Abstract
Aims: To shed light on the conflicting findings of the association between the methylenetetrahydrofolate reductase gene (MTHFR) 677C/T polymorphism and the risk of diabetic retinopathy (DR), a meta-analysis was conducted. Methods: A predefined search was performed on 1747 DR cases and 3146 controls from 18 published studies by searching electronic databases and reference lists of relevant articles. A random-effects or fixed-effects model was used to estimate the sizes of overall and stratification effects of the MTHFR 677C/T polymorphism on the risk of DR, as appropriate. Results: Risks were evaluated by odds ratios (OR) with 95% confidence intervals (95% CI). We found a significant association between the MTHFR 677C/T polymorphism and the risk of DR for each genetic model (recessive model: OR = 1.67; 95% CI: 1.19–2.40 and dominant model: OR = 1.71; 95% CI: 1.28–2.28; respectively). In stratified analysis; we further found that the Asian group with both types of diabetes mellitus (DM) showed a significant association with genetic models (recessive model: OR = 2.16; 95% CI: 1.75–2.60 and dominant model: OR = 1.98; 95% CI: 1.42–2.76; respectively). Conclusions: Our study suggested that the MTHFR 677C/T polymorphism may contribute to DR development, especially in Asian populations. Prospective and additional genome-wide association studies (GWAS) are needed to clarify the real role of the MTHFR gene in determining susceptibility to DR.
Keywords: MTHFR 677C/T, polymorphism, diabetic retinopathy, DM type, ethnicity
1. Introduction
Diabetic retinopathy (DR) is the leading cause of vision loss in adults aged 20–74 years [1]. With the increasing prevalence of diabetes, the number of instances of DR and vision-threatening DR (VTDR), has been estimated to rise to 191.0 million and 56.3 million, respectively, by 2030 [2]. DR is a complex trait involving polygenic, metabolic, and environmental influence. Known risk factors—most notably duration of diabetes and glycemic control—explain some, but not all, of the progression of DR [3,4,5]. There are diabetic patients that develop DR despite short durations of diabetes and/or excellent glycemic control, and other diabetic patients who do not develop DR in the face of long-standing diabetes and/or long-term hyperglycemia [6]. Therefore, given the complexity of the disease, genetic factors may explain some of the variation in the development of DR [7].
The gene encoding methylenetetrahydrofolate reductase (MTHFR, chromosome 1p36.3), which catalyzes the methylation of homocysteine to methionine [8], has been widely regarded as a genetic candidate for diabetes mellitus (DM). It has been demonstrated that the single nucleotide polymorphism (SNP) in MTHFR gene at nucleotide 677C/T(amino acid 222Ala/Val) can destroy the enzyme activity and cause hyperhomocysteinemia (HHcy) [9,10,11]. Because of such critical functional influence, it is readily postulated that the polymorphism of MTHFR 677C/T contributes to the development of DR, and a number of studies have addressed the role of the variation in the complex etiology of DR.
Numerous molecular epidemiological studies have been performed to estimate the relationship between the MTHFR 677C/T polymorphism and DR [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29], but the results remain inconclusive. Although several meta-analyses have been published [30,31], they still did not reach a consistent conclusion. To better shed light on these conflicting findings, we conducted a comprehensive meta-analysis on 18 published studies from 1996 to 2016, with 1747 diabetic retinopathy cases and 3146 controls relating the variant of the MTHFR 677C/T to the risk of developing DR.
2. Methods
This study was reported according to the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines and Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for reporting systematic reviews and meta-analyses. Study selection, data extraction, and quality assessment were completed independently by two investigators. Disagreement was resolved through discussion. If the discussion did not lead to a consensus, Professor Wang made the final decision.
2.1. Identification and Eligibility of Relevant Studies
We attempted to include all the studies that determined the genotype distribution of MTHFR 677C/T polymorphism in cases of diabetes retinopathy, and (i) in healthy controls or (ii) in diseased controls (subjects with diabetes and free of DR) in the meta-analysis. Cases were type 1 or 2 diabetic subjects with background, simple, advanced, or proliferative DR. The diseased control group consisted of subjects with diabetes and free of diabetic retinopathy disease, i.e., diabetes nephropathy.
We first identified studies by searching the electronic literature PubMed and Embase for relevant reports in English and CNKI for papers in Chinese (from January 1996 to April 2016, using the search terms “(MTHFR or methylenetetrahydrofolate reductase) and (diabetes or diabetic) and (retinopathy) and (gene or polymorphism or allele or genotype or variant or variation or mutation)” [30]. We chose articles which were conducted among human subjects. Eligible studies were then identified by further searching the studies on the association between MTHFR 677C/T polymorphism and diabetic retinopathy risk. We restricted attention to the studies that satisfied all of the following criteria: (1) studies related to the MTHFR polymorphism were determined regardless of sample size and study design (case-control, cross-sectional or cohort studies); (2) each genotype frequency was reported; and (3) there was sufficient information for extraction of data. If studies had partly overlapped subjects, only the one with a larger and/or latest sample size was selected for the analysis. Additional studies were identified by hands-on searches from references of original studies or review articles on this topic. According to these criteria, we finally included 18 papers in our meta-analysis.
2.2. Data Extraction and Conversion
Two investigators independently extracted data and reached a consensus on all of the items. Data extracted from these articles included the first author’s name, publication year, study design, ethnicity of population, DM type, clinical characteristics, and the number of cases and controls for MTHFR C677T genotypes. The frequencies of the alleles and the genotypic distributions were extracted or calculated for both cases and controls. We merged the original data into the control group or case group if the study did not provide corresponding data. For some studies without sufficient information for extraction of data, we tried to contact with the studies’ authors by sending emails from their articles to request missing data. In addition, it was tested whether the distribution of genotypes in the controls was consistent with Hardy–Weinberg equilibrium (HWE) for each study, and the frequency of the minor allele for MTHFR 677C/T polymorphism was calculated.
2.3. Quality Assessment and Study Stratification
We used the Newcastle–Ottawa scale (NOS) method to assess the observational studies that were included. The NOS is composed of 3 parts (8 entries): selection, comparability, and exposure. A quality item is given only one star for the study in selection and exposure, and a quality item is given at most two stars for the study in comparability. It is a semi-quantitative scale, and a score of 0–9 stars is assigned to each study. Studies whose scores were more than 6 stars were considered to be of relatively high quality [32]. The scores of included studies are shown in Table 1.
Table 1.
