Abstract
AIM
To investigate the expression characteristics of heterogeneous nuclear ribonucleoprotein H1 (HNRNPH1) mRNA and protein in cell lines and tissues of esophageal squamous cell carcinoma (ESCC).
METHODS
Western blotting was used to assess the expression of HNRNPH1 protein in seven ESCC cell lines and 30 paired fresh tissue specimens. The subcellular localization of HNRNPH1 was determined by immunofluorescence in ESCC cells. The RNA sequencing data from 87 patients with ESCC were obtained from the cancer genome atlas (TCGA), and the expression and clinical characteristics analysis of different transcript variants of HNRNPH1 were evaluated in this dataset. In addition, immunohistochemistry was carried out to detect the expression of HNRNPH1 protein in 125 patients.
RESULTS
The expression of HNRNPH1 protein varied across different ESCC cell lines. It was exclusively restricted to the nucleus of the ESCC cells. There are two transcript variants of the HNRNPH1 gene. Variant 1 was constitutively expressed, and its expression did not change during tumorigenesis. In contrast, levels of variant 2 were low in non-tumorous tissues and were dramatically increased in ESCC (P = 0.0026). The high levels of variant 2 were associated with poorer differentiated tumors (P = 0.0287). Furthermore, in paired fresh tissue specimens, HNRNPH1 protein was overexpressed in 73.3% (22/30) of neoplastic tissues. HNRNPH1 was significantly upregulated in ESCC, with strong staining in 43.2% (54/125) of tumor tissues and 22.4% (28/125) of matched non-cancerous tissues (P = 0.0005). Positive HNRNPH1 expression was significantly associated with poor tumor differentiation degree (P = 0.0337).
CONCLUSION
The different alternative transcript variants of HNRNPH1 exhibited different expression changes during tumorigenesis. Its mRNA and protein were overexpressed in ESCC and associated with poorer differentiation of tumor cells. These findings highlight the potential of HNRNPH1 in the therapy and diagnosis of ESCC.
Keywords: Heterogeneous nuclear ribonucleoprotein H1, Esophageal squamous cell carcinoma, Alternative transcript variants, Biomarker
Core tip: Heterogeneous nuclear ribonucleoprotein H1 (HNRNPH1) is an evolutionarily conserved splicing factor. It is involved in alternative splicing, polyadenylation, mRNA export, and translation. This study investigated the expression, localization, and clinical significance of HNRNPH1 in esophageal squamous cell carcinoma (ESCC). We found that this gene possesses two alternative transcript variants; one was constitutively expressed, while the other was regulated and dramatically increased in ESCC. HNRNPH1 protein was overexpressed in ESCC tissues. Strong HNRNPH1 levels were associated with poorer tumor differentiation and alternative splicing of apoptosis-related genes. These findings suggest that HNRNPH1 is a potential diagnostic biomarker and therapeutic target for ESCC.
INTRODUCTION
Esophageal cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer death in China[1]. It was estimated that 477900 new cases and 375000 deaths occurred in 2015, accounting for about 13.3% of all cancer deaths. The predominant histological subtype in China is esophageal squamous cell carcinoma (ESCC).
Under various environmental exposures and genetic factors, ESCC develops through progression from normal esophageal epithelium to dysplasia, early stage and then advanced stage esophageal carcinoma with an accumulation of numerous genetic and epigenetic abnormalities[2]. Currently, alternations in thousands of genes have been found in ESCC. Nascent transcripts that are produced by RNA polymerase II undergo precursor mRNA splicing to generate mature mRNAs. Heterogeneous nuclear ribonucleoproteins (hnRNPs) play critical roles in this process. In addition, hnRNPs act as trans-factors to regulate alternative splicing, gene expression, mRNA export, localization, translation, and stability[3]. In humans, the hnRNPs family consists of at least 20 abundant, major hnRNP proteins and other less abundant, minor hnRNP proteins[3]. Most of the major hnRNP proteins have been shown to be overexpressed in ESCC tissues by proteomic analysis[4-7]. Immunohistochemical staining showed that HNRNPB1 was a potential diagnostic marker for squamous cell carcinoma of various organs, including ESCC[8].
Our previous proteomic study found that a major hnRNP protein, HNRNPH1, was upregulated approximately 8.4-fold in ESCC. Its overexpression was also observed in another proteomic study in ESCC[5]. HNRNPH1 was first purified in 1994 as an abundant component of hnRNP complexes[9]. It is ubiquitously expressed in various human tissues and binds only to poly (rG) sequence[9,10]. HNRNPH1 was demonstrated to stimulate pre-mRNA cleavage and polyadenylation, and it is an important determinant of alternative splicing[11-17]. At present, the relevant reports about HNRNPH1 and cancer are still very limited. Upregulation of HNRNPH1 was found in pancreatic adenocarcinoma, hepatocellular carcinoma, gastric carcinoma, head and neck carcinomas, and colon cancer[18,19]. However, the aberrant expression of HNRNPH1 has not been verified and evaluated in large-scale clinical samples. In this study, we investigated the expression and clinical significance of HNRNPH1 mRNA and protein in ESCC using an RNA sequencing dataset from the cancer genome atlas (TCGA), western blotting, and immunohistochemical staining assays.
