Abstract
Pitx2 is a conserved homeodomain transcription factor that has multiple functions during embryonic development. Mutations in human PITX2 cause autosomal dominant Axenfeld-Rieger syndrome (ARS), characterized by congenital eye and tooth malformations. Pitx2−/− knockout mouse models recapitulate aspects of ARS, but are embryonic lethal. To date, ARS treatments remain limited to managing individual symptoms due to an incomplete understanding of PITX2 function. In addition to regulating eye and tooth development, Pitx2 is a target of a conserved Nodal (TGFβ) signaling pathway that mediates left-right (LR) asymmetry of visceral organs. Based on its highly conserved asymmetric expression domain, the Nodal-Pitx2 axis has long been considered a common denominator of LR development in vertebrate embryos. However, functions of Pitx2 during asymmetric organ morphogenesis are not well understood. To gain new insight into Pitx2 function we used genome editing to create mutations in the zebrafish pitx2 gene. Mutations in the pitx2 homeodomain caused phenotypes reminiscent of ARS, including aberrant development of the cornea and anterior chamber of the eye and reduced or absent teeth. Intriguingly, LR asymmetric looping of the heart and gut was normal in pitx2 mutants. These results suggest conserved roles for Pitx2 in eye and tooth development and indicate Pitx2 is not required for asymmetric looping of zebrafish visceral organs. This work establishes zebrafish pitx2 mutants as a new animal model for investigating mechanisms underlying congenital malformations in ARS and high-throughput drug screening for ARS therapeutics. Additionally, pitx2 mutants present a unique opportunity to identify new genes involved in vertebrate LR patterning. We show Nodal signaling—independent of Pitx2—controls asymmetric expression of the fatty acid elongase elovl6 in zebrafish, pointing to a potential novel pathway during LR organogenesis.
Keywords: Pitx2, Axenfeld-Rieger syndrome, Left-right asymmetry, Elovl6, Genome editing, Zebrafish
1. Introduction
Pitx2 (Paired-like homeodomain 2) is a member of the bicoid class of homeodomain transcription factors that has been highly conserved during evolution. Vertebrate Pitx2 genes encode multiple protein isoforms that have different N-termini but share a common C-terminal homeodomain that mediates DNA binding (Chaney et al., 2005). Loss-of-function mutations in the homeodomain of human PITX2 cause autosomal dominant Axenfeld-Rieger syndrome (ARS). PITX2 haploinsufficiency in ARS patients disrupts development of the anterior segment of the eye and tooth morphogenesis (Footz et al., 2009). Ocular defects found in ARS patients include malformation of the anterior chamber angle, central corneal thickness and iris atrophy with corectopia. A persistence of the endothelial layer on the angular structures and an anterior iris root insertion result in aqueous outflow defects. As a consequence, patients often have high intraocular pressure and are at risk for developing glaucoma that can lead to blindness (Chang et al., 2012; Shields et al., 1985). Dental abnormalities caused by PITX2 mutations include microdontia, hypodontia and enamel hypoplasia (Jena and Kharbanda, 2005). Additional craniofacial malformations such as hypertelorism, a broad flat nasal bridge and maxillary defects are associated with ARS (Antevil et al., 2009). ARS patients generally have a normal life span and treatment options are currently limited to managing individual symptoms, which require complex multidisciplinary approaches.
Animal models have played an important role in our understanding of Pitx2 function and underlying causes of ARS malformations. Targeting the Pitx2 homeodomain in knockout mouse models eliminated the activity of all Pitx2 isoforms and caused eye and tooth developmental defects in Pitx2−/− embryos that are consistent with ARS (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999). However, global loss of Pitx2 resulted in additional developmental malformations in the brain, visceral organs and body wall and was embryonic lethal. Most heterozygous Pitx2 knockout mice developed into normal adults, but ~10% developed ARS phenotypes that included small eyes, tooth defects and reduced body size (Gage et al., 1999). Additional mutant alleles have been used to study Pitx2 isoforms and gene dosage effects in mouse (Liu et al., 2001, 2002, 2003) and conditional alleles have helped define tissue-specific roles, such as the requirement for Pitx2 in neural crest cells for normal eye development (Evans and Gage, 2005). However, fundamental gaps in our understanding of Pitx2 functions and target genes have made it difficult to conceptualize therapeutic approaches for ARS patients. Thus, it will be critical to exploit existing knockout mouse models and develop new animal models to fully understand molecular functions of PITX2 and identify effective treatments for ARS.
One of the roles of Pitx2 during embryonic development is to function as a downstream effector of an ancient Nodal signaling pathway associated with generating morphological asymmetries in cnidarians (Watanabe et al., 2014), echinoderms (Duboc et al., 2005) and chordates (Boorman and Shimeld, 2002). In vertebrates, the secreted TGFβ-related ligand Nodal triggers asymmetric expression of a specific Pitx2 isoform, Pitx2c, in left lateral plate mesoderm during left-right (LR) patterning of the embryo (Hamada et al., 2002). Unlike Nodal, asymmetric Pitx2c expression was found to persist on left side of the developing heart and gut, where it is thought to regulate genes that mediate asymmetric morphogenesis of these organs. In chicken and frog embryos, misexpression of Pitx2 or Nodal in right lateral plate mesoderm reversed heart and gut looping (Levin et al., 1997; Ryan et al., 1998; Sampath et al., 1997) and blocking Pitx2 or Nodal function randomized looping direction (Toyoizumi et al., 2005; Yu et al., 2001). Further analyses in chick embryos revealed Pitx2c induces cellular condensations during gut looping (Davis et al., 2008; Welsh et al., 2013). Together, these findings suggested Nodal-Pitx2 signaling controls direction of asymmetric organ development. Consistent with this model, Nodal mouse mutants showed randomized heart and gut looping (Brennan et al., 2002). However, the initial looping of the heart and gut occurred normally in Pitx2 knockout mice (Gage et al., 1999; Lin et al., 1999; Lu et al., 1999). Abnormalities during subsequent steps of asymmetric organ morphogenesis in Pitx2 knockout mice, and mice specifically lacking Pitx2c expression (Liu et al., 2001; Shiratori et al., 2006), resulted in organ-specific LR defects that included lung isomerism, abnormal looping of the duodenum and cardiac malformations. Cardiac outflow tract defects were subsequently linked to Pitx2 functions in the second heart field that contributes to outflow tract development (Ai et al., 2006). These studies revealed that the roles of Pitx2 downstream of Nodal in LR patterning remain unclear and may not be completely conserved.
The zebrafish provides a useful model system to investigate gene function and evolution, and the small size and external development of the zebrafish embryo makes it ideal for high-throughput chemical screening (Zon and Peterson, 2005). Since pitx2 mutations have not been identified in genetic screens, we generated the first zebrafish pitx2 mutants via genome editing to gain insight into Pitx2 functions and develop a new ARS animal model. We found that mutations that truncate the zebrafish homeodomain, which is 100% identical to human PITX2, caused eye and tooth developmental defects consistent with phenotypes observed in ARS patients. Importantly, homozygous pitx2−/− zebrafish grow to adulthood and thereby provide a new model of adult ARS. Interestingly, analysis of the heart and gut looping in mutant embryos revealed Pitx2 is dispensable for correct LR asymmetric orientation of these organs. These results identify functions for Pitx2 in eye and tooth development that appear conserved from fish to mammals, and indicate Pitx2 is not required for LR asymmetric looping of visceral organs in zebrafish. Mutant pitx2 zebrafish are an important new disease model that can be used in chemical screens for therapeutic treatments of ARS and can be leveraged to discover new pathways that guide LR morphogenesis. To begin to identify new candidate molecules that may regulate LR development, we show asymmetric expression of the ELOVL fatty acid elongase 6 (elovl6) gene is controlled by LR Nodal signaling and is independent of Pitx2 function in zebrafish.
2. Material and methods
2.1. Zebrafish
Wild-type TAB zebrafish, obtained from Zebrafish International Resource Center, were used to generate pitx2 mutants and for microinjections. cftrpd1048 mutants were provided by the Bagnat lab (Duke University). All animal studies were approved by the Institutional Animal Care and Use Committee at SUNY Upstate Medical University.