Baseline characteristics of qualified studies included in the meta-analysis.
| Author (Ref *) | Year | Ethnicity | Design | Case | Control | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | Age (year) | DM Duration (year) | Definition | Sample Size | Age (year) | DM Duration (year) | Definition | HWE # | MAF & | NOS (Stars) | ||||
| Neugebauer, S. et al. [12] | 1997 | Japan | CS | 76 | 55.5 ± 7.9 | 16.5 ± 5.1 | DR | 36 | 50.5 ± 9.7 | 11.2 ± 4.2 | NCTDM | 0.67 | 0.26 | 7 |
| Fujita, H. et al. [13] | 1999 | Japan | CS | 105 | 60±12 | NR | PDR + DN | 68 | 62 ± 10 | NR | T2DM | 0.14 | 0.42 | 6 |
| Lauszus, F.F. et al. [14] | 2001 | Denmark | CS | 112 | NR | NR | DR | 1084 | NR | NR | T1DM | 0.53 | 0.29 | 5 |
| Wang, L.Q. et al. [15] | 2001 | China | CC | 62 | 62.50 ± 8.08 | 8.29 ± 6.40 | DR | 202 | 59.42 ± 14.87 for T2DM41.83 ± 17.10 for Healthy | 7.29 ± 5.80 for T2DM | Healthy + T2DM | 0.73 | 0.32 | 8 |
| Yang, G.Q. et al. [16] | 2001 | China | CC | 60 | 50.7 ± 12.1 | 1 (0.1–4) | DR | 231 | 51.1 ± 12.8 for T2DM with DN63.0 ± 8.8 for T2DM with NCD52.6 ± 14.9 for Healthy | 2 (1–4) for T2DM with DN14 (11–18) for T2DM with NCD | Healthy + T2DM | 0.73 | 0.44 | 8 |
| Maeda, M. et al. [17] | 2003 | Japan | CS | 51 | NR | NR | DR | 105 | NR | NR | T2DM | 0.06 | 0.37 | 5 |
| Santos, K.G. et al. [18] | 2003 | Brazil | CS | 99 | NA | NA | DR | 111 | NA | NA | T2DM | 0.98 | 0.39 | 6 |
| Sun, J. et al. [19] | 2003 | China | CC | 110 | 55.6 ± 6.7 | <5 | DR | 155 | 54.7 ± 7.1 for NDR42.3 ± 6.1 for Healthy | >10 for NDR | T2DM | 0.00 | 0.33 | 7 |
| Yoshioka, K. et al. [20] | 2003 | Japan | CS | 98 | NA | NA | DR | 268 | NA | NA | T2DM | 0.46 | 0.38 | 6 |
| Huang, D.F. et al. [21] | 2005 | China | CC | 50 | NR | NR | DR | 47 | NR | NR | Healthy | 0.96 | 0.26 | 5 |
| Yi, X.X. et al. [22] | 2005 | China | CC | 249 | 56.53 ± 10.45 | 5.9 ± 4 | DR | 65 | NR | NR | Healthy | 0.01 | 0.31 | 5 |
| Errera, F.I. et al. [23] | 2006 | Brazil | CC | 141 | 55.43 ± 15.33 | 18 ± 8.67 | DR | 107 | 66.11 ± 7.06 | NA | Healthy | 0.24 | 0.40 | 6 |
| Liu, D.M. et al. [24] | 2006 | China | CC | 44 | 51.9 ± 7.5 | NR | DR | 84 | 54.0 ± 13.2 | NA | Healthy | 0.01 | 0.29 | 5 |
| Maeda, M. et al. [25] | 2008 | Japan | CS | 75 | NA | NA | DR | 115 | NA | NA | T2DM | 0.06 | 0.36 | 5 |
| Ukinc, K. et al. [26] | 2009 | Turkey/ | CS | 25 | NA | NA | DR | 27 | NA | NA | T2DM | 0.10 | 0.24 | 5 |
| Ren, M. et al. [27] | 2011 | China | CC | 161 | 59.95 ± 10.55 | 11 | DR | 213 | 58.52 ± 12.61 | 7 | T2DM | 0.23 | 0.42 | 7 |
| Yigit, S. et al. [28] | 2013 | Turkey | CS | 81 | NA | NA | DR | 149 | NA | NA | T1DM + T2DM | 0.98 | 0.24 | 5 |
| Simoes, M.J. et al. [29] | 2014 | Portugal | CS | 148 | 58.5 ± 8.4 | 10.5 ± 5.3 | PDR | 79 | 61.8 ± 7.8 | 8.9 ± 4.8 | T2DM | 0.73 | 0.24 | 7 |
* The ref was referred to the reference numbers in this study; # Hardy–Weinberg equilibrium (HWE) test and & the minor allele frequency (MAF) were calculated in control group for each study. NR, not reported; NA, not available; CC, case-control; CS, cross-sectional; CH cohort; DR, diabetes retinopathy; PDR, proliferative diabetes retinopathy; NCD: non-complicated; DN, diabetes nephropathy;T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus; NCTDM, non-insulin dependent diabetes mellitus.
2.4. Meta-Analysis
Our meta-analysis evaluated the relationship between the MTHFR 677C/T polymorphism and the risk of DR for each study by odds ratio (OR) with 95% confidence intervals (95% CI). For all studies, we calculated the ORs for the: (i) separate pairwise comparisons; (ii) allele contrast; (iii) recessive model; and (iv) dominant model. In addition, we conducted stratification analysis by ethnicity and DM type. A sensitivity analysis, which examines the effect of excluding specific studies, was also performed [33]. Our meta-analysis was subjected to sensitivity analysis for studies with the controls not in HWE (p < 0.05).
The χ2-based Q statistic test was used to assess the heterogeneity, and it was considered significant for p < 0.05. Heterogeneity was quantified with the I2 metric, which is independent of the number of studies in the meta-analysis. I2 takes values between 0% and 100%, with higher values denoting greater degree of heterogeneity (I2 > 50% was considered significant) [34]. We used the fixed-effects model and the random-effects model based on the Mantel–Haenszel method and the DerSimonian and Laird method, respectively, to combine values from each of the studies. When the effects were assumed to be heterogeneous, the random-effects model was used; otherwise, the fix-effects model was more appropriate [35]. In addition, we further conducted meta-regression analyses to estimate the source of heterogeneity. Publication bias was assessed according to the Begg adjusted rank correlation test and the Egger regression asymmetry test [36,37]. All analysis was done using the Stata software (v.12.1) (StataCorp LP, College Station, TX, USA). All the p values were two-sided.
3. Results
3.1. Literature Search
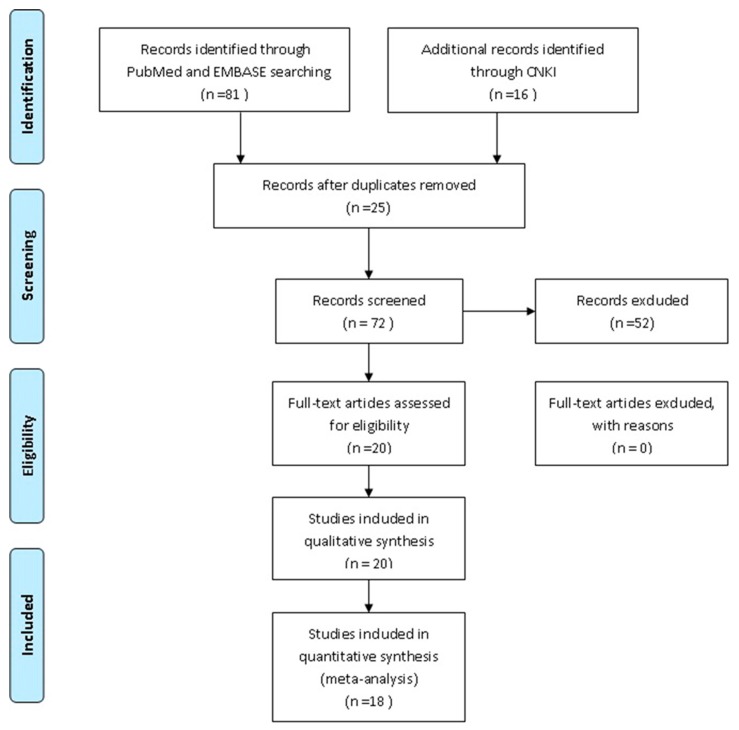
The study selection process is shown in Figure 1. A total of 97 articles (PubMed 35, Embase 46 and CNKI 16) were identified from the databases, and 25 duplicates were excluded using EndNote (X7). In addition, 52 articles were excluded based on a review of the titles and abstracts, and 20 full-text articles were assessed for eligibility; 2 articles were excluded due to could not provide each genotype frequency or other sufficient information for extraction of data. Finally, a total of 18 [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29] articles were included in this meta-analysis.