MATERIALS AND METHODS
Cell lines and cell cultures
The human ESCC cell lines KYSE30, KYSE140, KYSE170, KYSE180, KYSE410, and KYSE510 were the gifts from Dr. Y. Shimada at Hyogo College of Medicine. EC0156 was established by our laboratory[20]. EC0156 was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, NY, United States). The other cell lines were cultured in Roswell Park Memorial Institute (RPMI)1640 medium. All the cells were incubated at 37 °C in a humidified atmosphere of 5% CO2.
Clinical specimen collection and preparation
Surgical tissues from ESCC patients were collected after obtaining informed consent and approval from the Institutional Review Board of the Cancer Hospital of Chinese Academy of Medical Sciences (CAMS, Beijing, China). A total of 30 fresh tumor and paired adjacent non-tumor esophagus tissue samples were collected from patients (25 male, five female; median age, 58 ± 10 SD; range 32-72 years) undergoing resection from January 2005 to January 2009. All patients were diagnosed by two senior pathologists without chemo/radiotherapy before surgical operation. The tissue samples were collected and washed right after surgical resection. They were then snap-frozen in liquid nitrogen immediately and stored at -80 °C.
For immunohistochemical staining, 50 formalin-fixed, paraffin-embedded tissues specimens were collected from surgically resected ESCC in Cancer Hospital of CAMS from January 1999 to 2009. In addition, a tissue microarray that contained 75 ESCC cases were purchased from Shanghai Outdo Biotech Co., Ltd (Shanghai, China).
Protein extraction
Cells in the exponential phase of growth were harvested using a protein lysis buffer (pH 7.4) containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% nonidet P-40 (NP-40), 0.1% sodium dodecyl sulfate (SDS), and protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). In addition, subcellular protein extraction was performed using ProteoExtractTM Subcellular Proteome Extraction Kit (Calbiochem, Billerica, MA, United States) according to the manufacturer’s guidelines. Fresh tissue samples were homogenized, and the proteins were extracted using the protein lysis buffer described above. The protein content was determined by Coomassie Plus Protein Assay (Pierce, Rockford, IL, Untied States).
Western blot analysis
Approximately 15 μg of total proteins or subcellular proteins were diluted in Laemmli buffer containing 10% β-mercaptoethanol and boiled at 95 °C for 10 min. Samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride membranes. After blocking, the membranes were incubated with anti-HNRNPH1 (ab10374, Abcam, Cambridge, United Kingdom), anti-Lamin B, anti-AIF (Santa Cruz Biotech., Dallas, TX, Untied States), and anti-β-actin (Sigma-Aldrich, St. Louis, MO, United States) antibodies. Following intensive washing, the membranes were developed with horseradish peroxidase conjugated second antibodies (Jackson Immunoresearch Lab., West Grove, PA, United States) and visualized using an enhanced chemiluminescence system (Santa Cruz Biotech.). The upregulation or downregulation of HNRNPH1 was defined as a change in relative band intensity in tumors compared with their paired adjacent normal tissues.
Immunofluorescence staining
EC0156 cells were grown in 0.01% poly-l-Lysine coated slices for 24 h. After being fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with phosphate buffered saline (PBS) (pH 7.4), the cells were blocked with 1% bovine serum albumin (BSA) and 0.1% Triton X-100 for 30 min at room temperature. Washed cells were incubated for 30 min with rabbit anti-HNRNPH1 antibody. The cells were then incubated in the dark for 60 min with Alexa Fluor 488-conjugated goat anti-rabbit secondary IgG (Life Technologies, Carlsbad, CA, United States). The fluorescence signals were captured under a Nikon E400 fluorescence microscope (Nikon Instech Co., Tokyo, Japan).
Immunohistochemistry
The tissue array of paraffin-embedded ESCC and their matched adjacent normal tissues were incubated with HNRNPH1 or control antibodies. After washing with 1 × PBS, slices were reacted with the biotin-labeled second antibody and then visualized using an ultrasensitive streptavidin-peroxidases system (Maxim Biotech, Fuzhou, China). Semi-quantitative analysis of the HNRNPH1 immunoreaction was quantified as described previously[21]. A staining index was used in which 0 was considered negative, 1-4 was weak, and > 4 was considered strong expression.
TCGA RNA sequencing data mining and statistical analysis
The ESCC transcriptome dataset was obtained from TCGA. The normalized transcripts (isoforms) sequencing data from 11 non-tumor tissues and 87 tumor tissues were available. The expression of two HNRNPH1 transcripts and their clinical significance were analyzed. The Mann-Whitney U test was used to compare the RPKM (Reads per kilobase of transcript per million reads mapped) between the two groups. Spearman rank correlation analysis was used to calculate the correlation coefficient of the two transcripts. P values < 0.05 were considered significant. All analyses were performed using GraphPad prism 6.0 (GraphPad Software Inc., La Jolla, CA, United States).
