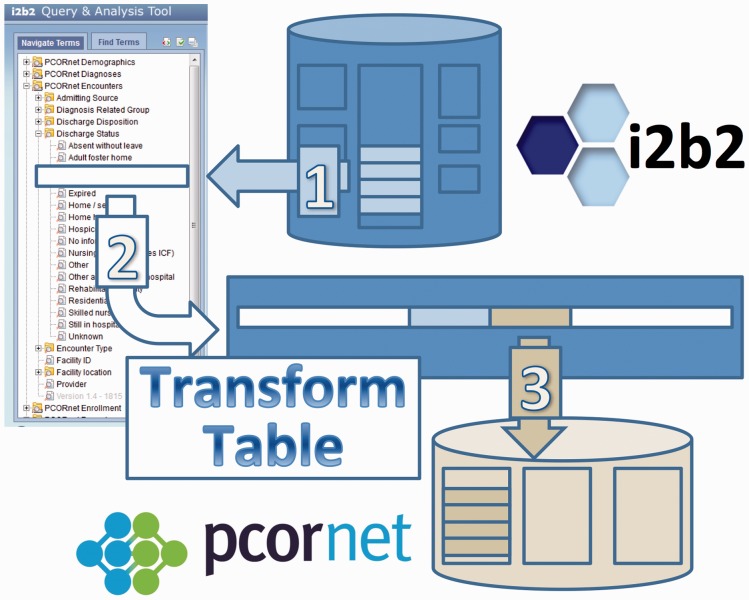
Figure 1:
The process of populating a new information model schema through look-ups mediated by the i2b2 ontology.
Extended Data
Our approach needed further specification for data in i2b2 that are more complex than simple entity-concept facts (eg, a diagnosis). In particular, patient facts might have associated sub-facts, such as numeric/text values or other, non-standard, attributes. For example:
Many laboratory tests and vital signs use continuous numeric values or text values to represent different results. For example, in PCORnet-CDM Version 1.0, blood pressure may contain the element “systolic,” which can contain an integer value.
Medications may have dosage, form, route, and other detailed data.
For the first case, there are standard columns in the i2b2 fact table that accommodate these values. i2b2’s ontology table contains metadata on these values, such as unit conversions. For the second case, which could be arbitrarily complex, i2b2 supports “linked facts” called “modifiers” to extend any fact. Therefore, some data in the PCORnet tables are populated through these i2b2 modifiers, which have path names that are mapped to the PCORnet-CDM in the same way we mapped simple facts to the CDM. For example, medication frequency is represented by a PCORI value set in the ontology (eg, RX_FREQUENCY: 01, meaning “every day”), which sites can modify to match their local codes.
We tested the ontology by developing test data and running a comprehensive set of test queries. This allowed us to test the model’s validity and its management of edge cases. For example, we tested edge cases in which: 1) CDM data are missing from i2b2, to make sure the correct CDM “missing value” is returned (rather than a query error); and 2) a large number of terms are mapped to a single element in the SCILHS ontology, to ensure that database errors do not occur.
Information Model Implementation Process
We developed the following process to implement a new i2b2 information model in a network of sites:
Consensus Building: For all the i2b2 sites on the new network, inventory the existing ontologies/terminologies needed to express the new information model. This allows a consensus of the various ways that an information model can be expressed in i2b2 to be developed.
Ontology Development: Develop a standard metadata ontology and default map for i2b2 sites, to expose their data in the new information model. This may be modified as the new network evolves and new versions are designed.
Validity Testing: Test the metadata ontology using both manufactured patient data and real patient data. This allows the validity, performance, and edge cases of the mapping to be tested.
Distribution to Network Sites: Publish the mapped ontology/terminology so that it becomes part of the standard ontologies of the i2b2 sites.
Local Mapping: For the new information model’s ontologies to function, the local codes need to be integrated into the mappings as, described in the Methods section, above.
Implementation Testing: The standard tests conducted for the validity testing are rerun at each site, to ensure quality control at all the sites, and are analyzed for the expected results.