2.2. Generation of pitx2 mutant zebrafish
pitx2HD and Pitx2c TALENs were designed using TAL Effector-Nucleotide Targeter 2.0 and assembled using golden gate assembly (Cermak et al., 2011). TALEN mRNAs were synthesized using the Ambion mMessage Machine kit and purified by LiCl precipitation. 150–200 pg of each TALEN mRNA was injected into wild-type zebrafish embryos at the one cell stage. Injected embryos were raised to adulthood and then outcrossed with wild-type fish to identify founders that transmitted mutations through the germ line. Mutations were identified via digestion of PCR products with SalI (pitx2HD TALEN) or BamHI (pitx2c TALEN) and confirmed by sequencing. The PCR primers used for the pitx2HD alleles were forward primer 5′-TGAAGCTTGTTCCTCTGC-3′ and Reverse primer 5′-AAAATTTAGGGTTATATCACATA-3′ and for the pitx2c allele were forward primer 5′ GGAGTG TCGCTTTTAGTGG-3′ and reverse primer 5′-ACTAGAGGCCATCGAAAGC-3′.
2.3.. mRNA synthesis and microinjection
For cloning of pitx2 cDNAs, total RNA was extracted from pooled embryos of pitx2HDsny15 intercross at 6 dpf and RT-PCR was used to amplify full-length pitx2c (primers: 5′-agatctatgacctctat-gaaggatcc 3′ and 5′-tctagattacaccggtctatccactg-3′). The product was inserted into a Topo vector (Invitrogen) and confirmed by sequencing. Confirmed pitx2c cDNAs were transferred into pCS2 vectors and then linearized for mRNA synthesis using the Sp6 mMessage mMachine kit (Ambion). 50 pg of wild-type or mutant pitx2c mRNA was injected into wild-type embryos at the 1-cell stage. Full-length elovl6 was amplified from total RNA from 1 dpf wild type embryos (primers: 5′-cgcggatccatgtcggtgctggcattg-3′ and ccgcctcgagttattggcttttcttggctgc-3′) and cloned using a pCS2 vector. mRNA was prepared the same as pitx2c and 300 pg was injected into 1-cell stage embryos.
2.4. Eye sectioning
5 day old zebrafish were euthanized and flash frozen in O.C.T (Sakura) and cryosectioned as described (Uribe and Gross, 2007). Sections for every 5 μm or 10 μm were collected for 5 dpf and 3 month fish respectively. Slides were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet (Sigma) 5 min at room temperature. Stained sections were dehydrated using a series of ethanol rinses, cleared with xylene and mounted in Poly-Mount medium (Polysciences).
2.5. Tooth staining
Staining of mineralized pharyngeal teeth was performed as described (Walker and Kimmel, 2007). Larvae at 5 dpf were fixed in 4% paraformaldehyde in PBS for 2 h. After 50% ethanol dehydration, embryos were stained with alizarin red solution (0.005% alizarin red and 60 mM MgCL2 in 70% ethanol) overnight. Embryos were cleared with 0.25% KOH in glycerol and imaged using a stereo microscope.
2.6. RNA in situ hybridizations
Whole mount RNA in situ hybridizations were conducted as described (Gao et al., 2010). In addition to previously described probes (cmlc2, foxa3 and spaw), pitx2c and elovl6 probes were made from corresponding cDNAs amplified from a zebrafish cDNA library (see primer sequences above) and cloned using a TOPO TA cloning Kit (invitrogen). elovl6 expression was analyzed in cftrpd1048 mutants (Navis et al., 2013) or embryos injected with Lgl2 morpholino (5′-gcccatgacgcctgaacctcttcat-3′) (Tay et al., 2013). gh1 (primers: 5′-atggctagagcattggtgc-3′ and 5′-tacagggtacagttggaatc-3′) and pomca (primers: 5′-atggtgaggggagtgaggatg-3′ and tcact-catccttcctcggttg-3′) cDNA fragments were cloned using a TOPO TA cloning kit with a pCRII-TOPO vector (Invitrogen) and used as templates for antisense probes. The areas of gh1 and pomca expression domains were measured using Zeiss AxioVision software. To genotype embryos following in situ hybridization analysis, DNA was extracted from individual embryos by boiling in 100 μl 50 mM NaOH and analyzed via PCR and restriction digest as described above.
2.7. Small molecule treatments
Embryos were cultured in 40 μM of SB-505142 (Sigma) in 0.2% DMSO between the tailbud and 18 somite stages. Embryos were then fixed at 18 somite stage for pitx2c or elovl6 in situ hybridizations. Control embryos were treated with 0.2% DMSO alone.
3. Results
3.1.. Generation of mutant pitx2 zebrafish
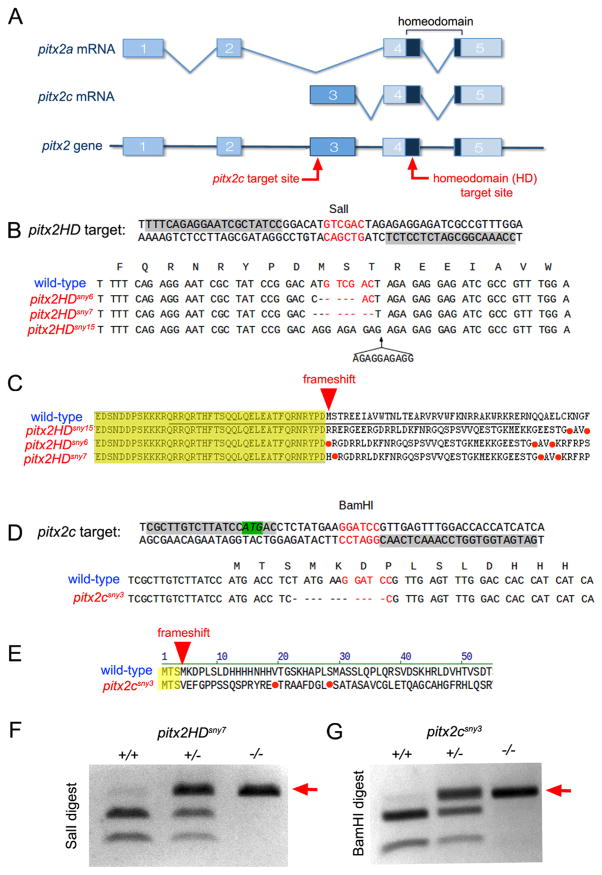
The zebrafish genome contains a single pitx2 gene that encodes two isoforms that correspond to human PITX2A and PITX2C. To disrupt zebrafish Pitx2 function, we used TALEN-mediated genome editing (Joung and Sander, 2013) to create small insertion or deletion (indel) mutations in the pitx2 gene. We targeted the homeodomain to disrupt both pitx2a and pitx2c or the first exon of pitx2c to generate pitx2c-specific mutations (Fig. 1A). Three different indel mutations in the homeodomain were recovered, which we refer to as pitx2HD alleles designated as pitx2HDsny6, pitx2HDsny7 and pitx2HDsny15 (Fig. 1B). All three pitx2HD mutations result in a frameshift and premature stop codon in the second helix of the homeodomain (Fig. 1C; Fig. S1A), which is predicted to eliminate DNA binding (Chaney et al., 2005) and result in loss of function of both Pitx2a and Pitx2c proteins. In addition, we recovered one pitx2c-specific mutation, designated as pitx2csny3 (Fig. 1D), that causes a frameshift after the third amino acid and a stop codon in the N-terminus of the protein (Fig. 1E). Both pitx2HD and pitx2c mutations were identified by loss of a restriction enzyme cut site in the target sequence, which provides an efficient assay for genotyping (Fig. 1F and G).
Fig. 1.