Figure 1.
Flow chart of the literature search.
3.2. Eligible Studies and Study Characteristics
The selected study baseline characteristics from the qualified studies included in the meta-analysis are provided in Table 1 and the frequencies on MTHFR C677T polymorphism allele/genotype prevalence are shown in Table 2. Of 18 studies, 12 studies (9 Asian, 2 Caucasian, and 1 American population) were based on type 2 DM (T2DM) including participants (case group; control group), and 6 studies (3 Caucasian and 3 Asian) was based onnon-T2DM (2 studies ([12,14]) with type 1 DM (T1DM), 2 ([21,24]) with un-defined DM type, and 2 ([23,28]) involved both T1DM and T2DM) including participants (case group; control group). Ten studies were case-control study design and 8 studies were cross-sectional study design.
Table 2.
The frequency of methylenetetrahydrofolate reductase gene (MTHFR) C677T polymorphism allele/genotype prevalence.
| Author (Ref) | Prevalence of MTHFR C677T Genotype | Prevalence of Allele Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||||
| Case | Control | Case | Control | Case | Control | Case | Control | Case | Control | |
| Neugebauer, S. et al. [12] | 26 | 20 | 38 | 13 | 12 | 3 | 90 | 53 | 62 | 19 |
| Fujita, H. et al. [13] | 31 | 20 | 57 | 39 | 17 | 9 | 119 | 79 | 91 | 57 |
| Lauszus, F.F. et al. [14] | 47 | 542 | 57 | 455 | 8 | 87 | 151 | 1539 | 73 | 629 |
| Wang, L.Q. et al. [15] | 8 | 94 | 27 | 86 | 27 | 22 | 43 | 274 | 81 | 130 |
| Yang, G.Q. et al. [16] | 8 | 75 | 33 | 111 | 19 | 45 | 49 | 261 | 71 | 201 |
| Maeda, M. et al. [17] | 18 | 37 | 20 | 58 | 13 | 10 | 56 | 132 | 46 | 78 |
| Santos, K.G. et al. [18] | 34 | 41 | 53 | 53 | 12 | 17 | 121 | 135 | 77 | 87 |
| Sun, J. et al. [19] | 33 | 82 | 46 | 45 | 31 | 28 | 112 | 209 | 108 | 101 |
| Yoshioka, K. et al. [20] | 33 | 100 | 50 | 132 | 15 | 36 | 116 | 332 | 80 | 204 |
| Huang, D.F. et al. [21] | 17 | 26 | 25 | 18 | 8 | 3 | 59 | 70 | 41 | 24 |
| Yi, X.X. et al. [22] | 68 | 35 | 110 | 19 | 71 | 11 | 246 | 89 | 252 | 41 |
| Errera, F.I. et al. [23] | 61 | 36 | 66 | 57 | 14 | 14 | 188 | 129 | 94 | 85 |
| Liu, D.M. et al. [24] | 18 | 47 | 16 | 25 | 10 | 12 | 52 | 119 | 36 | 49 |
| Maeda, M. et al. [25] | 31 | 43 | 28 | 62 | 16 | 10 | 90 | 148 | 60 | 82 |
| Ukinc, K. et al. [26] | 14 | 14 | 11 | 13 | 0 | 0 | 39 | 41 | 11 | 13 |
| Ren, M. et al. [27] | 26 | 77 | 78 | 95 | 57 | 41 | 130 | 249 | 192 | 177 |
| Yigit, S. et al. [28] | 38 | 85 | 30 | 55 | 13 | 9 | 106 | 225 | 56 | 73 |
| Simoes, M.J. et al. [29] | 69 | 45 | 60 | 30 | 19 | 4 | 198 | 120 | 98 | 38 |
| Total | 580 | 1419 | 805 | 1366 | 362 | 361 | 1965 | 4204 | 1529 | 2088 |
3.3. Summary Statistics
Data from 18 articles that investigated the association between the MTHFR 677C/T polymorphism and DR risk were included in the meta-analysis. The overall frequency (%) of minor D allele frequency (MAF) was 0.44/0.33 for cases and controls. The frequency of the MAF for each individual study polymorphism for controls is given in Table 1. All studies indicated that the distribution of genotypes in the controls was consistent with Hardy–Weinberg equilibrium except for 3 studies ([19,22,24]), indicating genotyping errors and/or population stratification [33]; therefore, a sensitivity analysis was performed by excluding these studies.
3.4. Main Results, Stratification, and Sensitivity Analyses
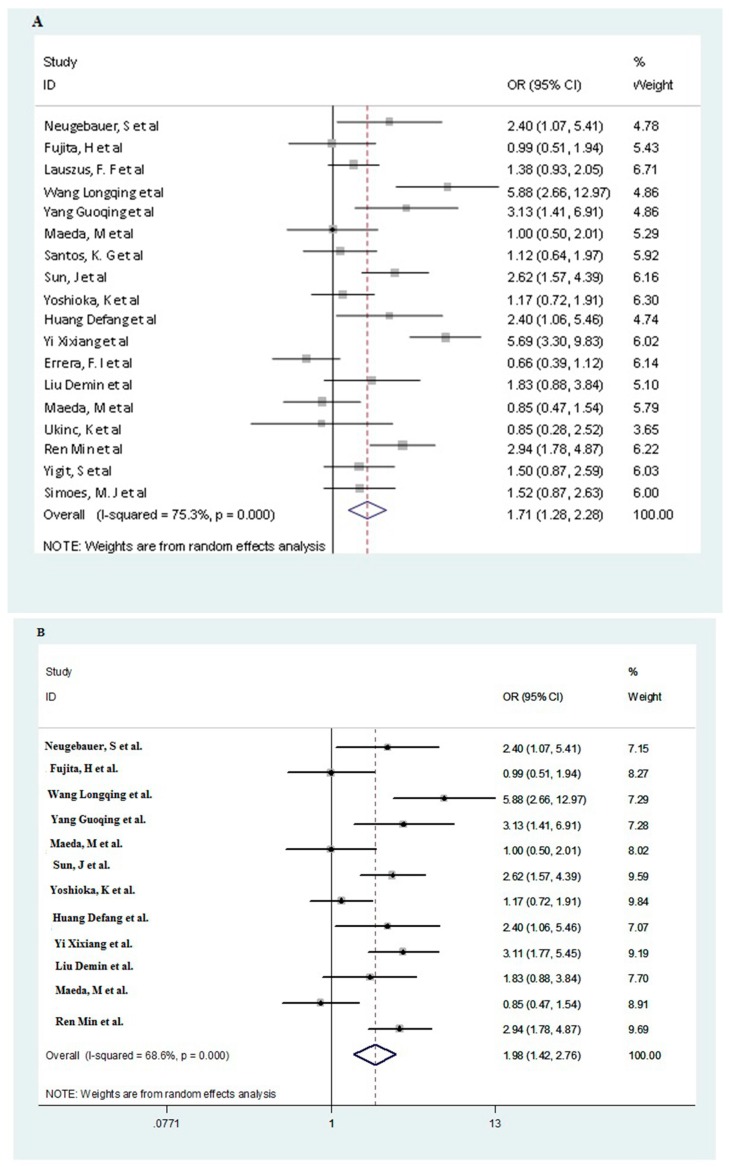
The estimation results of the relationship of MTHFR 677C/T polymorphism with DR are presented in Table 3. Figure 2 shows the overall results for the association between the polymorphism and the risk of DR (in dominant model).