From Information Model to SHRINE and Data Model
Once implementation and mapping is completed, two important use-cases are enabled. Within i2b2, sites can join a SHRINE network using the common information model at almost no cost. This enables on-the-fly querying of data within the network. Second, sites can “materialize” a data model from an i2b2 information model to support analytical programs outside of i2b2.
The high-level process to “materialize” a data model from an i2b2 information model is shown in Figure 2. The ontology path name is used to find existing data elements that will be translated into a materialized data model. These data are discovered in the i2b2 database by path name (Step 1) and matched to replacement codes in a transform table, again using the path name (Step 2). The data with replaced codes are then stored in a new database corresponding to the materialized information model. Continuing our previous example, the paths beginning with “\\PCORNET\Diagnosis\” would be searched for diagnosis data, the code “ODA:DIABM” would be found, this code would be matched to the path name “\\PCORNET\Diagnosis\09…\250,” and, in turn, the transform table would match this path name to a standardized ICD-9 code (“250.00”), to be stored in the output database.
Figure 2:
The PCORnet-CDM Version 1.0 data schema.
RESULTS
Eight of our SCILHS sites have implemented and mapped the SCILHS Information Model, joined the SCILHS SHRINE, and created PCORnet-CDM databases for DRN queries.
Mapping the Logical Information Model to Local Data
Our network utilized the six-step “Information Model Implementation Process,” described above, to develop and implement this information model at eight of our SCILHS sites over the past year. Each site linked each i2b2 fact to its appropriate path name in the PCORnet-CDM table.
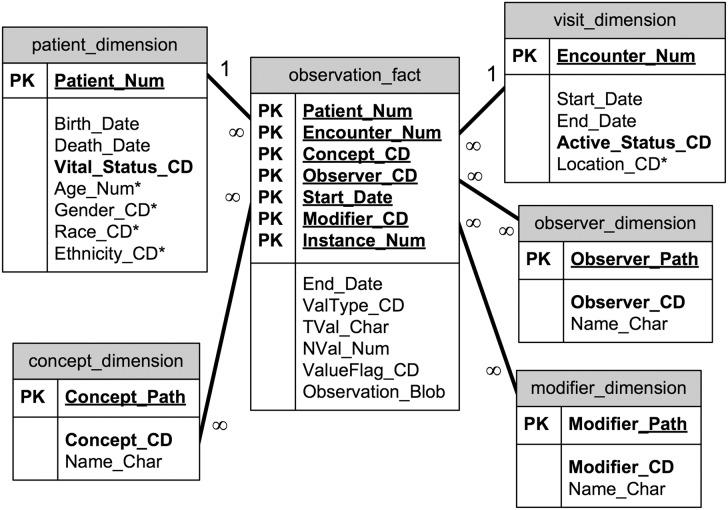
We identified two patterns during the process of developing our mapping process, dictated by where the i2b2 data elements reside. In i2b2’s augmented EAV-style schema (a star-schema), data can be stored in the observation-fact table or in a dimension table. The observation-fact table is the EAV table, in which “sparse” data usually resides. i2b2 also provides patient and visit dimension tables, which are traditional, column-oriented tables and are often more easily populated with data from existing electronic medical record or claims-type databases. The patient and visit tables often reflect the format of the source system tables. Unlike the i2b2 observation-fact table, the patient table does not require dates, making immutable items like birth or death dates easy to represent. The i2b2 schema is shown in Figure 3.
Figure 3:
The i2b2 star schema.
The two mapping patterns are as follows (and examples are shown in Table 1):
• When data are in the patient or visit dimension tables.
For these tables, a traditional entity-relational mapping, mediated by the i2b2 ontology, is possible. Each path points to a column and a set of values in the dimension tables. The transformation process copies and translates the data into the new schema. This can be seen in the first row of Table 1 – the “code” column for the patient ethnicity, “Hispanic,” is changed into a list of local codes representing that concept. The ontology release provides spreadsheets to assist local IT staff with mapping the local codes correctly.