RESULTS
Expression and localization of HNRNPH1 protein in ESCC cell lines
First, we observed the levels of HNRNPH1 protein in several ESCC cell lines. As shown in Figure 1A, HNRNPH1 expression varied across the different ESCC cells, with KYSE30, KYSE140, KYSE410, KYSE170, and EC0156 showing relatively high expression, whereas KYSKE180 and KYSE510 showing relatively low expression levels. Many members in the HNRNP family shuttle rapidly between the nucleus and cytoplasm. The shuttling capacity of HNRNPH1, however, remains unknown. Therefore, we next investigated the subcellular localization of HNRNPH1 via two methods. Immunofluorescence staining showed that it was localized in the nucleus but not the nucleolus (Figure 1B). Furthermore, western blotting analysis of subcellular protein showed that HNRNPH1 was strictly nuclear (Figure 1C). Thus, HNRNPH1 protein is ubiquitously expressed and exclusively sequestered to the nucleus in the ESCC cells.
Figure 1.
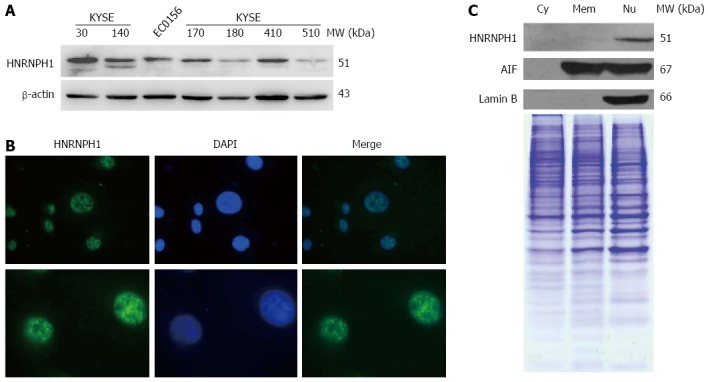
Expression and localization of heterogeneous nuclear ribonucleoprotein H1 in esophageal squamous cell carcinoma cells. A: Protein levels of HNRNPH1 were assessed by Western blots in seven ESCC cell lines. The β-actin protein was used as a loading control; B: Immunofluorescent visualization of HNRNPH1 in EC0156 cells; C: Subcellular protein levels of HNRNPH1 in EC0156 were assessed by western blots. Lamin B is a specific marker for nuclear proteins. AIF is a marker for intracellular membrane and nuclear proteins. Coomassie blue-stained SDS-PAGE gel was used as the loading control. AIF: Apoptosis inducing factor; HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC: Esophageal squamous cell carcinoma; SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis.
HNRNPH1 mRNAs are up-regulated in ESCC tissues
Based on the NCBI RNA reference sequences collection (RefSeq) database (hg19), the HNRNPH1 gene has two transcript variants, NM_001257293 (variant 1) and NM_005520 (variant 2). They are different in the 5’ untranslated region (UTR) region but encode the same protein (Figure 2A). Using the TCGA RNA sequencing gene isoforms data from ESCC patients (n = 87), we compared the abundance of these two variants between tumor and non-tumor tissues. In the non-tumorous tissues (n = 11), variant 1 was constitutively expressed, whereas most of the samples barely expressed variant 2. However, in the tumor tissues, the expression of variant 1 was not altered compared to control (P = 0.3211), whereas variant 2 was significantly up-regulated (P = 0.0026, Figure 2B). Because the samples in TCGA were comprised of different races, we compared the differences of variant 1 and 2 in Asians and Caucasians. Caucasians had slightly higher levels of HNRNPH1 than Asians, but there was no significant difference between the two races (Figure 2B). In addition, the expression of variant 1 was not correlated with that of variant 2 in tumor tissues (P = 0.1201, R = -0.1679; Figure 2C), suggesting that the two variants of HNRNPH1 are regulated by different mechanisms and display different expression characteristics.
Figure 2.
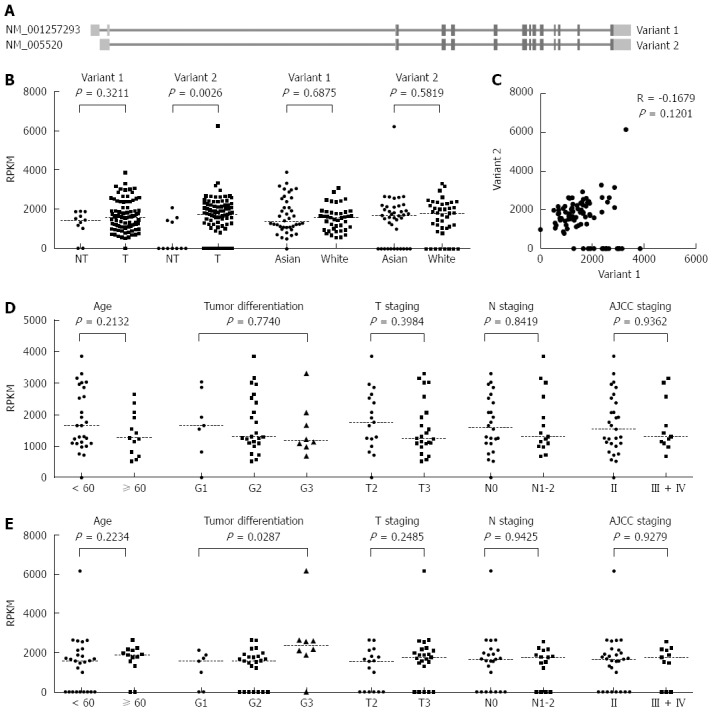
Expression and clinicopathological characteristics of heterogeneous nuclear ribonucleoprotein H1 mRNA presented in the cancer genome atlas RNA sequencing dataset. A: Transcript models for HNRNPH1 in hg19 visualized in the NCBI RefSeq. Human HNRNPH1 was encoded on the reverse strand. Two transcript variants were transcribed with different 5’UTR region. The light grey represents the untranslated region, while dark grey represents the coding region; B: These two transcripts had different expression patterns in ESCC. NT, non-tumor tissues (n = 11); T, tumor tissues (n = 87); C: The correlation analysis between variant 1 and 2 in the tumor tissues of ESCC (n = 87). Pearson correlation coefficients were calculated between their mRNA levels; D and E: The clinicopathological characteristics analysis of variant 1 (D) and variant 2 (E) expression in the Asian ESCC cases (n = 40). T staging, tumor invasive depth; N staging, lymph node metastasis; AJCC staging, the 7th edition cancer staging of American Joint Committee on Cancer. HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC:Esophageal squamous cell carcinoma; UTR: Untranslated region.