Table 3.
Summary ORs and heterogeneity results for associations between the MTHFR C677T polymorphism and DR.
| Subgroup | Genetic Model | Studies No (All/Sensitivity) | OR (95% CI) | p * | I2 (%) | OR se (95% CI) # |
|---|---|---|---|---|---|---|
| Overall | CT vs. CC | 18/15 | 1.46 (1.15–1.86) | 0.00 | 60.0 | 1.32 (1.02–1.70) |
| TT vs. CC | 18/15 | 2.45 (1.66–3.60) | 0.00 | 69.0 | 2.27 (1.44–3.58) | |
| Allele contrast | 18/15 | 1.52 (1.26–1.84) | 0.00 | 72.4 | 1.45 (1.17–1.79) | |
| Recessive model | 18/15 | 1.67 (1.19–2.40) | 0.00 | 71.9 | 1.87 (1.32–2.66) | |
| Dominant model | 18/15 | 1.71 (1.28–2.28) | 0.00 | 75.3 | 1.50 (1.13–1.98) | |
| Asian | CT vs. CC | 12/9 | 1.71 (1.22–2.39) | 0.00 | 64.9 | 1.52 (1.01–2.30) |
| TT vs. CC | 12/9 | 2.97 (2.06–4.29) | 0.02 | 51.5 | 3.07 (1.83–5.14) | |
| Allele contrast | 12/9 | 1.75 (1.42–2.18) | 0.00 | 67.3 | 1.69 (1.28–2.24) | |
| Recessive model | 12/9 | 2.16 (1.75–2.65) | 0.11 | 35.1 | 2.34(1.62–3.38) | |
| Dominant model | 12/9 | 1.98 (1.42–2.76) | 0.00 | 68.6 | 1.83 (1.20–2.81) | |
| Non-Asian | CT vs. CC | 6/6 | 1.15 (0.92–1.45) | 0.38 | 5.4 | 1.15 (0.92–1.45) |
| TT vs. CC | 6/6 | 1.33 (0.69–2.54) | 0.04 | 61.4 | 1.33 (0.69–2.54) | |
| Allele contrast | 6/6 | 1.14 (0.89–1.46) | 0.07 | 51.5 | 1.14 (0.89–1.46) | |
| Recessive model | 6/6 | 1.24 (0.69–2.23) | 0.05 | 57.7 | 1.24 (0.69–2.23) | |
| Dominant model | 6/6 | 1.18 (0.95–1.47) | 0.21 | 30.70 | 1.18 (0.95–1.47) | |
| T2DM | CT vs. CC | 12/10 | 1.50 (1.08–2.09) | 0.00 | 66.5 | 1.32 (0.93–1.86) |
| TT vs. CC | 12/10 | 2.68 (1.74–4.13) | 0.00 | 66.2 | 2.61 (1.50–4.53) | |
| Allele contrast | 12/10 | 1.59 (1.26–2.01) | 0.00 | 74.0 | 1.49 (1.14–1.96) | |
| Recessive model | 12/10 | 2.05 (1.47–2.84) | 0.01 | 56.9 | 2.10 (1.38–3.17) | |
| Dominant model | 12/10 | 1.72 (1.23–2.42) | 0.00 | 72.1 | 1.54 (1.06–2.24) | |
| Non-T2DM | CT vs. CC | 6/5 | 1.30 (1.02–1.66) | 0.12 | 43.5 | 1.32 (0.89–1.97) |
| TT vs. CC | 6/5 | 1.75 (0.93–3.27) | 0.05 | 55.9 | 1.70 (0.80–3.63) | |
| Allele contrast | 6/5 | 1.38 (1.02–1.88) | 0.02 | 64.5 | 1.34 (0.95–1.91) | |
| Recessive model | 6/5 | 1.38 (0.96–2.00) | 0.14 | 39.5 | 1.32 (0.89–1.97) | |
| Dominant model | 6/5 | 1.46 (1.00–2.13) | 0.04 | 57.4 | 1.42 (0.92–2.19) | |
| Asian with T2DM | CT vs. CC | 9/7 | 1.64 (1.07–2.50) | 0.00 | 73.8 | 1.40 (0.86–2.29) |
| TT vs. CC | 9/7 | 3.00 (1.92–4.68) | 0.01 | 63.8 | 2.99 (1.63–5.50) | |
| Allele contrast | 9/7 | 1.73 (1.33–2.26) | 0.00 | 76.0 | 1.64 (1.17–2.29) | |
| Recessive model | 9/7 | 2.21 (1.59–3.06) | 0.03 | 51.9 | 2.33 (1.52–3.57) | |
| Dominant model | 9/7 | 1.94 (1.27–2.94) | 0.00 | 76.7 | 1.72 (1.03–2.89) | |
| Asian with Non-T2DM | CT vs. CC | 3/2 | 1.99 (1.22–3.25) | 0.87 | 0.0 | 2.19 (1.19–4.01) |
| TT vs. CC | 3/2 | 2.80 (1.39–5.66) | 0.77 | 0.0 | 3.50 (1.28–9.62) | |
| Allele contrast | 3/2 | 1.86 (1.32–2.60) | 0.90 | 0.0 | 1.97 (1.28–3.05) | |
| Recessive model | 3/2 | 2.06 (1.06–4.02) | 0.87 | 0.0 | 2.38 (0.91–6.24) | |
| Dominant model | 3/2 | 2.17 (1.38–3.42) | 0.85 | 0.0 | 2.40 (1.35–4.28) | |
| Non-Asian with T2DM | CT vs. CC | 3/3 | 1.20 (0.81–1.76) | 0.79 | 0.0 | 1.20 (0.81–1.76) |
| TT vs. CC | 3/3 | 1.54 (0.43–5.46) | 0.08 | 68.2 | 1.54 (0.43–5.46) | |
| Allele contrast | 3/3 | 1.18 (0.90–1.56) | 0.25 | 27.0 | 1.18 (0.90–1.56) | |
| Recessive model | 3/3 | 1.37 (0.39–4.82) | 0.06 | 70.8 | 1.37 (0.39–4.82) | |
| Dominant model | 3/3 | 1.24 (0.86–1.80) | 0.58 | 0.0 | 1.24 (0.86–1.80) | |
| Non-Asian with Non-T2DM | CT vs. CC | 3/3 | 1.09 (0.69–1.71) | 0.09 | 58.0 | 1.09 (0.69–1.71) |
| TT vs. CC | 3/3 | 1.24 (0.49–3.14) | 0.03 | 71.9 | 1.24 (0.49–3.14) | |
| Allele contrast | 3/3 | 1.13 (0.76–1.68) | 0.02 | 73.3 | 1.13 (0.76–1.68) | |
| Recessive model | 3/3 | 1.21 (0.54–2.72) | 0.05 | 66.9 | 1.21 (0.54–2.72) | |
| Dominant model | 3/3 | 1.13 (0.69–1.83) | 0.05 | 66.7 | 1.13 (0.69–1.83) |
# OR se: Sensitivity analysis OR for HWE; * Test for heterogeneity: random-effects model was used when p value for heterogeneity test <0.05 and I2 > 50%; otherwise, fixed-effects model was used; T2DM, type 2 diabetes mellitus;Non-T2DM, type 1 diabetes mellitus or unknown DM type or mixed DM type; Non-Asian, Caucasian and African-American population.