• When data are in the observation-fact table.
- This approach has two sub-patterns.
- When there is a 1:1 mapping between local codes and the PCORnet Information Model.When the raw i2b2 data use the same terminology as the PCORnet-CDM (eg, ICD-9), the ontology entry’s referenced code is simply renamed to match the particular coding mechanism used at the site. For example, in Table 1, the value in the code column “ICD9:250.1” is replaced with “PHSICD9:2501.”
- When there is an n:1 mapping between local codes and the PCORnet Information Model.In this case, new rows are added to the ontology table, reflecting local codes that are children of the standard element. This is shown in the final three rows of Table 1, in which two new children are created under the “Hispanic” path name. The i2b2 mapping program can generate mappings in this format. Then, because of the dimensional-query design of i2b2, these child nodes are automatically retrieved by queries for the parent.
Table 1:
The Three Patterns for Mapping an Information Model (Ontology) Table to Local Codes, for the PCORnet Ontology
| Pattern | Name | Path | Code | Description |
|---|---|---|---|---|
| Dimension table | White | \PCORI\DEMOGRAPHIC\RACE\05\ | Default: “05” | To map data in the patient and encounter dimensions, augment the value list in the dimension code column with local values. |
| Local mapping: “his/white,” “white,” “mid.eastern” | ||||
| Observation-fact table (1:1 mapping) | Diabetes mellitus | \PCORI\DIAGNOSIS\09\001-999.99\240-279.99\249-259.99\250.10\ | Default: ICD9:250.10 | If local codes are different but have a 1:1 mapping with the PCORnet-CDM, change the base code column to match the local code. |
| Local mapping: PHSICD9:25010 | ||||
| Observation-fact table (n:1 mapping) | Hispanic | \PCORI\DEMOGRAPHIC\HISPANIC\Y\ | ETHNICITY: HISPANIC | If a PCORnet code maps to several local codes, create these as children underneath that PCORnet code. Queries will automatically gather all the local codes. |
| \PCORI\DEMOGRAPHIC\HISPANIC\Y\Hispanic\ | ETH:HISP | |||
| \PCORI\DEMOGRAPHIC\HISPANIC\Y\Latino\ | ETH:LAT |
The “Default” codes are those distributed with the ontology. The “Local mapping” codes are examples of changes made to the default ontology by a site, to reflect their own codes. These local mappings are italicized.
SHRINE and the Physical PCORnet-CDM
This mapping supports both i2b2 SHRINE queries and the generation of PCORnet DRN databases.
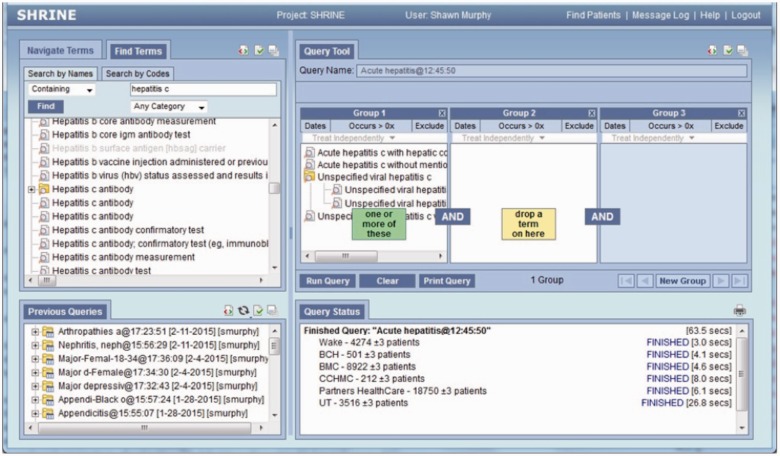
The eight sites that have implemented the SCILHS Information Model have joined the SCILHS Hub, which allows users to perform on-the-fly federated querying across the network from the SHRINE software. An example SHRINE query, for patients with hepatitis C, is shown in Figure 4.
Figure 4:
An example query for hepatitis C patients, performed using the SCILHS CDM.