Genome editing of pitx2 in zebrafish. (A) Intron and exon (boxes) structure of zebrafish pitx2a and pitx2c transcripts and the pitx2 gene. The homeodomain encoded by exons 4–5 is dark blue. Red arrows indicate TALEN target sites. (B) Sequence of the pitx2HD target site and pitx2HD alleles. TALEN binding sites are highlighted gray and a SalI restriction site in the spacer is red. pitx2HD alleles are aligned with the wild-type sequence; deleted nucleotides are indicated by dashes. pitx2HDsny15 has an insertion. (C) Predicted peptide sequences for wild-type and pitx2HD alleles. A red arrow indicates a frameshift and a red dot indicates a premature stop codon. The HD sequence is underlined. (D) Sequence of the pitx2c target site and pitx2c allele. The pitx2c start codon is highlighted green and a BamHI restriction site is red. The mutant pitx2csny3 allele is aligned with the wild-type sequence. (E) The predicted peptide sequence of the pitx2csny3 allele revealed a frameshift after the third amino acid in Pitx2c. (F, G) Genotyping results using loss of restriction digest to identify mutant pitx2HD (F) or pitx2c (G) alleles (arrows). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
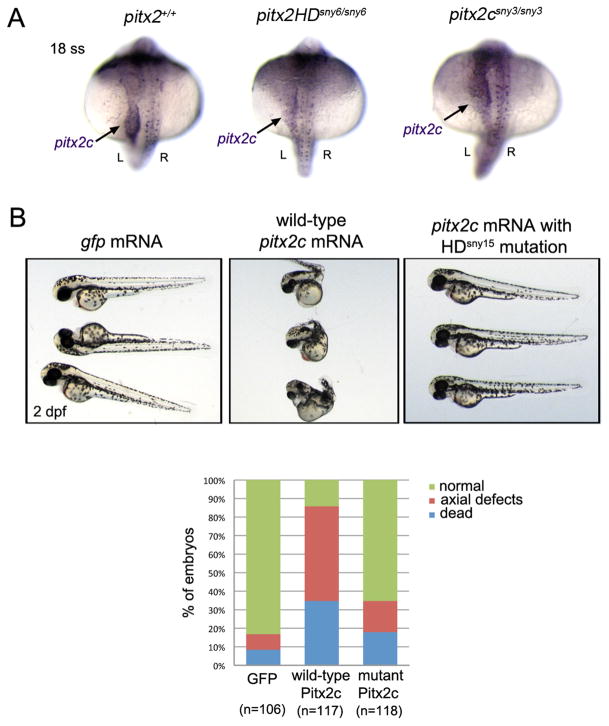
To characterize the effect of these mutations on pitx2 expression, we first analyzed pitx2 transcripts in mutant embryos. RNA in situ hybridizations revealed pitx2 mRNAs were present in mutants, including asymmetric expression of pitx2c in left lateral plate mesoderm (Fig. 2A). RT-PCR and sequencing confirmed only mutant pitx2 mRNAs were present in the homozygous mutants (Fig. S1B). Attempts to detect zebrafish Pitx2 protein using commercially available antibodies raised against human PITX2 were not successful in immunohistochemistry or Western blot experiments, so we cloned the mutant pitx2HDsny15 transcripts for functional analysis. Similar to previous observations (Essner et al., 2000), injecting wild-type pitx2a or pitx2c mRNA into wild-type embryos resulted in severe malformations that included axial defects (Fig. 2B). In contrast, most embryos injected with mutant pitx2c mRNA that contained the HDsny15 frameshift mutation developed normally (Fig. 2B). These results suggested pitx2HD mutations were loss-of-function alleles.
Fig. 2.
Functional analysis of mutant pitx2HD transcripts. (A) RNA in situ hybridizations revealed normal pitx2 mRNA expression in wild-type, pitx2HD−/− and pitx2c−/− embryos, including asymmetric pitx2c in lateral plate mesoderm (arrow) at the 18 somite stage (18 ss). L=left; R=right. (B) Microinjection of wild-type pitx2c mRNA into wild-type embryos resulted in severe developmental defects or lethality at 2 dpf, which was not frequently observed in controls injected with gfp mRNA. Most embryos injected with pitx2c mRNA containing the HDsny15 mutation developed normally. n=number of embryos analyzed (pooled from three experiments).
We next characterized development of embryos that inherited the edited pitx2 alleles. Expression of pitx2a and pitx2c is not maternally supplied (Fig. S2), but both isoforms are expressed during gastrulation in mesendoderm and/or prechordal plate (Essner et al., 2000). These early expression domains suggested loss-of-function might cause early patterning defects in the embryo. However, all offspring from pitx2HDsny7/+ parents were grossly indistinguishable through the first 5 days of development (Fig. 3A). Genotyping individual embryos revealed the expected Mendelian ratio of heterozygous and homozygous genotypes. Normal gross morphology was also observed in pitx2HDsny6/sny6 and pitx2HDsny15/sny15 embryos at 5 days post-fertilization (dpf) (Fig. S3A). Homozygous mutant pitx2HD fish survived to adulthood at variable rates (Table S1). Adult pitx2HD fish were smaller overall than wild-type siblings (Fig. 3B) and had variable eye defects that appeared to affect the iris and cornea (Fig. 3C, Fig. S3B). Bilateral eye phenotypes were observed in nearly all pitx2HDsny7 (n=17/17), pitx2HDsny6 (n=4/4) and pitx2HDsny15 (n=5/6) affected embryos. Intriguingly, 1/6 mutant pitx2HDsny15 embryos had a defect in only one eye. Similar to pitx2HD mutants, embryos homozygous for the pitx2c-specific mutation (pitx2csny3/sny3) were indistinguishable from wild-type at 5 dpf (Fig. 3A) and survived to adulthood. However, adult pitx2csny3/sny3 fish were the same size as wild-type siblings (Fig. 3B) and eye phenotypes were not observed (Fig. 3C). Interestingly, based on external morphological features, we have recovered only male pitx2csny3/sny3 adults (10 homozygous mutant males out of 57 adults genotyped). Male sex was confirmed by the presence of testes (Fig. S4) and outcrosses with wild-type females resulted in viable embryos (7/16 crosses were successful) indicating pitx2csny3/sny3 males are fertile. This finding raises the possibility that Pitx2c may be involved in zebrafish sex determination or gonad development as described in chicken (Guioli and Lovell-Badge, 2007; Rodriguez-Leon et al., 2008).
Fig. 3.
Pitx2HD mutants grow to adulthood and develop eye defects. (A) Normal gross appearance of wild-type, pitx2HDsny7/sny7 and pitx2csny3/sny3 fish at 5 dpf. (B) Homozygous pitx2HD mutants reached adulthood, but were smaller than heterozygous siblings and had malformed eyes (arrow). Adult homozygous pitx2c mutants were indistinguishable from wild-type siblings. (C) Eye and craniofacial phenotypes in adult fish. Ocular defects of the iris and cornea (arrow) were observed in pitx2HD mutants, but not pitx2c mutants.
3.2. pitx2HD mutant zebrafish develop defects associated with ARS
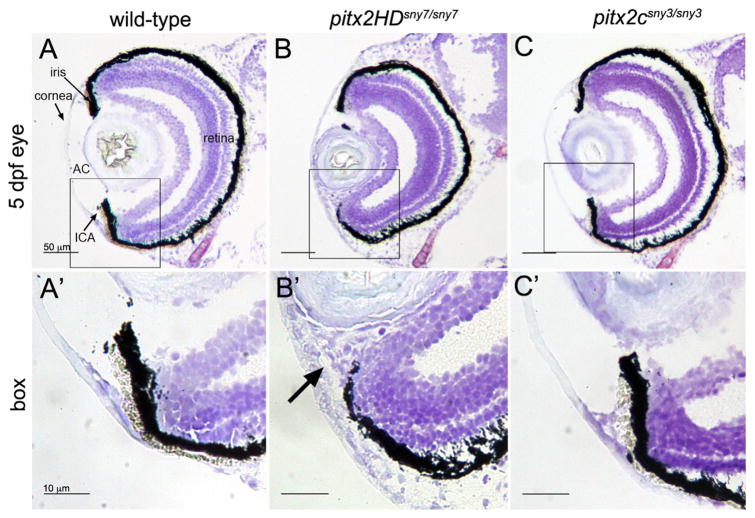
Patients with PITX2 homeodomain mutations at the same amino acid as our pitx2HD alleles developed ARS phenotypes, including malformations of the anterior segment of the eye (Yin et al., 2014). To investigate eye phenotypes observed in pitx2HD mutant zebrafish, we analyzed eye morphology during embryogenesis. At 5 dpf, sectioned of homozygous mutant pitx2HD sny7 eyes (identified by genotyping) were smaller and malformed relative to wild-type eyes (Fig. 4A and B). We observed a reduced anterior chamber, an increased thickness of the mesenchyme surrounding the cornea and malformation of the iridocorneal angle (Fig. 4B′). The same anterior segment defects were present in mutant pitx2HDsny6 embryos (Fig. S5A), but were not observed in pitx2csny3 mutants (Fig. 4C, C′). Higher resolution images indicated that the layers of the retina were intact in pitx2HD and pitx2c mutants (Fig. S6). These findings are reminiscent of eye phenotypes in ARS patients and Pitx2 homeodomain mutant mouse embryos (Gage et al., 1999; Lu et al., 1999), suggesting the role for Pitx2 in development of the anterior segment of the eye is conserved from zebrafish to mammals.