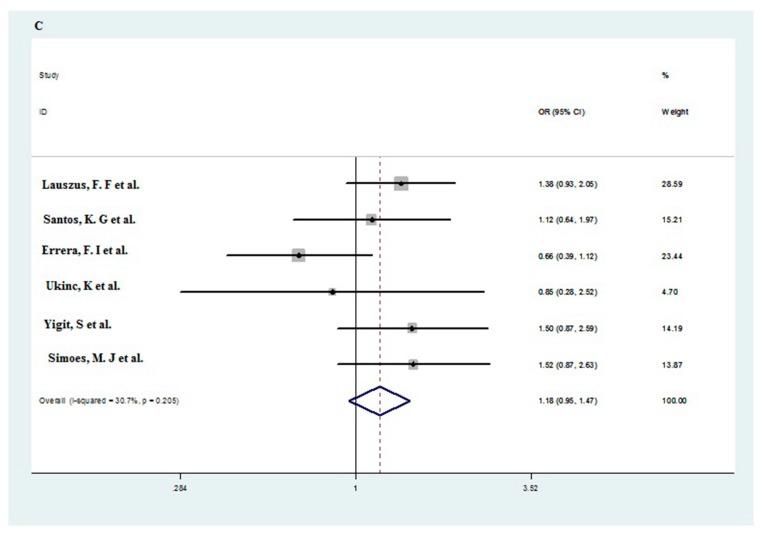
Figure 2.
Oddsratios (ORs; log scale) of diabetic retinopathy (DR) associated with MTHFR 677C/T polymorphism for dominant genetic model. The graph shows individual and pooled estimates for different ethnic groups (A for all studies; B for Asian group; C for Non-Asian group).
As it shown in Table 3, the overall analysis found a significant association between the MTHFR 677C/T polymorphism and the risk of DR for all genetic models (CT vs. CC: OR = 1.46, 95% CI: 1.15–1.86; TT vs. CC:OR = 2.45, 95% CI: 1.66–3.60; Allele contrast: OR = 1.52, 95% CI: 1.26–1.84; recessive model: OR = 1.67, 95% CI: 1.19–2.40 and dominant model: OR = 1.71, 95% CI: 1.28–2.28, respectively).
In stratified analysis by ethnicity and DM type, we further detected that the Asian group, T2DM group showed significant associations for all genetic models (CT vs. CC: OR = 1.71, 95% CI: 1.22–2.39 for Asian group, OR = 1.50, 95% CI: 1.08–2.09 for T2DM group, respectively; TT vs. CC:OR = 2.97, 95% CI: 2.06–4.29 for Asian group, OR = 2.68, 95% CI: 1.74–4.13 for T2DM group, respectively; Allele contrast: OR = 1.75, 95% CI: 1.42–2.18 for Asian group, OR = 1.59, 95% CI: 1.26–2.01 for T2DM group, respectively; recessive model: OR = 2.16, 95% CI: 1.75–2.65 for Asian group, OR = 2.05, 95% CI: 1.47–2.84 for T2DM group, respectively and dominant model: OR = 1.98, 95% CI: 1.42–2.76 for Asian group, OR = 1.72, 95% CI: 1.23–2.42 for T2DM group, respectively). In addition, we further found significant associations for all genetic models in both Asian group with T2DM and non-T2DM (See Table 3). However, we did not find any significant effects for different genetic models in other subgroups. Further sensitivity analysis for HWE almost did not alter the pattern of results in both overall analysis and subgroup analysis (See Table 3).
3.5. Source of Heterogeneity and Publication Bias
From Table 3, we found that the heterogeneity between studies was observed in overall comparisons as well as subgroup analyses. We estimated the source of heterogeneity in dominant genetic models of the variant allele by ethnicity (Asian or non-Asian), DM type (T2DM or non-T2DM), HWE (in HWE or not), and study design (case-control or cross-sectional design) by meta-regression analyses. It revealed that DM type and HWE factors could not significantly influence between-study heterogeneity in genetic model for the polymorphism MTHFR 677C/T: DM type (p = 0.36) and HWE (p = 0.06). However, we found that ethnicity and study design factors might be the source of heterogeneity between studies: ethnicity (p = 0.03 and contributed 26.8% source of heterogeneity) and study design (p = 0.01 and contributed 52.1% source of heterogeneity). In addition, we further found that study design might be the major source of heterogeneity between studies for the Asian DM group (p = 0.001 and contributed 100% source of heterogeneity).
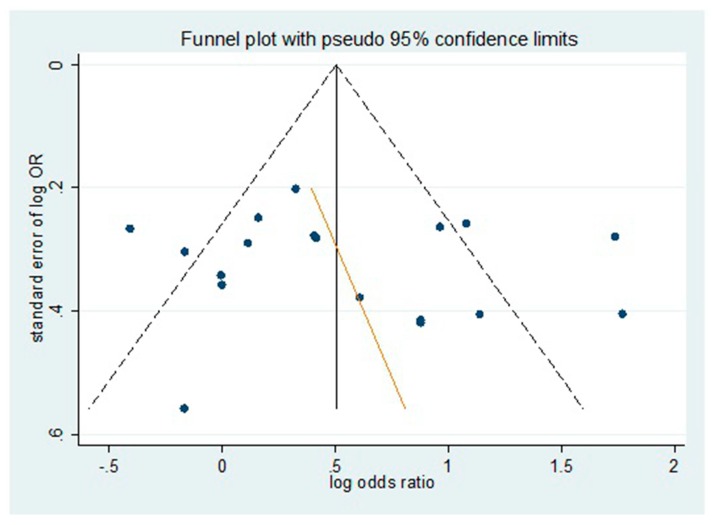
The potential presence of publication bias was estimated by using a funnel plot by evaluating log–odds ratio for the genotype TT + CT versus CC against the reciprocal of its standard error (Figure 3). As is shown, we failed to observe any significant funnel asymmetry which could indicate publication bias. We further conducted the Egger regression asymmetry test and the Begg adjusted rank correlation tests to estimate the publication bias of included literatures in the meta-analysis. As shown in Table 4, no publication bias was found for the polymorphism and risk of DR in both dominant and recessive genetic models.
Figure 3.
Evaluation of publication bias for all studies using funnel plots. No significant funnel asymmetry was observed which could indicate publication bias.
Table 4.
The results of publication bias test by Egger and Begg test.
| Subgroup | Egger Test | Begg Test | ||
|---|---|---|---|---|
| Dominant | Recessive | Dominant | Recessive | |
| all study | 0.59 | 0.48 | 0.65 | 0.48 |
| T2DM | 0.91 | 0.94 | 0.84 | 0.64 |
| Non-T2DM | 0.28 | 0.16 | 0.06 | 0.26 |
| Asian | 0.62 | 0.91 | 0.63 | 0.54 |
| Non-Asian | 0.57 | 0.10 | 1.00 | 0.46 |
T2DM, type 2 diabetes mellitus;Non-T2DM, type 1 diabetes mellitus or unknown DM type or mixed DM type; Non-Asian, Caucasian and African-American population.