Furthermore, we investigated the clinicopathological significance of variant 1 and 2 mRNA levels in Asians. No correlation between variant 1 and clinical features was observed (Figure 2D), whereas the levels of variant 2 were higher in poorly differentiated tumors (P = 0.0287; Figure 2E). Moreover, all of the cases were dichotomized into two groups, a high level group and a low level group, based on the median RPKM values in the tumor tissues. There was no significant relationship between variant 2 expression and overall survival of ESCC patients (Figure 3). Overall, it seems that the variant 1 of HNRNPH1 is constitutively expressed, whereas the expression of variant 2 is modulated. Variant 2 expression was more associated with tumorigenesis in ESCC than variant 1 expression.
Figure 3.
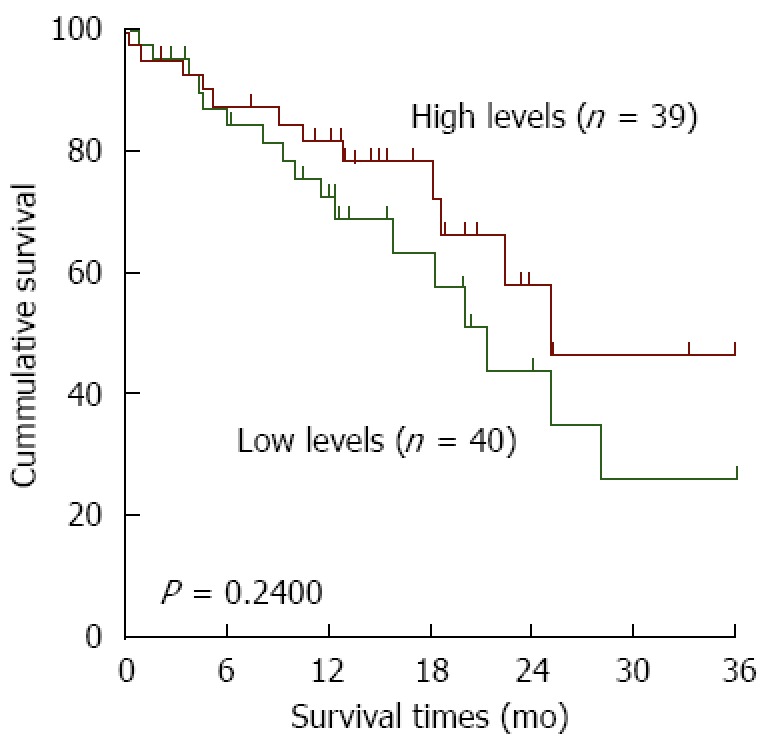
Kaplan-Meier curves of esophageal squamous cell carcinoma patients with low and high levels of variant 2 transcript of heterogeneous nuclear ribonucleoprotein H1 (n = 79). All of the cases were dichotomized into two groups, high level group and low level group, by the median RPKM values in tumor tissues. Log-rank test was used to compare the survival curves (P = 0.2400). HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC:Esophageal squamous cell carcinoma.
HNRNPH1 protein is overexpressed in ESCC tissues
To confirm the observation at the mRNA level, the protein expression of HNRNPH1 in ESCC patients was analyzed using western blotting and immunohistochemical staining assays. First, western blotting results revealed that HNRNPH1 was overexpressed in 73.3% (22/30) of neoplastic tissues compared to non-tumorous esophageal mucosal tissues (Figure 4A and B).
Figure 4.