This mapping was also used to create the PCORnet database tables required by the PCORnet DRN. SCILHS supplied a transform table that connects path names to standardized data elements for the CDM data model. SCILHS further developed a Structured Query Language (SQL) script to execute the transformation process outlined in Figure 2, driven by the ontology and transform table. The eight SCILHS sites used this script to transform a portion of their data into the CDM table structure. Figure 5 illustrates the specific process of using these mappings to transform the data into the PCORnet model. The steps of the process are as follows:
For each table in the PCORnet data model, the appropriate set of ontology paths are searched to find data in the fact table. Matching facts in the database are identified by their local codes. In the example shown in Figure 5, the ontology table is searched for codes that match the path “\PCORI\DIAGNOSIS\09…\250.10\,” and “ODA:DIADM” and “PHSICD9:25010” are found. Facts with those codes are extracted from the database.
The facts are translated into standard codes through a transform table, which defines standardized CDM codes for each pathname. In the example shown in Figure 5, the transform table is searched for the same path used to extract the codes. The transform table specifies that, for this path, the code “250.10” is to be inserted into the DX column in the Diagnosis table, and the code “09” is to be inserted into DX_TYPE.
The standardized data are copied into the CDM table structure. In the example shown in Figure 5, the correct row is inserted into the Diagnosis table.
Figure 5:
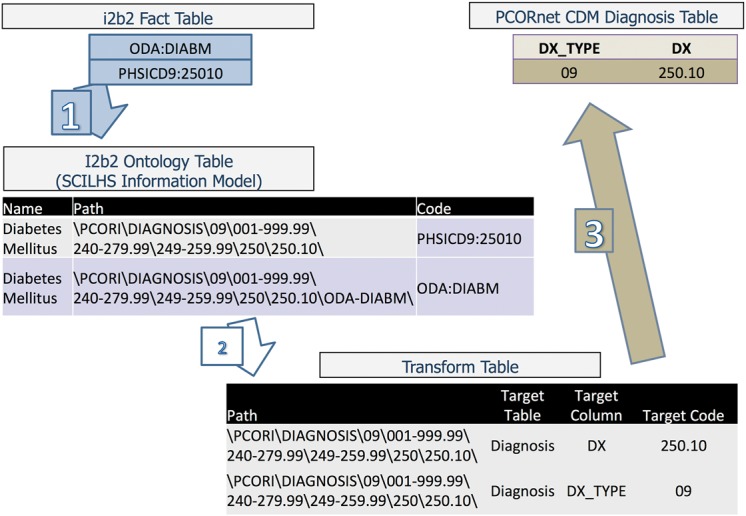
The transformation process for PCORnet, from i2b2 into the CDM.
The eight participating SCILHS sites executed this SQL script on their mapped i2b2 warehouse and were able to automatically generate a PCORnet-CDM database. The PCORnet Coordinating Center sends DRN queries as SQL programs that execute against these databases. Seven sites have successfully responded to at least one DRN query using the generated database. (The eighth site is not presently participating in the PCORnet DRN.)
DISCUSSION
Different analytical programs may require different data formats in order to run. Although enabling the interchange of data between diverse normalized data models is important, few networks are designed with this in mind. The general approach to exchanging data between different networks is to invent new data schemas and software (or adapt existing ones) for every network. Partially, this method is justified, because different networks have different purposes and, therefore, need to conform to disparate use-cases. However, many networks have been oriented around the need to bring data together, and, for that reason, it becomes more difficult to continue to justify a developing completely new approach for each new data model.
Herein, we hypothesize that uniting standardized information models using the flexible i2b2 approach could positively impact the interoperability of disparate networks and data interchange between networks by decoupling the information model from the data model. We have established a process and methodology to adapt i2b2 to new information models and to utilize those models to transform i2b2 data into the various schemas required by various network and analytical processes. The advantage of this method is its ability to scale the participation of multiple i2b2 sites into a new network. Inherent to this method is a specific understanding of how i2b2 elements at each site relate to the new information model. Each site’s ETL process remains unchanged, and semantic meaning is disambiguated by the well-defined mappings.