4. Discussion
The common environmental risk factors of DR include hyperglycemia, hypertension, hyperlipidemia, obesity, duration of diabetes, puberty, and pregnancy [2]. However, despite having a long-term hyperglycemia, we often found that some diabetics develop retinopathy, whereas others do not. Because such known environmental factors do not fully explain this, researchers have sought the answer in genetic factors. The enzyme MTHFR methylates homocysteine to generate methionine, and its dysfunction can lead to HHcy [38]. Studies reported that HHcy induces endothelial dysfunction and arterial stiffness [39], and has also been associated with atherosclerosis [40] and retinopathy in both T1DM and T2DM patients [41,42]. The MTHFR C677T polymorphism leads to an Ala222Val substitution in the N-terminal catalytic domain of the enzyme, which reduces enzyme activity [43,44,45]. In addition, several recent genome-wide association studies (GWAS) have confirmed the association between MTHFR C677T genotype and homocysteine levels in healthy populations [46,47]. Numerous investigations into the potential role of MTHFR as a susceptibility gene for DR have been conducted over the past decades, with controversial results. Early meta-analyses attempted to reconcile these findings, but attempts to draw definite conclusions have been hindered by limited data, particularly when examining specific patient subgroups and increased relative studies [30,31].
In the current meta-analysis, the effect of separate pairwise comparisons, allele contrast, and the effects of the dominant and recessive genetic models were estimated. Subgroup analysis by ethnicity and DM type, and sensitivity for studies not in HWE, were performed. In addition, we further evaluated the source of heterogeneity and publication bias of included literatures. It is worth mentioning that we found study design might be the major source of heterogeneity between studies. To provide better power to detect smaller effect sizes, studies related to the MTHFR polymorphism were determined regardless of sample size and study design (case-control, cross-sectional, or cohort studies), however, different study designs may generate potential effects on the meta-analysis. For example, the cohort study design may obtain a much more powerful research conclusion than the case-control study design, and the cross-sectional study design could obtain less powerful research conclusion than the case-control study design. Therefore, the overall effect of all studies with different study designs might be deviated from the real effect, to some extent.
Our meta-analysis obtained several critical different conclusions from the previous reports [30,31]. In Zintzaras et al. [31] report, they just included 5 studies with 435 DR cases and 620 controls, which provides relatively poor power to detect smaller effect sizes. In addition, although they found a marginal association between C677T and the risk of developing DR, the results of overall analysis were less significant after conducting sensitivity analysis by excluding the study with the control not in HWE. Niu et al. [30] just performed the meta-analysis with 8 studies including 1599 subjects and they did not observe significant association with DR in heterozygous genotypic comparison (CT vs. CC) or in dominant model. In addition, they did not perform meta-regression analysis to identify the sources of between-study heterogeneity and found publication bias for comparison. However, from the present meta-analysis of 18 studies—reported from 1996 to 2016 and comprising 4893 subjects—we not only found the main effects for MTHFR C677T polymorphism on DR risk with all studies in all genetic models, without any publication bias, but further sensitivity analysis for HWE also did not alter the pattern of results in the overall analysis. From the stratification analysis by ethnicity and DM type, we also found that the MTHFR C677T polymorphism was significantly associated with DR risk in T2DM and Asian group, especially in Asian group with T2DM and non-T2DM.These findings may indicate that genetic factors may have more impact on Asian population.
We conducted a comprehensive meta-analysis on 18 published studies with 1747 DR cases and 3146 controls relating to the mutation of the MTHFR C677T to the risk of DR, which can provide better power to detect smaller effect sizes. Its strength was based on the accumulation of published data giving greater information to detect significant differences. In order to estimate the power of the study, we used the Power and Precision 4 software to conduct the power calculation by respectively accumulating the frequency of MTHFR 677T allele in case (0.44) and control (0.33) groups from all studies, and the result showed the power of our study is over 80%.
Despite the clear strengths of our study, some limitations merit serious consideration. First, non-English/Chinese, non-indexed, and non-published literature were not reviewed in our meta-analysis, thus might introduce some bias [48]. Second, only the unadjusted pooled ORs were calculated, because data for possible confounding factors that influence the estimates of associations (e.g., age, sex, body mass index) were not provided. Third, sampling variability and stratification in genetic association studies could be a possible confounding factor on the role of genetic markers. In addition, the risk effect may depend on the interaction with other risk factors: diabetes duration, HbA1c, blood pressure, total serum cholesterol, control of diabetes, and body mass index, all of which modulate the development of DR [49]. Furthermore, small numbers of individuals and inadequate information of lifestyle factors and dietary intake by the published studies limited our statistic power to fully investigate the gene-environment interactions [35]. Therefore, further well-designed large studies, particularly referring to GWAS and gene-environment interactions are warranted to confirm the real contribution of these polymorphisms to DR susceptibility and might further elucidate the genetics of DR.
Although there are several previous GWAS relevant to DR [50,51,52,53], the limitations in GWAS studies are still inconsistency and low reproducibility in different populations [2]. Several reasons may explain this [7]: (i) since the genetic effects of DR might be much modest than other diseases, it requires large sample sizes to identify the real role of genetic factors using GWAS, however, the sample sizes of previous studies on DR GWAS studies were relatively modest; (ii) the diagnosis of DR is clinically complicated with different types of DM, a case with slight retinopathic changes may not be assigned as a DR case, but some of the DR GWAS used case definitions that include such patients which may generate the bias. In addition, different population and heterogeneous phenotypes of DR patients, as well as poor characterization of controls, also affect the consistency of GWAS studies. Therefore, it is still very important to conduct the meta-analysis to estimate the variant of the MTHFR C677T to the risk of DR.
5. Conclusions
In summary, our present meta-analysis finds a relationship between DR and MTHFR C677T polymorphism, especially in Asian groups. Prospective and additional GWAS are needed to clarify the real role of the MTHFR gene in the development of DR.
Acknowledgments
This work was supported by the Surface Project of the Nanjing Medical University (2012NJMU255).