Fig. 4.
Malformations of the anterior segment of the eye in Pitx2HD mutant embryos. (A–C) Cryosections of the eye from 5 dpf wild-type (A), pitx2HDsny7/sny7 (B) and pitx2csny3/sny3 (C) fish stained with crystal violet. AC=anterior chamber; ICA=iridocorneal angle. Scale bars=50 μm. (A′–C′) Enlarged view of box 1 in A–C showing anterior segment structures. In pitx2HD mutants (B′), the anterior chamber was reduced and the iridocorneal angle (arrow) was malformed. Scale bars=10 μm.
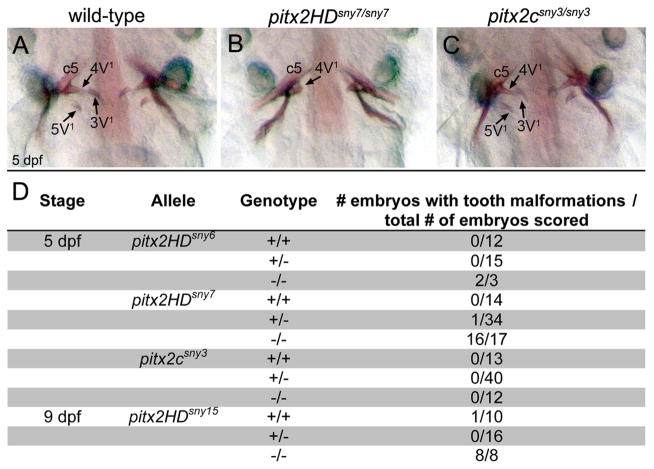
In addition to ocular defects, Pitx2 homeodomain mutations cause dental hypoplasia, which manifests as small, missing or malformed teeth in ARS patients (Dressler et al., 2010; Semina et al., 1996) and knockout mice (Lin et al., 1999; Lu et al., 1999) Zebrafish lack oral teeth but have pharyngeal teeth that have all the features of teeth in other vertebrates (Yelick and Schilling, 2002). At 5 dpf, three pairs of mineralized teeth, denoted as 3V1, 4V1 and 5V1, are attached to fifth branchial arches (Van der Heyden and Huysseune, 2000; Wise and Stock, 2010). Using alizarin red to stain developing teeth at 5 dpf, we found aberrant tooth morphogenesis in pitx2HD mutants. Compared to normal tooth development in wild-type siblings (Fig. 5A), the 3V1 and 5V1 teeth were absent and 4V1 teeth were malformed and smaller in pitx2HD mutants (Fig. 5B, D; Fig S5B). Similar tooth morphogenesis defects were observed at 9 dpf (Fig. 5D), indicating tooth development is not just delayed. In contrast to pitx2HD mutants, pitx2csny3/sny3 embryos had normal teeth (Fig. 5C, D). Taken together, these results suggest a conserved role for Pitx2 in vertebrate tooth development.
Fig. 5.
Tooth morphogenesis is disrupted in Pitx2HD mutants. (A–C) Ventral views of alizarin red staining of teeth at 5 dpf in wild-type (A), pitx2HDsny7/sny7 (B) and pitx2csny3/sny3 (C) fish. c5: fifth ceratobranchial; 4V1, 3V1 and 5V1 designate individual teeth. (D) Quantification of tooth defects observed in pitx2HDsny7 and pitx2csny fish at 5 dpf. Tooth malformations were defined as a reduced number of teeth and incomplete tooth formation as shown in (B).
To begin to test whether pitx2HD mutants develop additional phenotypes found in ARS patients, we analyzed pituitary development. Abnormalities associated with pituitary defects are found in ARS patients (Chang et al., 2012; Tumer and Bach-Holm, 2009) and pitx2 knockout mice (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Suh et al., 2002). To assess the pituitary in zebrafish pitx2 mutants, RNA in situ hybridizations were performed using markers of different cell types in the pituitary, which included gh1 (growth hormone 1) that marks somatotropes and pomca (proopiomelanocortin a) that marks corticotropes and melanotropes (Herzog et al., 2003). By quantifying these expression domains at 5 dpf, we found that the area of both gh1 and pomca staining was reduced in pitx2HD+/− and pitx2HD−/− embryos as compared to wild-type siblings (Fig. S7). These results suggest a reduction of somatotropes, corticotropes and melanotropes in pitx2HD−/− embryos and a slight reduction of somatotropes and corticotropes in pitx2HD+/− embryos relative to wild-type. This analysis provides evidence that some zebrafish pituitary phenotypes are Pitx2 dose-dependent, as observed in mammals. In pitx2 knockout mice, a dose-dependent effect was reported that moderately reduced Gh staining in mutants, but unlike pitx2HD zebrafish, Pomc expression was not changed (Suh et al., 2002). These results suggest Pitx2 has a function in zebrafish pituitary development that may be different than in mouse and provide the foundation for future work analyzing Pitx2 functions in the pituitary.
3.3.. The heart and gut undergo normal left-right asymmetric looping and placement in pitx2 mutants
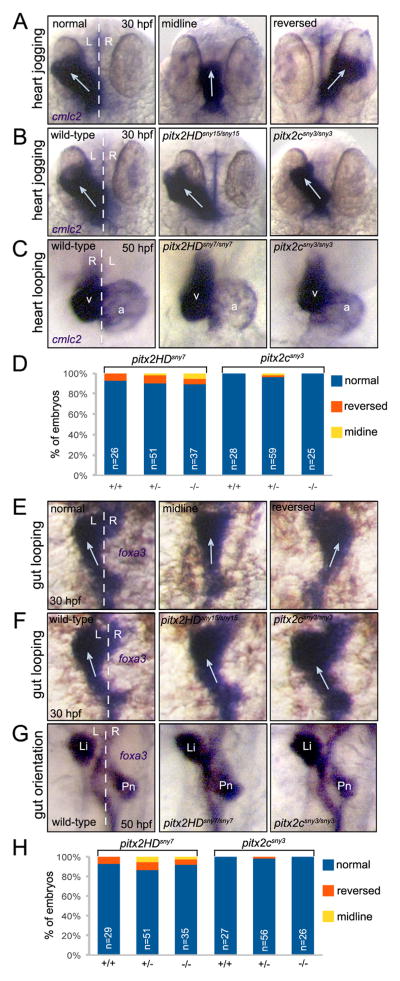
We next wanted to determine the role of Pitx2 in establishing LR asymmetry of visceral organs in zebrafish. The zebrafish Nodal homolog southpaw (spaw) is asymmetrically expressed in left lateral plate mesoderm and is required for asymmetric pitx2c expression (Long et al., 2003). Homozygous spaw mutants—in which pitx2 expression is absent in lateral plate mesoderm—have LR patterning defects: heart looping was altered in 30% of mutant embryos and gut looping was abnormal in 50% of the mutants (Noel et al., 2013). This suggested Pitx2 may function downstream of Nodal/Spaw to regulate asymmetric morphogenesis of the zebrafish heart and gut. To test this, we first analyzed asymmetric heart development in pitx2 mutants via RNA in situ hybridizations using the heart-specific marker cmlc2. At 30 hours post-fertilization (hpf), the developing heart tube undergoes asymmetric movements referred to as heart ‘jogging’ (Chen et al., 1997). The heart typically jogs to the left in wild-type embryos, but jogging can occur along the midline or to the right (Fig. 6A) when LR patterning cues are disrupted. Analysis of pitx2HD and pitx2c mutants revealed normal leftward jogging of the heart at 30 hpf (Fig. 6B; Table S2). Since pitx2 mutants could not be distinguished from wild-type siblings, each embryo was genotyped after RNA in situ analysis. Following asymmetric jogging, the heart tube undergoes rightward looping as observed in other vertebrate embryos. We visualized asymmetric looping of the zebrafish heart tube at 50 hpf and found heart morphology and looping direction in pitx2HD and pitx2c mutants were similar to wild-type siblings (Fig. 6C and D; Table S2). Examination of the heart in live embryos did not reveal gross defects in heart formation or function through 5 dpf, which is consistent with survival of pitx2HD and pitx2c mutants to adulthood. These results indicated loss of Pitx2 function did not alter cardiac jogging or looping morphogenesis.