Essentially, after a single ETL process and mapping effort, both an i2b2 SHRINE node and CDM database can be simultaneously created at each site for almost no additional cost. Because clinical systems need to be accessed only once to support all three processes, the complex technical and regulatory challenges inherent to ETL processes (see the Background and Significance section) are minimized. Additionally, supporting new information models for other use cases will be an incremental process, because large portions of the mappings can be reused. For example, the Accrual to Clinical Trials Network is implementing an information model that builds off of (and is compatible with) the CDM Information Model. For these reasons, we expect that our approach will be easier to maintain than traditional ETL tools for sites that seek to support multiple data models.
Eight SCILHS sites have vetted this methodology and implemented it using our PCORnet-CDM i2b2 ontology, resulting in a SCILHS network that, at present, can perform research on 10 million patients’ data. The SCILHS Hub is actively being used to perform cohort-finding queries within the network using the CDM. The SCILHS sites have established a mutual trust relationship, which enables our investigators to perform ad hoc exploratory prep-to-research queries without individual Institutional Review Board approval. This enables rapid identification of patient cohorts with specific conditions, to determine research feasibility using SCILHS. Additionally, seven sites have successfully responded to a DRN query using the database generated by the PCORnet-CDM data model transformation. For SCILHS, we plan to continue this approach at the remaining four sites in our network and to expand the ontology to support the next release of CDM. Additionally, several other i2b2-based PCORnet networks are adopting this approach using compatible information models.
The disadvantage of this method is the complexity of the mappings. The mapping tools were developed in an attempt to help reduce this complexity. Furthermore, before any ETL process, quality assurance reports can be run on the mappings, to make sure that ETL code will run on top of a sound base. Notably, this interoperability methodology does nothing to ensure data quality. Raw electronic health record data is rife with missing data, duplicate patient identifiers, and patients with such sparse data that effective research on them is impossible. Work is underway to combine the data transformation process with a data quality process, to ensure that patients included in the data transform also have sufficient longitudinal data for effective research and to identify data gaps.
Although the i2b2 data model has the ability to represent arbitrarily complex attributes in the data as modifiers, many SCILHS sites did not have all of the data attributes specified by the PCORnet-CDM in pre-existing i2b2 databases. This problem will exist when a previous information model does not match the new information model, essentially representing different choices in data granularity made in different national initiatives. Initially, SCILHS’s data was missing many of these modifiers, including blood pressure position and medication frequency. New ETL programming will be needed to obtain these values. Often, only a small “tweak” is needed, rather than developing ETL programming from scratch. Nonetheless, this is a potential source of difficulty.
We believe this approach could drive other interoperability applications by defining multiple reusable ontologies. For example, a Fast Healthcare Information Resource-based ontology could be used to drive data exchange for future SMART applications.13 Or, an OMOP ontology could enable transformations to support the Observational Health Data Sciences and Informatics’ (OHDSI) large-scale data analytics tools.14 Previously, a Consolidated Clinical Document Architecture ontology was developed for Query Health, and this approach could be used to bolster its utility for data exchange.15
CONCLUSION
This work demonstrates our ability to generate new physical databases for new network-based analyses without the need to create analysis-specific data extraction programs from the electronic health record. By harmonizing sites to a common i2b2 ontology, new information models can be automatically generated across the SCILHS network without further terminology translation. Researchers can query the SCILHS network directly in i2b2 using SHRINE and can simultaneously participate in networks that support other information models, such as PCORnet. The ability to quickly translate and materialize the i2b2 data into new physical schema provides a quick win for participating institutions without imposing new data extraction requirements.
ACKNOWLEDGEMENTS
Thanks to Maryan Zirkle, SCILHS’s PCORI Program Officer, for making helpful suggestions to improve the readability of this manuscript. Thanks to the members of the i2b2 and SCILHS teams who have made this work possible: Marc Natter, Douglas MacFadden, Sarah Weiler, and Lori Philips. Thanks to Russ Waitman and Dan Conolly at Kansas University Medical, with whom we collaborated to develop the i2b2 ontology for PCORnet. Finally, thanks to the innumerable technical staff at our sites that have performed the heavy lifting on vetting our tools and methodology.
