Abstract
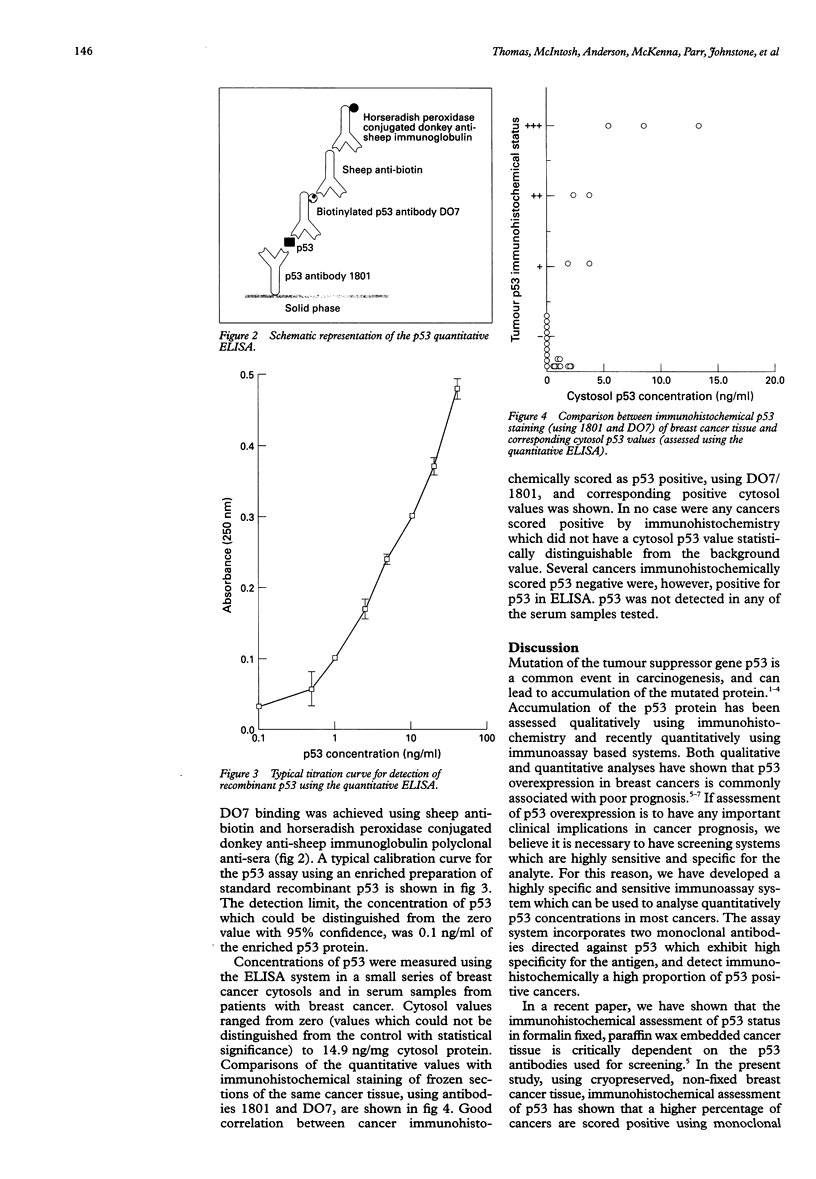
AIM: To develop a highly sensitive and specific enzyme linked immunosorbent assay (ELISA) system for analysis of p53 protein in cancer lysates. METHODS: The anti-p53 monoclonal antibodies DO7, 1801, BP53.12, and 421, and anti-p53 polyclonal antiserum CM1 were assessed by immunohistochemistry and western blot analysis to identify those most suitable for determining p53 status of cancer cells. Antibodies with desired characteristics were used to develop a non-competitive sandwich type ELISA system for analysis of p53 expression in cancer cytosols. Using the ELISA, p53 protein concentrations were measured in a small series of breast cancers, and the quantitative values compared with p53 immunohistochemical data of the same cancers. RESULTS: DO7 and 1801 gave the most specific and reliable results on immunohistochemistry and western blot analysis. Using these two antibodies, a non-competitive sandwich type ELISA system was developed to analyse p53 quantitatively. Analysis of the breast cancer series showed a good correlation between immunohistochemistry and the ELISA-tumours were generally positive using both techniques. Discrepancies were noted however: some cancers were immunohistochemically negative but ELISA positive. One explanation for this may be that the ELISA is more sensitive than immunohistochemistry. CONCLUSION: The p53 ELISA system is a non-competitive double monoclonal antibody sandwich method, using DO7 and 1801 which have been shown to be highly specific for p53 protein by immunohistochemistry and western blot analysis. The lower threshold of the assay is 0.1 ng/ml analyte in an enriched recombinant p53 preparation. As p53 is now regarded as a protein associated with prognosis in breast and other cancers, the assay may have clinical applications.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banks L., Matlashewski G., Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986 Sep 15;159(3):529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- Elledge R. M., Fuqua S. A., Clark G. M., Pujol P., Allred D. C., McGuire W. L. Prognostic significance of p53 gene alterations in node-negative breast cancer. Breast Cancer Res Treat. 1993;26(3):225–235. doi: 10.1007/BF00665800. [DOI] [PubMed] [Google Scholar]