Fig. 6.
Direction of asymmetric heart and gut morphogenesis is normal in pitx2 mutants. (A–C) RNA in situ hybridizations using cmlc2 probes labels the developing heart tube. (A) Selected wild-type embryos that show the potential outcomes of heart jogging at 30 hpf, which include normal left-sided jogging, jogging along the midline and reversed jogging. (B) Representative images of heart jogging in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of heart jogging. (C) Normal asymmetric heart looping in wild-type and pitx2HD and pitx2c mutant embryos placed the ventricle (v) to the right of the atrium (a) at 50 hpf. (D) Heart looping direction was scored as either normal, reversed or along the midline for individual embryos at 50 hpf and the genotype (+/+, +/− or −/−) was then determined for each embryo. (E–G) foxa3 in situ hybridizations marked the developing gut tube. (E) Potential outcomes of gut looping at 30 hpf include normal left-sided looping, remaining at the midline and reversed looping. (F) Direction of gut looping was similar in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of gut looping. (G) Normal asymmetric placement of the liver (Li) and pancreas (Pn) at 50 hpf. (H) Gut asymmetry was scored at 50 hpf and then each embryo was genotyped. No statistical differences in heart looping (D) or gut asymmetry (H) were identified using single factor one way ANOVA. White dashed lines represent the midline; L=left; R=right; n=number of embryos analyzed.
Next, asymmetric development of the gut was analyzed using foxa3 RNA in situ hybridization probes that mark the developing gut tube. Leftward looping of the zebrafish gut occurs between 26 and 30 hpf and is driven by movements of adjacent lateral plate mesoderm (Horne-Badovinac et al., 2003). Similar to the heart, the usual left-sided gut looping can remain at the midline or be reversed to the right (Fig. 6E) if embryo laterality is perturbed. When analyzed at 30 hpf, we found no differences in gut looping among wild-type, pitx2HD and pitx2c mutant embryos (Fig. 6F; Table S3). At 50 hpf, normal asymmetric looping of the gut places the liver to the left of the midline and the pancreas to the right (Fig. 6G). The orientation of these organs was reversed in nearly 50% of spaw mutants (Noel et al., 2013). However, consistent with normal gut looping, asymmetric positioning of the liver and pancreas developed normally in pitx2HD and pitx2c mutants (Fig. 6G and H; Table S3). At 7 dpf, visualization of gut auto-fluorescence in living embryos confirmed the LR orientation of visceral organs was unaffected in pitx2HD and pitx2c larvae (Fig. S8). RNA in situ analysis of the Nodal-Pitx2 cascade in lateral plate mesoderm revealed left-sided expression of spaw and pitx2c was expressed correctly in both pitx2HD and pitx2c mutants (Fig. S9), indicating early LR patterning of these embryos is normal. Taken together, these results indicate Pitx2 is dispensable for LR orientation of the zebrafish heart and gut and suggest molecules other than Pitx2 function downstream of Nodal to regulate heart and gut looping morphogenesis.
3.4.. Asymmetric expression of elovl6 is linked to left-right patterning information generated by Kupffer’s vesicle
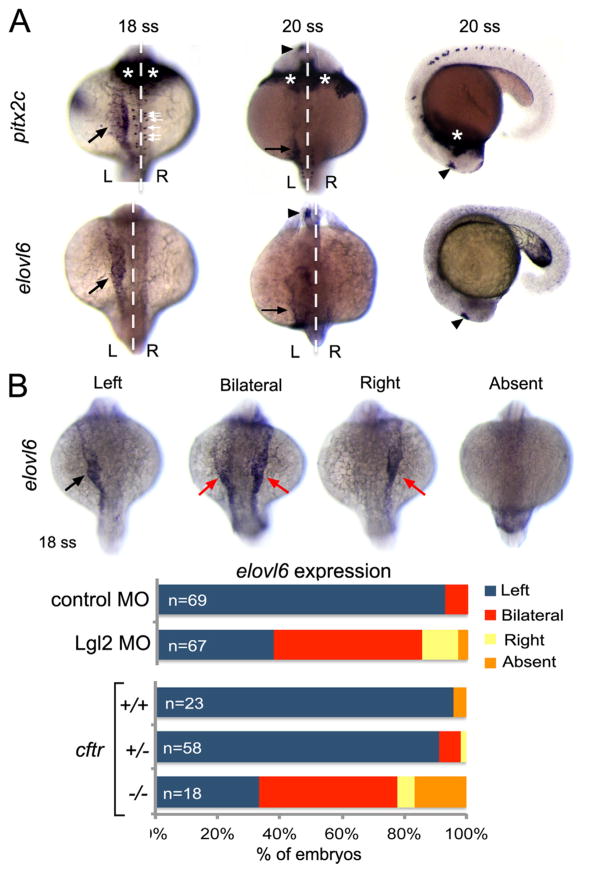
Our finding that Pitx2 is not required to establish heart or gut laterality in zebrafish led us to search for new Nodal-responsive genes that may function in LR development. We found that the fatty acid elongase elovl6, which lies immediately adjacent to pitx2 on chromosome 14, was identified in a large-scale RNA in situ screen to be asymmetrically expressed during LR patterning stages (Thisse et al., 2001). Similar to pitx2c, we found asymmetric elovl6 mRNA expression in what appears to be left lateral plate mesoderm at the 18 somite stage and expression in the left brain at the 20 somite stage (Fig. 7A). To test whether asymmetric elovl6 expression changes when LR patterning is perturbed, we used previously characterized approaches to alter asymmetric Nodal/Spaw signaling. In zebrafish, correct left-sided asymmetric gene expression depends on the function of motile cilia in Kupffer’s vesicle (KV) that generate a directional fluid flow that is required to orient the LR body axis (Essner et al., 2005; Kramer-Zucker et al., 2005). We first analyzed elovl6 expression in embryos injected with antisense morpholinos that knockdown expression of the Lethal giant larvae (Lgl2) protein. Lgl2 depletion disrupts KV development and results in bilateral spaw expression in ~50% of the embryos (Tay et al., 2013). Similarly, we observed bilateral elovl6 in ~50% of Lgl2 depleted embryos (Fig. 7B). In a few cases, elovl6 expression was right-sided or absent. We also tested elovl6 expression in cftrpd1048 mutant embryos, in which KV fails to develop and spaw expression is also predominantly bilateral (Navis et al., 2013). Again, we observed altered elovl6 expression that was primarily bilateral in cftrpd1048 mutants that were confirmed by genotyping (Fig. 7B). Thus, similar to Nodal/Spaw signaling, correct asymmetric expression of elovl6 depends on LR patterning information generated by Kupffer’s vesicle.
Fig. 7.
Asymmetric expression of elovl6 depends on Kupffer’s vesicle. (A) RNA in situ hybridizations revealed similar asymmetric expression patterns for pitx2c and evolv6 in left lateral plate mesoderm (black arrows) and brain (arrowheads) at 18–20 somite stages (ss). pitx2c mRNA was detected in hatching gland (asterisks) and neurons (white arrows), whereas elovl6 was not expressed in these domains. White dashed line is the midline; L=left and R=right. (B) Representative images are shown of left-sided, bilateral, right-sided and absent elovl6 expression in lateral plate mesoderm (arrows) at 18 somite stage. The graph shows quantification of asymmetric elovl6 expression profiles in control, Lgl2 depleted and genotyped cftrpd1048 mutant embryos.