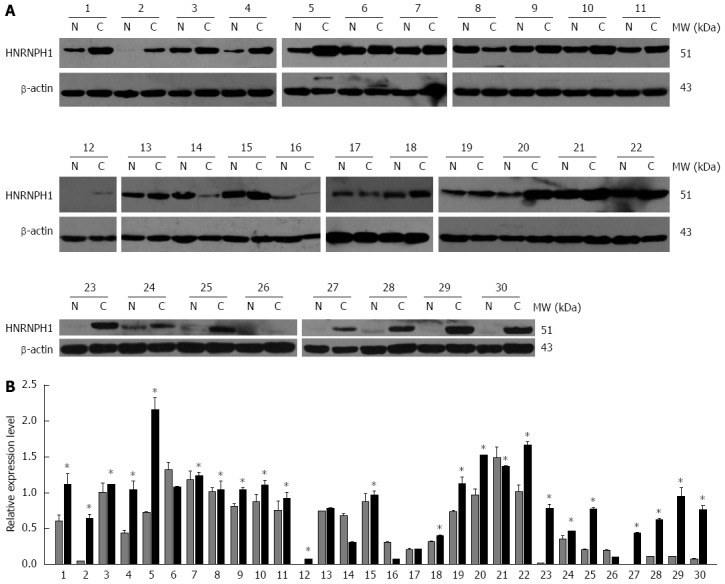
Western blotting analysis of heterogeneous nuclear ribonucleoprotein H1 in paired esophageal squamous cell carcinoma specimens. A: Representative western blotting images of tumor (C) and matched adjacent non-tumor esophageal mucosal tissues (N) from 30 patients with ESCC. β-actin protein levels are shown as a loading control. The patients were coded from 1 to 30. B: Densitometric analysis of 30 ESCC cases. The gray and black bars represent the relative band intensity of HNRNPH1 in non-tumor (N) or tumor (C) tissues. Each data point represents the mean ± SD derived from three independent experiments. The asterisks mark the cases that overexpressed HNRNPH1 in tumor tissues. HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1.
Immunohistochemistry in 125 paired ESCC and non-neoplastic esophageal mucosa was used to evaluate the expression of HNRNPH1 in more detail. The staining of HNRNPH1 was confined to the nuclei in both tumor and non-tumor cells (Figure 5). HNRNPH1 was strongly stained in 43.2% (54/125) of tumor tissues and in 22.4% (28/125) of matched non-cancerous tissues. In addition, HNRNPH1 expression was weak in 17.6% (22/125) of tumors, while its expression was weak in 35.2% (44/125) of normal esophageal epithelia. The upregulation of HNRNPH1 in tumor tissues was statistically significant (P = 0.0005). The correlations between the clinicopathologic characteristics of ESCC patients and the expression of HNRNPH1 in their tumors are summarized in Table 1. Its high expression in ESCC correlated significantly with the poorer tumor differentiation degree (P = 0.0337). However, no correlation was found between its expression and age, gender, and lymph node metastasis (P > 0.05). These trends were consistent with the results demonstrated at the mRNA level. Taken together, these findings suggest that HNRNPH1 is overexpressed in ESCC, and its high expression is associated with worse biological behaviors of tumors.
Figure 5.
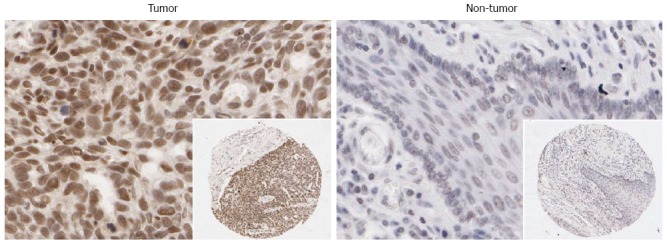
Representative immunohistochemistry staining of heterogeneous nuclear ribonucleoprotein H1 in the tumor and non-tumor tissues from a esophageal squamous cell carcinoma patient. HNRNPH1 was mainly localized to the nuclei. The images are shown at high magnification (× 400), and the lower right panels are magnification × 40. HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC: Esophageal squamous cell carcinoma.
Table 1.
Expression of heterogeneous nuclear ribonucleoprotein H1protein and its clinical significance in 125 esophageal squamous cell carcinomaspecimens n (%)
| Characteristics | All cases (n) |
HNRNPH1 |
P value1 | ||
| Negative | Weak | Strong | |||
| Tissues | |||||
| Normal | 125 | 53 (42.4) | 44 (35.2) | 28 (22.4) | 0.0005 |
| Cancer | 125 | 49 (39.2) | 22 (17.6) | 54 (43.2) | |
| Age (yr) | 0.5422 | ||||
| ≥ 60 | 71 | 26 (36.6) | 16 (22.5) | 29 (40.8) | |
| < 60 | 54 | 23 (42.6) | 6 (11.1) | 25 (46.3) | |
| Gender | 0.9615 | ||||
| Male | 90 | 34 (37.8) | 17 (18.9) | 39 (43.3) | |
| Female | 35 | 15 (42.9) | 5 (14.2) | 15 (42.9) | |
| Tumor differentiation | 0.0652 | ||||
| Well | 29 | 15 (51.7) | 5 (17.2) | 9 (31.1) | |
| Moderately | 72 | 26 (36.1) | 16 (22.2) | 30 (41.7) | |
| Poorly | 24 | 8 (33.3) | 1 (4.2) | 15 (62.5) | |
| Tumor differentiation | 0.0337 | ||||
| Well + moderately | 101 | 41 (40.6) | 21 (20.8) | 39 (38.6) | |
| Poorly | 24 | 8 (33.3) | 1 (4.2) | 15 (62.5) | |
| Lymph Node Metastasis2 | 0.1839 | ||||
| Present | 42 | 14 (33.3) | 9 (21.4) | 19 (45.2) | |
| Not present | 36 | 18 (50.0) | 7 (19.4) | 11 (30.6) | |
The strong expression of HNRNPH1 was compared with negative and weak expression;
Only 78 cases had information on lymph node metastasis. HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC: Esophageal squamous cell carcinoma.