Author Contributions
Conceived and designed the experiments: Furu Wang and Zhifeng Wu. Performed the experiments: Shasha Luo. Analyzed the data: Furu Wang and Chao Shi. Contributed reagents/materials/analysis tools: Furu Wang and Chao Shi. Wrote the paper: Shasha Luo and Furu Wang.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Cheung N., Mitchell P., Wong T.Y. Diabetic retinopathy. Lancet. 2010;376:124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 2.Ting D.S., Cheung G.C., Wong T.Y. Diabetic retinopathy: Global prevalence, major risk factors, screening practices and public health challenges: A review. Clin. Exp. Ophthalmol. 2015;44:206–277. doi: 10.1111/ceo.12696. [DOI] [PubMed] [Google Scholar]
- 3.Klein R., Klein B.E., Moss S.E., Linton K.L. The beaver dam eye study. Retinopathy in adults with newly discovered and previously diagnosed diabetes mellitus. Ophthalmology. 1992;99:58–62. doi: 10.1016/S0161-6420(92)32011-1. [DOI] [PubMed] [Google Scholar]
- 4.Klein R., Klein B.E., Moss S.E., Cruickshanks K.J. Relationship of hyperglycemia to the long-term incidence and progression of diabetic retinopathy. Arch. Intern. Med. 1994;154:2169–2178. doi: 10.1001/archinte.1994.00420190068008. [DOI] [PubMed] [Google Scholar]
- 5.Klein R., Klein B.E., Moss S.E., Cruickshanks K.J. The wisconsin epidemiologic study of diabetic retinopathy: Xvii. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998;105:1801–1815. doi: 10.1016/S0161-6420(98)91020-X. [DOI] [PubMed] [Google Scholar]
- 6.Nathan D.M. Long-term complications of diabetes mellitus. N. Engl. J. Med. 1993;328:1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 7.Cho H., Sobrin L. Genetics of diabetic retinopathy. Curr.Diabetes Rep. 2014;14:515. doi: 10.1007/s11892-014-0515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch G.N., Loscalzo J. Homocysteine and atherothrombosis. N. Engl. J. Med. 1998;338:1042–1050. doi: 10.1056/NEJM199804093381507. [DOI] [PubMed] [Google Scholar]
- 9.DAngelo A., Selhub J. Homocysteine and thrombotic disease. Blood. 1997;90:1–11. [PubMed] [Google Scholar]
- 10.Engbersen A.M., Franken D.G., Boers G.H., Stevens E.M., Trijbels F.J., Blom H.J. Thermolabile 5,10-methylenetetrahydrofolate reductase as a cause of mild hyperhomocysteinemia. Am. J. Hum. Genet. 1995;56:142–150. [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman G., Goldschmidt N., Friedlander Y., Ben-Yehuda A., Selhub J., Babaey S., Mendel M., Kidron M., Bar-On H. A common mutation a1298c in human methylenetetrahydrofolate reductase gene: Association with plasma total homocysteine and folate concentrations. J. Nutr. 1999;129:1656–1661. doi: 10.1093/jn/129.9.1656. [DOI] [PubMed] [Google Scholar]
- 12.Neugebauer S., Baba T., Kurokawa K., Watanabe T. Defective homocysteine metabolism as a risk factor for diabetic retinopathy. Lancet. 1997;349:473–474. doi: 10.1016/S0140-6736(05)61185-3. [DOI] [PubMed] [Google Scholar]
- 13.Fujita H., Narita T., Meguro H., Ishii T., Hanyu O., Suzuki K., Kamoi K., Ito S. No association between mthfr gene polymorphism and diabetic nephropathy in Japanese type II diabetic patients with proliferative diabetic retinopathy. J. Diabetes Complicat. 1999;13:284–287. doi: 10.1016/S1056-8727(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 14.Lauszus F.F., Gron P.L., Klebe J.G. Association of polymorphism of methylene-tetrahydro-folate-reductase with urinary albumin excretion rate in type 1 diabetes mellitus but not with preeclampsia, retinopathy, and preterm delivery. Acta Obstet. Gynecol. Scand. 2001;80:803–806. doi: 10.1034/j.1600-0412.2001.080009803.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Wang J., Xue Y., Chen Y., Zou Y. Relationship between methylenetetrahydrofolate reductase gene polymorphism and diabetic retinopathy. Chin. J. Ocul. Fundus Dis. 2001;17:31–33. [PubMed] [Google Scholar]
- 16.Yang G., Lu J., Pan C. Study on the relationship between n5, 10-methylenetetrahydrofolate reductase gene polymorphism and the susceptibility to microangiopathy in type 2 diabetes mellitus. Chin. J. Endocrinol. Metab. 2001;17:36–39. [Google Scholar]
- 17.Maeda M., Yamamoto I., Fukuda M., Nishida M., Fujitsu J., Nonen S., Igarashi T., Motomura T., Inaba M., Fujio Y., et al. Mthfr gene polymorphism as a risk factor for diabetic retinopathy in type 2 diabetic patients without serum creatinine elevation. Diabetes Care. 2003;26:547–548. doi: 10.2337/diacare.26.2.547-a. [DOI] [PubMed] [Google Scholar]
- 18.Santos K.G., Tschiedel B., Schneider J., Souto K., Roisenberg I. Diabetic retinopathy in euro-brazilian type 2 diabetic patients: Relationship with polymorphisms in the aldose reductase, the plasminogen activator inhibitor-1 and the methylenetetrahydrofolate reductase genes. Diabetes Res. Clin. Pract. 2003;61:133–136. doi: 10.1016/S0168-8227(03)00112-8. [DOI] [PubMed] [Google Scholar]
- 19.Sun J., Xu Y., Zhu Y., Lu H., Deng H., Fan Y., Sun S., Zhang Y. The relationship of methylenetetrahydrofolate reductase gene polymorphism and plasma homocysteine levels in type 2 diabetes mellitus patients with diabetic retinopathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:131–134. [PubMed] [Google Scholar]
- 20.Yoshioka K., Yoshida T., Takakura Y., Kogure A., Umekawa T., Toda H., Yoshikawa T. No association between the mthfr gene polymorphism and diabetic retinopathy in type 2 diabetic patients without overt nephropathy. Diabetes Care. 2003;26:1947–1948; author reply 1948. doi: 10.2337/diacare.26.6.1947. [DOI] [PubMed] [Google Scholar]
- 21.Huang D.F., Cao H., Mao L. The relationship of homocysteine, methylenetetrahydrofolate reductasegene polymorphism and diabetic retinopathy. J. Chin. Microcirc. 2005;9:229–231. [Google Scholar]
- 22.Yi X., Yu Y., Zhang Y. The study on tcm syndrome differentiation of DR and the correlativity between its syndromes and types and gene polymorphism. J. Tradit. Chin. Ophthalmol. 2005;15:125–128. [Google Scholar]
- 23.Errera F.I., Silva M.E., Yeh E., Maranduba C.M., Folco B., Takahashi W., Pereira A.C., Krieger J.E., Passos-Bueno M.R. Effect of polymorphisms of the mthfr and apoe genes on susceptibility to diabetes and severity of diabetic retinopathy in Brazilian patients. Braz. J. Med. Biol. Res. 2006;39:883–888. doi: 10.1590/S0100-879X2006000700005. [DOI] [PubMed] [Google Scholar]
- 24.Liu D., Fan X., Sun Y., Yu D., Zhang J. Study on the relationship between homocys teine &n5,10-methylenetetrahydrofolate reductase and diabetic retinopathy. Tianjin Med. J. 2006;34:4–6. [Google Scholar]
- 25.Maeda M., Yamamoto I., Fukuda M., Motomura T., Nishida M., Nonen S., Fujio Y., Kasayama S., Azuma J. Mthfr gene polymorphism is susceptible to diabetic retinopathy but not to diabetic nephropathy in japanese type 2 diabetic patients. J. Diabetes Complicat. 2008;22:119–125. doi: 10.1016/j.jdiacomp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Ukinc K., Ersoz H.O., Karahan C., Erem C., Eminagaoglu S., Hacihasanoglu A.B., Yilmaz M., Kocak M. Methyltetrahydrofolate reductase c677t gene mutation and hyperhomocysteinemia as a novel risk factor for diabetic nephropathy. Endocrine. 2009;36:255–261. doi: 10.1007/s12020-009-9218-7. [DOI] [PubMed] [Google Scholar]