3.5. Asymmetric expression of elovl6 depends on Nodal signaling but not Pitx2
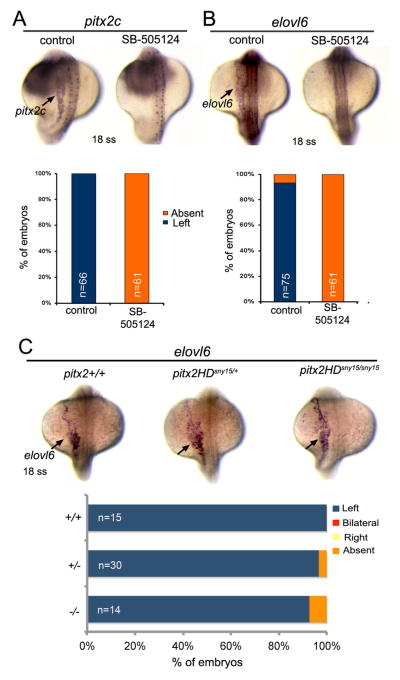
To determine whether asymmetric elovl6 expression requires Nodal/Spaw signaling, we used the small molecule inhibitor SB-505124 that effectively blocks TGFβ signaling in zebrafish (Lenhart et al., 2013). As expected, SB-505124 treatments between the bud stage and 18 somite stage eliminated asymmetric expression of pitx2c in lateral plate mesoderm (Fig. 8A), which is a known target of Nodal/Spaw signaling. Similarly, SB-505124 treatments abolished asymmetric elovl6 expression in lateral plate (Fig. 8B), suggesting elovl6 is a new Spaw target gene. To test whether elovl6 expression depends on Pitx2 transcriptional activity in lateral plate mesoderm, we assessed elovl6 mRNA in pitx2 mutants. Similar to wild-type siblings, elovl6 expression was present in left lateral plate mesoderm in pitx2HDsny15/sny15 mutants (Fig. 8C). Together, these analyses indicate asymmetric elovl6 expression requires Nodal/Spaw signaling but is independent of Pitx2. To begin to test Elovl6 function, we injected mRNA encoding full-length Elovl6 into wild-type embryos. These embryos showed severe malformations—including axial defects—that precluded analysis of heart and gut looping (Fig. S10). These results suggest Elovl6 is functional in the early zebrafish embryo. Future studies will need to address whether elovl6 functions downstream of Nodal signaling to impact LR patterning of the heart and gut.
Fig. 8.
Asymmetric expression of elovl6 requires Nodal signaling but does not depend on Pitx2. (A, B) Analysis of asymmetric pitx2c (A) or elovl6 (B) expression (arrows) at the 18 somite stage (ss) in control embryos and embryos treated with SB-505124 to block Nodal signaling. The graphs indicate the percentage of embryos with normal left-sided expression or absent expression. (C) Asymmetric elovl6 expression (arrows) in wild-type and pitx2HD mutant embryos. n=number of embryos analyzed.
4. Discussion
Here we present the initial characterization of the first zebra-fish mutant pitx2 alleles. Our results show zebrafish Pitx2 is required for proper development of the anterior segment of the eye and morphogenesis of teeth, which likely represent conserved functions for Pitx2 in vertebrate embryos. Since eye and tooth phenotypes in zebrafish pitx2HD mutants are similar to malformations observed in ARS patients, these zebrafish provide an important new animal model that can be used for investigating Pitx2 functions during early embryogenesis and for high-throughput screening of small molecules for ARS therapeutics. Intriguingly, although pitx2c is asymmetrically expressed in left lateral plate mesoderm, our analyses indicate Pitx2 is not required for asymmetric looping of the zebrafish heart and gut. This finding indicates Pitx2 is not an essential downstream effector of Nodal signaling during looping of visceral organs in zebrafish. The mutant pitx2 zebrafish background presents a new opportunity to characterize novel molecules and pathways that impact organ LR asymmetry.
Disrupting the zebrafish Pitx2 homeodomain resulted in tooth hypoplasia and ocular defects, which are features of ARS patients with heterozygous PITX2 mutations. In contrast to humans, eye and tooth phenotypes were only observed in homozygous pitx2HD−/− zebrafish. In previous work, transient knockdown of Pitx2 expression using morpholino oligonucleotides caused defects in eye, craniofacial and asymmetric brain development (Bohnsack et al., 2012; Garric et al., 2014; Liu and Semina, 2012). pitx2HD mutants developed eye phenotypes that were consistent with Pitx2 morpholino injected embryos, but did not develop other defects (e.g. pericardial edema) and could survive to adulthood. Defects in the iridocorneal angle observed in pitx2HD mutants are reminiscent of characteristic anterior chamber angle defects in ARS patients (Tumer and Bach-Holm, 2009) that commonly develop glaucoma in late childhood or early adulthood. In addition, we uncovered a requirement for Pitx2 in development of zebrafish pharyngeal teeth, which is consistent with pitx2 expression during tooth formation (Jackman et al., 2004). However, the function of Pitx2 in zebrafish tooth development may be different than in the mouse. In mice, Fgf8 and Bmp4 are expressed during tooth morphogenesis and it has been found that Ffg8 expression in the oral ectoderm is diminished and Bmp4 is expanded in pitx2−/− mice (Lin et al., 1999; Liu et al., 2003; Lu et al., 1999). In zebrafish, Bmp4 is dispensable for tooth development (Wise and Stock, 2010) and Fgf8 is not expressed in tooth-forming regions at any stage (Jackman et al., 2004). It will be interesting to compare and contrast Pitx2 functions in mouse and zebrafish models in future work. Our analysis of pituitary development also suggests differential effects in mouse and fish pitx2 mutants. These differences provide an opportunity to explore the evolution of Pitx2 functions. Interestingly, zebrafish with a pitx2c-specific mutation grew to adulthood with no apparent morphological abnormalities, but we have only recovered males. In birds, Pitx2 has been implicated in asymmetric gonad morphogenesis (Guioli and Lovell-Badge, 2007; Rodriguez-Leon et al., 2008), making it tempting to speculate that Pitx2c may have a role in gonad development in zebrafish. However, since we recovered a single pitx2c mutant allele we cannot rule out off-target effects of the TALEN mutagenesis. Thus, further studies are needed to investigate the possible role of Pitx2c in zebrafish gonad development.
In contrast to mouse Pitx2−/− knockouts that are embryonic lethal, pitx2HD−/− zebrafish can survive to adulthood. It is interesting to consider why the pitx2 knockout phenotype is less severe in zebrafish. Our results suggest Pitx2 has essential functions in the mouse embryo—such as establishing LR asymmetry in the heart and gut—that are not conserved in zebrafish. Additional problems that likely contribute to embryonic lethality, including body wall closure defects, are also specific to the mouse. It is also possible that compensatory mechanisms exist in pitx2HD−/− zebrafish that are absent in mice. This could include the presence of redundant transcription factors and/or the upregulation of factors to take the place of Pitx2 in some tissues. Nonetheless, adult pitx2HD−/− zebrafish provide an important new animal model that can be exploited to develop new strategies to treat or prevent eye and tooth diseases associated with ARS. The zebrafish is well suited for efficient high-throughput screening of thousands of small molecules, and mutant alleles have been successfully used to identify compounds that suppress disease phenotypes. In addition to using pitx2 mutant to screen for compounds that treat congenital eye and tooth malformations, Pitx2 has been implicated in other processes, such as myogenesis (Knopp et al., 2013; L’Honore et al., 2007), cardiac arrhythmia (Tao et al., 2014) and cancer (Vinarskaja et al., 2013). Although more work is needed to investigate these and other phenotypes, pitx2 mutant zebrafish may provide a screening platform for drug discovery for a broad range of maladies. Additionally, combining mutant alleles with zebrafish transgenes that label specific cell types provides a useful approach to identifying genes that are misregulated in a particular organ or tissue of the mutant fish. Since defects in neural crest development likely contribute to eye, tooth and craniofacial malformations in ARS patients and zebrafish pitx2 mutants, transgenic zebrafish that label neural crest cells (Kwak et al., 2013) will allow real-time tracking and transcriptome analyses of neural crest cells in pitx2 mutant embryos. This approach provides an opportunity to characterize Pitx2−/− cell behaviors and identify cell-specific target genes of Pitx2 to get at molecular mechanisms of Pitx2 function. Discovery of new genes and pathways downstream of Pitx2 may suggest new therapeutic targets.