HNRNPH1 regulates cell proliferation and apoptosis-related genes in ESCC
Previously, it was shown that HNRNPH1 modulates the alternative splicing of apoptotic mediators Bcl-x and A-raf[18]. To clarify further the biological significance of HNRNPH1 in ESCC, we performed a correlation analysis between variant 2 of HNRNPH1 and the alternative transcripts of these genes. In addition, the cell proliferation marker MKI67, which encodes Ki-67 protein, was included. As shown in Figure 6A and B, variant 2 was positively correlated with the expression of MKI67 (Rho = 0.3101, P = 0.0035) and the prominent transcript of A-raf (Rho = 0.2787, P = 0.0090). Moreover, variant 2 was inversely correlated with the pro-apoptotic transcript Bcl-X [Bcl-X (S), Rho = -0.2349, P = 0.0285; Figure 6C]. Therefore, overexpression of variant 2 of HNRNPH1 contributes to cell growth and anti-apoptosis in ESCC.
Figure 6.
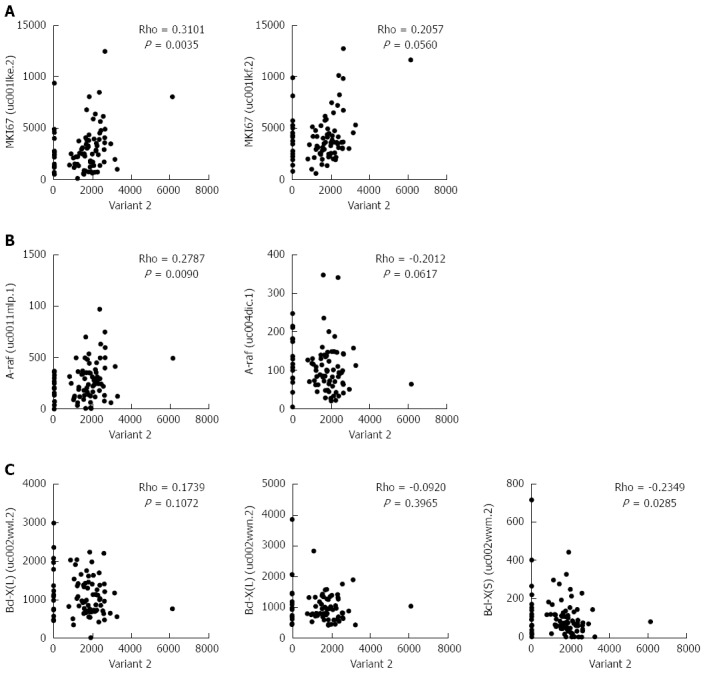
Expression levels of transcript variant 2 of heterogeneous nuclear ribonucleoprotein H1 were associated with the alternative splicing of proliferation- and apoptosis-related genes in esophageal squamous cell carcinoma. A: The correlation analysis between variant 2 and the two transcripts of MKI67 gene; B: The correlation analysis between variant 2 and the A-raf gene; C: The correlation analysis between variant 2 and the Bcl-X gene. HNRNPH1: Heterogeneous nuclear ribonucleoprotein H1; ESCC: Esophageal squamous cell carcinoma.
DISCUSSION
The human HNRNPH1 gene is mapped to the reverse strand at 5q35.3. It encodes two transcript variants based on the annotation of NCBI RefSeq hg19. Variant 1 possesses 14 exons, whereas variant 2 has 13 exons (Figure 2A). They are differed at the 5’UTR region but encode the same protein. Until now, there were no studies describing the expression characteristics of these two variants. Our results showed that variant 1 is constitutively expressed, and it is not regulated by the disordered signal transduction networks during carcinogenesis. Meanwhile, variant 2 is barely expressed in normal tissues. These results are consistent with previous findings that have shown that the expression of HNRNPH1 was unaffected by treatment with two second messengers and seven cytokines in normal human keratinocytes[10].In addition, we found that variant 2 is significantly overexpressed in ESCC and correlated with poorer tumor differentiation. These results suggest that the transcript variant 1 of HNRNPH1 is responsible for maintaining its invariable intracellular levels, whereas variant 2 is regulated and responds to tumorigenesis. We have identified a new mechanism for the gene expression regulation of HNRNPH1.