- Hassapoglidou S., Diamandis E. P., Sutherland D. J. Quantification of p53 protein in tumor cell lines, breast tissue extracts and serum with time-resolved immunofluorometry. Oncogene. 1993 Jun;8(6):1501–1509. [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Horne G. M., Anderson J. J., Tiniakos D. G., McIntosh G. G., Thomas M. D., Angus B., Henry J. A., Lennard T. W., Horne C. H. p53 protein as a prognostic indicator in breast carcinoma: a comparison of four antibodies for immunohistochemistry. Br J Cancer. 1996 Jan;73(1):29–35. doi: 10.1038/bjc.1996.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaya K., Tsuda H., Hiraide H., Tamaki K., Tamakuma S., Fukutomi T., Mukai K., Hirohashi S. Nuclear p53 immunoreaction associated with poor prognosis of breast cancer. Jpn J Cancer Res. 1991 Jul;82(7):835–840. doi: 10.1111/j.1349-7006.1991.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. P. Cancer. p53, guardian of the genome. Nature. 1992 Jul 2;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Levesque M. A., Diamandis E. P., Yu H., Sutherland D. J. Quantitative analysis of mutant p53 protein in breast tumor cytosols and study of its association with other biochemical prognostic indicators in breast cancer. Breast Cancer Res Treat. 1994;30(2):179–195. doi: 10.1007/BF00666062. [DOI] [PubMed] [Google Scholar]
- Nigro J. M., Baker S. J., Preisinger A. C., Jessup J. M., Hostetter R., Cleary K., Bigner S. H., Davidson N., Baylin S., Devilee P. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989 Dec 7;342(6250):705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- Stephen C. W., Helminen P., Lane D. P. Characterisation of epitopes on human p53 using phage-displayed peptide libraries: insights into antibody-peptide interactions. J Mol Biol. 1995 Apr 21;248(1):58–78. doi: 10.1006/jmbi.1995.0202. [DOI] [PubMed] [Google Scholar]
- Stephen C. W., Lane D. P. Mutant conformation of p53. Precise epitope mapping using a filamentous phage epitope library. J Mol Biol. 1992 Jun 5;225(3):577–583. doi: 10.1016/0022-2836(92)90386-x. [DOI] [PubMed] [Google Scholar]
- Vojtesek B., Fisher C. J., Barnes D. M., Lane D. P. Comparison between p53 staining in tissue sections and p53 proteins levels measured by an ELISA technique. Br J Cancer. 1993 Jun;67(6):1254–1258. doi: 10.1038/bjc.1993.234. [DOI] [PMC free article] [PubMed] [Google Scholar]