- 27.Min R. Master Thesis. Tianjin Medical University; Tianjin, China: 2011. Study on Risk Factors and Susceptibility Genes of Diabetic Retinopathy in Patients with Type 2 Diabetes Mellitus. [Google Scholar]
- 28.Yigit S., Karakus N., Inanir A. Association of mthfr gene c677t mutation with diabetic peripheral neuropathy and diabetic retinopathy. Mol. Vis. 2013;19:1626–1630. [PMC free article] [PubMed] [Google Scholar]
- 29.Simoes M.J., Lobo C., Egas C., Nunes S., Carmona S., Costa M.A., Duarte T., Ribeiro L., Faro C., Cunha-Vaz J.G. Genetic variants in icam1, ppargc1a and mthfr are potentially associated with different phenotypes of diabetic retinopathy. Ophthalmologica. 2014;232:156–162. doi: 10.1159/000365229. [DOI] [PubMed] [Google Scholar]
- 30.Niu W., Qi Y. An updated meta-analysis of methylenetetrahydrofolate reductase gene 677c/t polymorphism with diabetic nephropathy and diabetic retinopathy. Diabetes Res. Clin. Pract. 2012;95:110–118. doi: 10.1016/j.diabres.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Zintzaras E., Chatzoulis D.Z., Karabatsas C.H., Stefanidis I. The relationship between c677t methylenetetrahydrofolate reductase gene polymorphism and retinopathy in type 2 diabetes: A meta-analysis. J. Hum. Genet. 2005;50:267–275. doi: 10.1007/s10038-005-0250-z. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y., Chen Y., Cao X., Liu C., Xie Y. Association between plasma homocysteine status and hypothyroidism: A meta-analysis. Int. J. Clin. Exp. Med. 2014;7:4544–4553. [PMC free article] [PubMed] [Google Scholar]
- 33.Zintzaras E., Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J. Clin. Epidemiol. 2008;61:634–645. doi: 10.1016/j.jclinepi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 35.Wang F., Fang Q., Yu N., Zhao D., Zhang Y., Wang J., Wang Q., Zhou X., Cao X., Fan X. Association between genetic polymorphism of the angiotensin-converting enzyme and diabetic nephropathy: A meta-analysis comprising 26,580 subjects. J. Renin-Angiotensin-Aldosterone Syst. JRAAS. 2012;13:161–174. doi: 10.1177/1470320311417655. [DOI] [PubMed] [Google Scholar]
- 36.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 38.Goyette P., Sumner J.S., Milos R., Duncan A.M., Rosenblatt D.S., Matthews R.G., Rozen R. Human methylenetetrahydrofolate reductase: Isolation of cdna, mapping and mutation identification. Nat. Genet. 1994;7:195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 39.Doupis J., Eleftheriadou I., Kokkinos A., Perrea D., Pavlatos S., Gonis A., Katsilambros N., Tentolouris N. Acute hyperhomocysteinemia impairs endothelium function in subjects with type 2 diabetes mellitus. Exp. Clin. Endocrinol. Diabetes. 2010;118:453–458. doi: 10.1055/s-0030-1248290. [DOI] [PubMed] [Google Scholar]
- 40.Akalin A., Alatas O., Colak O. Relation of plasma homocysteine levels to atherosclerotic vascular disease and inflammation markers in type 2 diabetic patients. Eur. J. Endocrinol. 2008;158:47–52. doi: 10.1530/EJE-07-0470. [DOI] [PubMed] [Google Scholar]
- 41.Hoogeveen E.K., Kostense P.J., Eysink P.E., Polak B.C., Beks P.J., Jakobs C., Dekker J.M., Nijpels G., Heine R.J., Bouter L.M., et al. Hyperhomocysteinemia is associated with the presence of retinopathy in type 2 diabetes mellitus: The hoorn study. Arch. Intern. Med. 2000;160:2984–2990. doi: 10.1001/archinte.160.19.2984. [DOI] [PubMed] [Google Scholar]
- 42.Goldstein M., Leibovitch I., Yeffimov I., Gavendo S., Sela B.A., Loewenstein A. Hyperhomocysteinemia in patients with diabetes mellitus with and without diabetic retinopathy. Eye. 2004;18:460–465. doi: 10.1038/sj.eye.6700702. [DOI] [PubMed] [Google Scholar]
- 43.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J., den Heijer M., Kluijtmans L.A., van den Heuvel L.P., et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 44.Weisberg I., Tran P., Christensen B., Sibani S., Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol. Genet. Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 45.Weisberg I.S., Jacques P.F., Selhub J., Bostom A.G., Chen Z., Curtis Ellison R., Eckfeldt J.H., Rozen R. The 1298a→c polymorphism in methylenetetrahydrofolate reductase (MTHFR): In vitro expression and association with homocysteine. Atherosclerosis. 2001;156:409–415. doi: 10.1016/S0021-9150(00)00671-7. [DOI] [PubMed] [Google Scholar]
- 46.Pare G., Chasman D.I., Parker A.N., Zee R.R., Malarstig A., Seedorf U., Collins R., Watkins H., Hamsten A., Miletich J.P., et al. Novel associations of cps1, mut, nox4, and dpep1 with plasma homocysteine in a healthy population: A genome-wide evaluation of 13,974 participants in the women’s genome health study. Circ. Cardiovasc. Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange L.A., Croteau-Chonka D.C., Marvelle A.F., Qin L., Gaulton K.J., Kuzawa C.W., McDade T.W., Wang Y., Li Y., Levy S., et al. Genome-wide association study of homocysteine levels in filipinos provides evidence for cps1 in women and a stronger mthfr effect in young adults. Hum. Mol. Genet. 2010;19:2050–2058. doi: 10.1093/hmg/ddq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egger M., Zellweger-Zahner T., Schneider M., Junker C., Lengeler C., Antes G. Language bias in randomised controlled trials published in English and German. Lancet. 1997;350:326–329. doi: 10.1016/S0140-6736(97)02419-7. [DOI] [PubMed] [Google Scholar]
- 49.Yau J.W., Rogers S.L., Kawasaki R., Lamoureux E.L., Kowalski J.W., Bek T., Chen S.J., Dekker J.M., Fletcher A., Grauslund J., et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012;35:556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y.P., Hallman D.M., Gonzalez V.H., Klein B.E., Klein R., Hayes M.G., Cox N.J., Bell G.I., Hanis C.L. Identification of diabetic retinopathy genes through a genome-wide association study among Mexican-Americans from Starr County, Texas. J. Ophthalmol. 2010;2010:73–74. doi: 10.1155/2010/861291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grassi M.A., Tikhomirov A., Ramalingam S., Below J.E., Cox N.J., Nicolae D.L. Genome-wide meta-analysis for severe diabetic retinopathy. Hum. Mol. Genet. 2011;20:2472–2481. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y.C., Lin J.M., Lin H.J., Chen C.C., Chen S.Y., Tsai C.H., Tsai F.J. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011;118:642–648. doi: 10.1016/j.ophtha.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 53.Sheu W.H., Kuo J.Z., Lee I.T., Hung Y.J., Lee W.J., Tsai H.Y., Wang J.S., Goodarzi M.O., Klein R., Klein B.E., et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum. Mol. Genet. 2013;22:3165–3173. doi: 10.1093/hmg/ddt161. [DOI] [PMC free article] [PubMed] [Google Scholar]