HNRNPH1 protein has three RNA binding domains, a glycine-tyrosine-arginine-rich (GYR) domain and a C-terminus glycine-rich domain. The central GYR domain is responsible for its nuclear localization[22]. HNRNPH1 can shuttle between the nucleus and cytoplasm[22], and strong cytoplasmic staining was observed in some cases of pancreatic, rectal, liver, gastric, and lung cancer[19]. In head and neck cancer, however, HNRNPH1 was only overexpressed in the nuclei[18]. Our results showed that HNRNPH1 was restricted to the nucleus in ESCC. Therefore, the intracellular localization of HNRNPH1 may be tissue or cell type-dependent.
HNRNPH1 is an evolutionarily conserved splicing factor, and it plays a dual role in the activation and inhibition of pre-mRNA processing, polyadenylation, mRNA export, and translation. It can act as a component in the intronic splicing enhancer complex to stimulate gene splicing, including c-src, MAP kinase activating death domain (MADD), and macrophage stimulating 1 receptor (MST1R)[13,17]. In addition, it can be recruited to the exonic splicing silencer to regulate alternative splicing of tropomyosin, collagen-like tail subunit (COLQ), muscle nicotinic acetylcholine receptor alpha subunit, and fibroblast growth factor receptor 2[11,12,16,23,24]. HNRNPH1 can cooperate with other hnRNP proteins to stimulate polyadenylation through a direct interaction with poly (A) polymerase[25]. Moreover, it has been shown to bind some mRNAs to inhibit their nuclear export[26]. In amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), the RNA foci that result from GGGGCC (G4C2) intronic repeat expansion within C9ORF72 sequester HNRNPH1 and other RNA binding proteins, leading to neurotoxicity[27]. In view of these functions, aberrant expression of HNRNPH1 will alter cell phenotypes. Apoptosis was significantly activated when HNRNPH1 was depleted. Experiments showed that inhibition of HNRNPH1 induced apoptosis by activation of caspase-3, poly(ADP-ribose)polymerase (PARP) cleavage, and 10-fold increases in DNA fragmentation, whereas its transient overexpression was protective against etoposide-induced apoptosis[18]. The anti-apoptotic role of HNRNPH1 may result from its substrates, A-raf and Bcl-X[18,28]. Bcl-X encodes two isoforms with different functions. The longer isoform Bcl-X (L), which is translated from transcripts uc002wwl.2 and uc002wwn.2, acts as an apoptotic inhibitor, whereas the shorter form Bcl-X (S), which is translated from transcript uc002wwm.2, acts as an apoptotic activator. It also maintains p53 pre-mRNA 3’-end processing to contribute to p53-mediated apoptosis[15]. In this study, we observed that the mRNAs and proteins of HNRNPH1 are all increased in ESCC. These increases in HNRNPH1 levels promote cell proliferation and inhibit apoptosis partially via upregulating the pro-proliferative and anti-apoptotic transcripts of MKI67 and A-raf and restraining the pro-apoptotic transcripts of Bcl-X. These findings indicate that it may enhance the chemoresistance of ESCC cells.
In conclusion, our results demonstrated that there are two transcript variants of HNRNPH1. One is constitutively expressed, and the other is regulated. The regulated mRNA variant led to the overexpression of HNRNPH1 protein in ESCC. Its expression was restricted to the nucleus and was associated with poorer differentiation of tumor cells. Our study is the first to investigate the aberrant expression of HNRNPH1 in ESCC and highlights the potential of HNRNPH1 in the therapy and diagnosis of ESCC.
COMMENTS
Background
Esophageal squamous cell carcinoma (ESCC) is the third most commonly diagnosed cancer and the fourth leading cause of cancer death in China.Currently, thousands of gene alternations have been found in ESCC. One of these genes encodes heterogeneous nuclear ribonucleoprotein H1 (HNRNPH1), an evolutionarily conserved splicing factor that is involved in alternative splicing, polyadenylation, mRNA export, and translation.
Research frontiers
Previous proteomic studies have found that HNRNPH1 was upregulated approximately 8.4-fold in ESCC. However, data regarding the association between HNRNPH1 and cancer are still very limited.
Innovations and breakthroughs
This study is the first to investigate the expression, localization, and clinical significance of HNRNPH1 in cell lines and clinical specimens of ESCC using RNA sequencing dataset, western blotting, immunofluorescence, and immunohistochemistry staining.
Applications
There are two alternative transcript variants of HNRNPH1; one was constitutively expressed, while the other was regulated and dramatically increased in ESCC. HNRNPH1 protein was also overexpressed in ESCC tissues. Strong HNRNPH1 levels were significantly associated with poorer tumor differentiation, suggesting that it may be a potential diagnostic biomarker and therapeutic target in ESCC.
Terminology
Heterogeneous nuclear ribonucleoproteins (hnRNPs) play critical roles in precursor mRNA splicing, alternative splicing, gene expression, mRNA export, localization, translation, and stability regulation. In humans, the hnRNP family consists of at least 20 abundant, major hnRNP proteins and other less abundant, minor hnRNP proteins. HNRNPH1 is an ubiquitously expressed major hnRNP protein thatbinds only to the poly (rG) sequence.