Based on the highly conserved asymmetric expression of Nodal-Pitx2 axis in left lateral plate mesoderm, we predicted Pitx2 would be required for asymmetric development of visceral organs in zebrafish. However, we did not observe defects in LR asymmetry of the heart or gut in zebrafish pitx2 mutants. In Pitx2 mutant mice, heart looping direction is also normal and cardiac LR defects largely encompass defects in septation of the outflow tract that is not conserved in the zebrafish heart. Analysis of zebrafish spaw (Nodal) mutants (Noel et al., 2013), in which asymmetric pitx2c expression is lost in lateral plate mesoderm, revealed predominantly normal cardiac looping. However, in contrast to mild effects on the heart, loss of spaw resulted in complete randomization of gut laterality. Since we observed normal gut asymmetries in zebrafish pitx2 mutants, this indicates molecules other than Pitx2 mediate signaling downstream of Spaw/Nodal during zebrafish gut LR development. In mouse and chick embryos, gut looping depends on the mechanical forces generated from the dorsal mesentery that attaches to the gut (Savin et al., 2011). Pitx2c expression in the left dorsal mesentery controls Wnt signaling to generate asymmetric cellular condensations that mediate gut looping (Davis et al., 2008; Welsh et al., 2013). The gut looping process in zebrafish results from the asymmetric migration of the neighboring lateral plate mesoderm (Horne-Badovinac et al., 2003). Thus, differences in anatomy provide a plausible explanation for how zebrafish heart and gut LR morphogenesis could be independent of Pitx2.
If Pitx2 is not required for asymmetric looping of visceral organs in zebrafish, what are alternative Nodal pathway target molecules? We found that elovl6, which is adjacent to pitx2 on chromosome 14, is asymmetrically expressed with pitx2 and that normal left-sided elovl6 expression depends on Kupffer’s vesicle function and Nodal signaling. Taking advantage of our pitx2 mutants, we determined asymmetric elovl6 expression is independent of Pitx2 function. The close proximity of pitx2 and elovl6 suggests left-sided expression of these genes is controlled by a Nodal-responsive enhancer on chromosome 14. Intriguingly, Elovl6 is asymmetrically expressed opposite of Pitx2 in the right dorsal mesentery in the chicken embryo during gut looping (Welsh et al., 2015). It is not clear whether asymmetric Elovl6 expression in chicken is Nodal-dependent, but studies in chicken and mouse indicate chromatin conformation at the Pitx2 locus plays an important role in directing left-sided expression of Pitx2 and right-sided expression of downstream genes Enpep and Elovl6 (Welsh et al., 2015). In contrast to chicken and mouse, the zebrafish pitx2 locus lacks an enpep gene and the pitx2 and elovl6 genes are very close together. The distinct arrangement of genes at pitx2 loci provides a likely explanation for the differential regulation of elovl6 expression in zebrafish and chicken.
The findings of asymmetric elovl6 expression in chicken (right-sided) and zebrafish (left-sided) raise the question of whether Elovl6 plays roles during LR development. Elovl6 belongs to a family of rate-limiting enzymes that catalyze the synthesis of saturated and monounsaturated fatty acids and have been implicated in lipid metabolism related diseases (Matsuzaka and Shimano, 2009). Elovl6 may be involved in the synthesis of fatty acid molecules that participate in cell signaling. In zebrafish, sphingosine-1-phosphate receptors are required for myocardial migration and endoderm convergence (Kupperman et al., 2000; Ye and Lin, 2013), indicating regulation of fatty acids plays important roles during heart and gut morphogenesis. To test whether Elovl6 impacts LR asymmetry in zebrafish we first conducted gain-of-function experiments by expressing elovl6 ubiquitously (bilaterally) in the zebrafish embryo. These injections caused severe malformations and, due to the possibility of secondary effects, it was not possible to cleanly determine effects on visceral organ asymmetry. We also took a loss-of-function approach using the CRISPR-Cas9 system (Gagnon et al., 2014), which we have used to successfully generate mutations in several zebrafish genes. However, attempts to edit the elovl6 locus (using 5 different guide RNAs) have thus far been unsuccessful. This suggests alternative approaches (e.g. TALENs) may be needed to generate elovl6 mutations. Thus, additional work is still needed to test whether Eovl6 plays a role in establishing LR asymmetry in vertebrate embryos.
Taken together, these results establish a new animal model of ARS, provide new insight on the conservation of Pitx2 functions in vertebrate embryogenesis and raise the possibility that previously unrecognized molecules regulate zebrafish LR asymmetry. We propose pitx2 mutant zebrafish will be a useful model for elucidating molecular mechanisms of Pitx2 functions in embryos and adults, understanding the basis of congenital eye and tooth malformations in ARS patients and ultimately identifying new drugs to treat or prevent ARS symptoms.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Heart, Lung and Blood Institute Grant R01HL095690 to J.D.A.
We thank H. Hu, M. Yu for eye sections and E. Solessio and Y. Umino for discussion of eye malformations and the Bagnat lab for providing ctfr mutants. S. Adhikary provided excellent technical support and animal care.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.06.010.
Footnotes
Competing interests
No competing interests declared.
Author contributions
Y.J. and J.D.A. designed experiments. Y.J. and S.M.B. performed experiments. Y.J., S.M.B. and J.D.A. analyzed data and prepared the manuscript.
References
- Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, et al. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antevil J, Umakanthan R, Leacche M, Brewer Z, Solenkova N, Byrne JG, Greelish JP. Idiopathic mitral valve disease in a patient presenting with Axenfeld-Rieger syndrome. J Heart Valve Dis. 2009;18:349–351. [PubMed] [Google Scholar]
- Bohnsack BL, Kasprick DS, Kish PE, Goldman D, Kahana A. A zebrafish model of axenfeld-rieger syndrome reveals that pitx2 regulation by retinoic acid is essential for ocular and craniofacial development. Investig Ophthalmol Vis Sci. 2012;53:7–22. doi: 10.1167/iovs.11-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman CJ, Shimeld SM. Pitx homeobox genes in Ciona and amphioxus show left-right asymmetry is a conserved chordate character and define the ascidian adenohypophysis. Evol Dev. 2002;4:354–365. doi: 10.1046/j.1525-142x.2002.02021.x. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney BA, Clark-Baldwin K, Dave V, Ma J, Rance M. Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry. 2005;44:7497–7511. doi: 10.1021/bi0473253. [DOI] [PubMed] [Google Scholar]
- Chang TC, Summers CG, Schimmenti LA, Grajewski AL. Axenfeld-Rieger syndrome: new perspectives. Br J Ophthalmol. 2012;96:318–322. doi: 10.1136/bjophthalmol-2011-300801. [DOI] [PubMed] [Google Scholar]
- Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Davis NM, Kurpios NA, Sun X, Gros J, Martin JF, Tabin CJ. The chirality of gut rotation derives from left-right asymmetric changes in the architecture of the dorsal mesentery. Dev Cell. 2008;15:134–145. doi: 10.1016/j.devcel.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerdelmann T, Kojetin DJ, Baird-Titus JM, Solt LA, Burris TP, Rance M. Structural and biophysical insights into the ligand-free Pitx2 homeodomain and a ring dermoid of the cornea inducing homeodomain mutant. Biochemistry. 2012;51:665–676. doi: 10.1021/bi201639x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler S, Meyer-Marcotty P, Weisschuh N, Jablonski-Momeni A, Pieper K, Gramer G, Gramer E. Dental and craniofacial anomalies associated with Axenfeld-Rieger syndrome with PITX2 mutation. Case Rep Med. 2010;2010:621984. doi: 10.1155/2010/621984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development. 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Evans AL, Gage PJ. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum Mol Genet. 2005;14:3347–3359. doi: 10.1093/hmg/ddi365. [DOI] [PubMed] [Google Scholar]