CONTRIBUTORS
J.G.K. designed and developed the SCILHS PCORnet Information Model and guidance documents for implementation, and led the development of the PCORnet-CDM data transformation script. A.A. designed, then developed a portion of the i2b2 to PCORnet-CDM data transform script. S.N.M. is the Director of Research Computing and Information Systems at Partners Healthcare and is the chief architect of i2b2. He conceived of the methodology and provided guidance during the design and implementation of the SCILHS PCORnet Information Model and data transform tool. V.A.R. is the network project manager for SCILHS and oversaw the sites’ implementation of the ontology and transformation. K.D.M. and S.N.M. lead the Harvard SCILHS network and guide the networks’ development from both a technical and research perspective. J.G.K. and S.N.M. co-wrote the manuscript, with significant input from A.A. The other authors provided feedback and details. All authors gave final approval of the version of the manuscript to be published.
FUNDING
This work was funded through a Patient-Centered Outcomes Research Institute Award (CDRN-1306-04608) for development of the National Patient-Centered Clinical Research Network, known as PCORnet, and by R01GM104303 from the National Institute of General Medical Sciences, NIH.
COMPETING INTERESTS
None.
DISCLAIMER
The statements presented in this publication are solely the responsibility of the author(s) and do not necessarily represent the views of the Patient-Centered Outcomes Research Institute, its Board of Governors or Methodology Committee, or other participants in PCORnet.
REFERENCES
- 1.Manion FJ, Robbins RJ, Weems WA, Crowley RS. Security and privacy requirements for a multi-institutional cancer research data grid: an interview-based study. BMC Med Inform Decis Mak. 2009;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.What is eTRIKS. http://www.etriks.org. Accessed 29 July 2015. [Google Scholar]
- 3.McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: enabling nationally scalable multi-site disease studies. PLoS ONE. 2013;8:e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(4):576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity-attribute-value database. J Am Med Inform Assoc. 1998;5:511–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandl KD, Kohane IS, McFadden D, et al. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): Architecture. J Am Med Inform Assoc.. 2014;21(4):615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis LH, Weiner MG, Boudreau DM, et al. Design considerations, architecture, and use of the Mini-Sentinel distributed data system. Pharmacoepidemiol Drug Safety. 2012;21:23–31. [DOI] [PubMed] [Google Scholar]
- 9.Abend AH, Mandel A, Palchuk MB. Techniques for federating queries across different ontologies in i2b2. AMIA Annual Meeting Proceedings. 2011:1668. [Google Scholar]
- 10.Connolly DW. PCORnet Common Data Model in i2b2. Bitbucket. https://bitbucket.org/DanC/pcornet-dm. Accessed 1 March 2015. [Google Scholar]
- 11.Phillips L. NCBO Extraction Tool version 2.0. NCBO Ontology Tools for i2b2. https://community.i2b2.org/wiki/display/NCBO/NCBO+Extraction+Tool+version+2.0. Accessed 4 December 2014.. [Google Scholar]
- 12.Klann JG. SCILHS PCORnet Common Data Model Ontology. i2b2 Wiki. https://community.i2b2.org/wiki/display/SCILHS. Accessed 27 February 2015. [Google Scholar]
- 13.FHIR: A New HL7 Draft Standard May Boost Web Services Development. http://www.healthcare-informatics.com/article/top-ten-tech-trends-catching-fhir. Accessed 2 February 2015. [Google Scholar]
- 14.OHDSI | Observational Health Data Sciences and Informatics. http://www.ohdsi.org. Accessed 29 July 2015.
- 15.Klann JG, Buck MD, Brown J, et al. Query Health: standards-based, cross-platform population health surveillance. J Am Med Inform Assoc. 2014;21(4):650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]