Peer-review
This manuscript clearly demonstrated that HNRNPH1 is overexpressed in human ESCC and is associated with poor differentiation. Overexpression of HNRNPH1 was associated with alternative splicing of some proliferation and apoptosis related genes. Overall, the manuscript is well organized, and the data are convincing. However, the functional consequences of HNRNPH1 overexpression in human ESCC still need further exploration.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: All routine clinic biopsy specimens and blood samples from the patients were taken after informed consent and ethical permission was obtained for participation in the study.
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Data sharing statement: Technical appendix and dataset available from the corresponding author. Participants gave informed consent for data sharing. No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: March 28, 2016
First decision: May 12, 2016
Article in press: June 2, 2016
P- Reviewer: Chen XL S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Wang CH
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Li SQ, Li F, Xiao Y, Wang CM, Tuo L, Hu J, Yang XB, Wang JS, Shi WH, Li X, et al. Comparison of long noncoding RNAs, microRNAs and messenger RNAs involved in initiation and progression of esophageal squamous cell carcinoma. Mol Med Rep. 2014;10:652–662. doi: 10.3892/mmr.2014.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan NJ, Gao CF, Wang CS, Zhao G, Lv JJ, Wang XL, Chu GH, Yin J, Li DH, Chen X, et al. Identification of the up-regulation of TP-alpha, collagen alpha-1(VI) chain, and S100A9 in esophageal squamous cell carcinoma by a proteomic method. J Proteomics. 2012;75:3977–3986. doi: 10.1016/j.jprot.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Pawar H, Kashyap MK, Sahasrabuddhe NA, Renuse S, Harsha HC, Kumar P, Sharma J, Kandasamy K, Marimuthu A, Nair B, et al. Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery. Cancer Biol Ther. 2011;12:510–522. doi: 10.4161/cbt.12.6.16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du XL, Hu H, Lin DC, Xia SH, Shen XM, Zhang Y, Luo ML, Feng YB, Cai Y, Xu X, et al. Proteomic profiling of proteins dysregulted in Chinese esophageal squamous cell carcinoma. J Mol Med (Berl) 2007;85:863–875. doi: 10.1007/s00109-007-0159-4. [DOI] [PubMed] [Google Scholar]
- 7.Kashyap MK, Harsha HC, Renuse S, Pawar H, Sahasrabuddhe NA, Kim MS, Marimuthu A, Keerthikumar S, Muthusamy B, Kandasamy K, et al. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther. 2010;10:796–810. doi: 10.4161/cbt.10.8.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sueoka E, Sueoka N, Goto Y, Matsuyama S, Nishimura H, Sato M, Fujimura S, Chiba H, Fujiki H. Heterogeneous nuclear ribonucleoprotein B1 as early cancer biomarker for occult cancer of human lungs and bronchial dysplasia. Cancer Res. 2001;61:1896–1902. [PubMed] [Google Scholar]
- 9.Matunis MJ, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honoré B, Rasmussen HH, Vorum H, Dejgaard K, Liu X, Gromov P, Madsen P, Gesser B, Tommerup N, Celis JE. Heterogeneous nuclear ribonucleoproteins H, H’, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J Biol Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 11.Masuda A, Shen XM, Ito M, Matsuura T, Engel AG, Ohno K. hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum Mol Genet. 2008;17:4022–4035. doi: 10.1093/hmg/ddn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decorsière A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3’-end processing and function during DNA damage. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman MA, Azuma Y, Nasrin F, Takeda J, Nazim M, Bin Ahsan K, Masuda A, Engel AG, Ohno K. SRSF1 and hnRNP H antagonistically regulate splicing of COLQ exon 16 in a congenital myasthenic syndrome. Sci Rep. 2015;5:13208. doi: 10.1038/srep13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, Pan YX, Cartegni L. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011;30:4084–4097. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauch J, O’Neill E, Mack B, Matthias C, Munz M, Kolch W, Gires O. Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells by regulating A-Raf transcription. Cancer Res. 2010;70:1679–1688. doi: 10.1158/0008-5472.CAN-09-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honoré B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H’ show differential expression in normal and selected cancer tissues. Exp Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Zhou L, Huang J, Liu F, Yu J, Zhan Q, Zhang L, Zhao X. Role of Smac in determining the chemotherapeutic response of esophageal squamous cell carcinoma. Clin Cancer Res. 2011;17:5412–5422. doi: 10.1158/1078-0432.CCR-11-0426. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Sun YL, Xu Y, Liu F, Wang LS, Zhao XH. Expression and phosphorylation of stathmin correlate with cell migration in esophageal squamous cell carcinoma. Oncol Rep. 2013;29:419–424. doi: 10.3892/or.2012.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Dusen CM, Yee L, McNally LM, McNally MT. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol Cell Biol. 2010;30:2552–2562. doi: 10.1128/MCB.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohail M, Cao W, Mahmood N, Myschyshyn M, Hong SP, Xie J. Evolutionarily emerged G tracts between the polypyrimidine tract and 3’ AG are splicing silencers enriched in genes involved in cancer. BMC Genomics. 2014;15:1143. doi: 10.1186/1471-2164-15-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millevoi S, Decorsière A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3’ end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DH, Langlois MA, Lee KB, Riggs AD, Puymirat J, Rossi JJ. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]