- Footz T, Idrees F, Acharya M, Kozlowski K, Walter MA. Analysis of mutations of the PITX2 transcription factor found in patients with Axenfeld-Rieger syndrome. Investig Ophthalmol Vis Sci. 2009;50:2599–2606. doi: 10.1167/iovs.08-3251. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wang G, Amack JD, Mitchell DR. Oda16/Wdr69 is essential for axonemal dynein assembly and ciliary motility during zebrafish embryogenesis. Dev Dyn. 2010;239:2190–2197. doi: 10.1002/dvdy.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garric L, Ronsin B, Roussigne M, Booton S, Gamse JT, Dufourcq P, Blader P. Pitx2c ensures habenular asymmetry by restricting parapineal cell number. Development. 2014;141:1572–1579. doi: 10.1242/dev.100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guioli S, Lovell-Badge R. PITX2 controls asymmetric gonadal development in both sexes of the chick and can rescue the degeneration of the right ovary. Development. 2007;134:4199–4208. doi: 10.1242/dev.010249. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Herzog W, Zeng X, Lele Z, Sonntag C, Ting JW, Chang CY, Hammerschmidt M. Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev Biol. 2003;254:36–49. doi: 10.1016/s0012-1606(02)00124-0. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Rebagliati M, Stainier DY. A cellular framework for gut-looping morphogenesis in zebrafish. Science. 2003;302:662–665. doi: 10.1126/science.1085397. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Jena AK, Kharbanda OP. Axenfeld-Rieger syndrome: report on dental and craniofacial findings. J Clin Pediatr Dent. 2005;30:83–88. doi: 10.17796/jcpd.30.1.v1732398454r0244. [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- Knopp P, Figeac N, Fortier M, Moyle L, Zammit PS. Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev Biol. 2013;377:293–304. doi: 10.1016/j.ydbio.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406:192–195. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- Kwak J, Park OK, Jung YJ, Hwang BJ, Kwon SH, Kee Y. Live image profiling of neural crest lineages in zebrafish transgenic lines. Mol Cells. 2013;35:255–260. doi: 10.1007/s10059-013-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, Camper S, Drouin J. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307:421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Lenhart KF, Holtzman NG, Williams JR, Burdine RD. Integration of nodal and BMP signals in the heart requires FoxH1 to create left-right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet. 2013;9:e1003109. doi: 10.1371/journal.pgen.1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Pagan S, Roberts DJ, Cooke J, Kuehn MR, Tabin CJ. Left/right patterning signals and the independent regulation of different aspects of situs in the chick embryo. Dev Biol. 1997;189:57–67. doi: 10.1006/dbio.1997.8662. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu W, Lu MF, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- Liu Y, Semina EV. pitx2 Deficiency results in abnormal ocular and craniofacial development in zebrafish. PLoS One. 2012;7:e30896. doi: 10.1371/journal.pone.0030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Matsuzaka T, Shimano H. Elovl6: a new player in fatty acid metabolism and insulin sensitivity. J Mol Med. 2009;87:379–384. doi: 10.1007/s00109-009-0449-0. [DOI] [PubMed] [Google Scholar]
- Navis A, Marjoram L, Bagnat M. Cftr controls lumen expansion and function of Kupffer’s vesicle in zebrafish. Development. 2013;140:1703–1712. doi: 10.1242/dev.091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel ES, Verhoeven M, Lagendijk AK, Tessadori F, Smith K, Choorapoikayil S, den Hertog J, Bakkers J. A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat Commun. 2013;4:2754. doi: 10.1038/ncomms3754. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leon J, Rodriguez Esteban C, Marti M, Santiago-Josefat B, Dubova I, Rubiralta X, Izpisua Belmonte JC. Pitx2 regulates gonad morphogenesis. Proc Natl Acad Sci USA. 2008;105:11242–11247. doi: 10.1073/pnas.0804904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Sampath K, Cheng AM, Frisch A, Wright CV. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development. 1997;124:3293–3302. doi: 10.1242/dev.124.17.3293. [DOI] [PubMed] [Google Scholar]
- Savin T, Kurpios NA, Shyer AE, Florescu P, Liang H, Mahadevan L, Tabin CJ. On the growth and form of the gut. Nature. 2011;476:57–62. doi: 10.1038/nature10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Shields MB, Buckley E, Klintworth GK, Thresher R. Axenfeld-Rieger syndrome. A spectrum of developmental disorders. Surv Ophthalmol. 1985;29:387–409. doi: 10.1016/0039-6257(85)90205-x. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Yashiro K, Shen MM, Hamada H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development. 2006;133:3015–3025. doi: 10.1242/dev.02470. [DOI] [PubMed] [Google Scholar]
- Suh H, Gage PJ, Drouin J, Camper SA. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–337. doi: 10.1242/dev.129.2.329. [DOI] [PubMed] [Google Scholar]
- Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, Kaminski HJ, Martin JF. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7:23–32. doi: 10.1161/CIRCGENETICS.113.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay HG, Schulze SK, Compagnon J, Foley FC, Heisenberg CP, Yost HJ, Abdelilah-Seyfried S, Amack JD. Lethal giant larvae 2 regulates development of the ciliated organ Kupffer’s vesicle. Development. 2013;140:1550–1559. doi: 10.1242/dev.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) 2001 [Google Scholar]
- Toyoizumi R, Ogasawara T, Takeuchi S, Mogi K. Xenopus nodal related-1 is indispensable only for left-right axis determination. Int J Dev Biol. 2005;49:923–938. doi: 10.1387/ijdb.052008rt. [DOI] [PubMed] [Google Scholar]
- Tumer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet. 2009a;17:1527–1539. doi: 10.1038/ejhg.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe RA, Gross JM. Immunohistochemistry on cryosections from embryonic and adult zebrafish eyes. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4779. pdb prot4779. [DOI] [PubMed] [Google Scholar]
- Van der Heyden C, Huysseune A. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae) Dev Dyn. 2000;219:486–496. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Vinarskaja A, Schulz WA, Ingenwerth M, Hader C, Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urol Oncol. 2013;31:622–627. doi: 10.1016/j.urolonc.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Schmidt HA, Kuhn A, Hoger SK, Kocagoz Y, Laumann-Lipp N, Ozbek S, Holstein TW. Nodal signalling determines biradial asymmetry in Hydra. Nature. 2014;515:112–115. doi: 10.1038/nature13666. [DOI] [PubMed] [Google Scholar]
- Welsh IC, Thomsen M, Gludish DW, Alfonso-Parra C, Bai Y, Martin JF, Kurpios NA. Integration of left-right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev Cell. 2013;26:629–644. doi: 10.1016/j.devcel.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh IC, Kwak H, Chen FL, Werner M, Shopland LS, Danko CG, Lis JT, Zhang M, Martin JF, Kurpios NA. Chromatin architecture of the Pitx2 locus requires CTCF- and Pitx2-dependent asymmetry that mirrors embryonic gut laterality. Cell Rep. 2015;13:337–349. doi: 10.1016/j.celrep.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SB, Stock DW. bmp2b and bmp4 are dispensable for zebrafish tooth development. Dev Dyn. 2010;239:2534–2546. doi: 10.1002/dvdy.22411. [DOI] [PubMed] [Google Scholar]
- Ye D, Lin F. S1pr2/Galpha13 signaling controls myocardial migration by regulating endoderm convergence. Development. 2013;140:789–799. doi: 10.1242/dev.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelick PC, Schilling TF. Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med. 2002;13:308–322. doi: 10.1177/154411130201300402. [DOI] [PubMed] [Google Scholar]
- Yin HF, Fang XY, Jin CF, Yin JF, Li JY, Zhao SJ, Miao Q, Song FW. Identification of a novel frameshift mutation in PITX2 gene in a Chinese family with Axenfeld-Rieger syndrome. J Zhejiang Univ Sci B. 2014;15:43–50. doi: 10.1631/jzus.B1300053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, St Amand TR, Wang S, Li G, Zhang Y, Hu YP, Nguyen L, Qiu MS, Chen YP. Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development. 2001;128:1005–1013. doi: 10.1242/dev.128.6.1005. [DOI] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.