Abstract
Genetic variations mapping to 3’ untranslated regions (3’UTRs) may overlap with microRNA (miRNA) binding sites, therefore potentially interfering with translation inhibition or messenger RNA (mRNA) degradation. The aim of this study was to investigate whether single nucleotide polymorphisms (SNPs) located within the 3’UTRs of six candidate genes and predicted to interfere with miRNA ligation could account for disease-relevant differential mRNA levels. Focusing on pemphigus foliaceus (PF) – an autoimmune blistering skin condition with unique endemic patterns – we investigated if nine 3’UTR SNPs from the CD1D, CTLA4, KLRD1, KLRG1, NKG7, and TNFSF13B genes differentially expressed in PF were disease-associated. The heterozygous genotype of the KLRG1 rs1805672 polymorphism was associated with increased predisposition to PF (A/G vs. A/A: P=0.038; OR=1.60), and a trend for augmented susceptibility was observed for carriers of the G allele (P=0.094; OR=1.44). In silico analyses suggested that rs1805672 G allele could disrupt binding of miR-584-5p, and indicated rs1805672 as an expression Quantitative Trait Locus (eQTL), with an effect on KLRG1 gene expression. Dual-luciferase assay showed that miR-584-5p mediated approximately 50% downregulation of the reporter gene’s activity through the 3’UTR of KLRG1 harboring rs1805672 A allele (vs. miRNA-negative condition, P=0.006). This silencing relationship was lost after site-directed mutation to G allele (vs. miRNA-negative condition, P=0.391; vs. rs1805672 A allele, P=0.005). Collectively, these results suggest that a disease-associated SNP located within the 3’UTR of KLRG1 directly interferes with miR-584-5p binding, allowing for KLRG1 mRNA differential accumulation, which in turn may contribute to pathogenesis of autoimmune diseases, such as pemphigus.
Keywords: KLRG1, rs1805672, gene expression, eQTL, 3’UTR, miR-584-5p
1. Introduction
Microribonucleic acids, or simply microRNAs (miRNAs), are short non-coding RNAs with a median size of 22 nucleotides (nt) in humans, once in their mature and posttranscriptional regulatory form (1,2). The genes of these tiny molecules map to intergenic or intronic regions, where they are supposed to have their own promoter or share it with the host gene, respectively – i.e., intronic miRNAs are expected to be co-expressed with their host gene (3,4). The mode by which miRNAs posttranscriptionally regulate gene expression through 3’ untranslated region (3’UTR)-binding of targeted messenger RNAs (mRNAs) is considered an RNA interference mechanism and leads to mRNA degradation or translation inhibition (1). This sequence complementarity involves the so called miRNA seed sequence, a 6 or 7 nt 3’UTR-perfect matching region at the 5’-end of miRNAs that is known to be essential for effective miRNA function (1). Interestingly, a new type of seed sequence located within the central region of miRNAs has been described (5), adding more complexity to the miRNA mode of action (6). Regardless of where the seed sequence is located, it is clear that 3’UTR polymorphisms overlapping with seed sequences can interfere with miRNA ligation, potentially disrupting or creating miRNA binding sites (7).
Through gene expression profiles of complex traits, researchers can unveil new candidate genes for association studies and understand how genetic variation modulates normal and pathological conditions. Since our publication of the CD4+ T cell genome-wide gene expression profile of pemphigus foliaceus (8), we have been interested in exploring how genetic polymorphisms influence gene transcription and mRNA stability and if these polymorphisms could be contributing to disease susceptibility (9).
Pemphigus is a chronic autoimmune blistering skin disease identified by the presence of IgG autoantibodies (autoAb) directed against adhesion molecules of keratinocytes. Historically, the disease has been characterized by the presence of autoAb against two desmosomal cadherins, desmoglein (Dsg) 1 and/or Dsg3, believed to specify two main forms of the disease. Pemphigus foliaceus (PF) is identified by the presence of anti-Dsg1 auto-Ab and superficial blistering. The other main form, pemphigus vulgaris (PV), is recognized by the presence of anti-Dsg3 or anti-Dsg3 and anti-Dsg1 autoAb followed by suprabasal blistering of mucous membranes or by cutaneous and mucous involvement, respectively (10). Additional self-antigens have been identified in pemphigus besides desmogleins: multiple other desmosomal and non-desmosomal cadherins, such as E-cadherin, and a large variety of human proteins that specifically react with pemphigus IgG (11,12). The traditional understanding that anti-Dsg1 and/or anti-Dsg3 autoAb cause the loss of keratinocyte adhesion leading to blister formation (acantholysis) is being challenged and pemphigus is now more likely to be understood as a disease mediated by “apoptolysis” leading to keratinocyte shrinkage/detachment (13,14).
In epidemiologic terms, both main forms of pemphigus occur worldwide and sporadically. PF, however, has a form geographically restricted in/to certain geographic regions, being the single known autoimmune disease with endemic features. Endemic pemphigus foliaceus (EPF) occurs in geographic areas of South America, such as in Brazil, where it was described in 1903 (15). Brazilian pemphigus, also known as fogo selvagem (“wild fire”, in literal translation), was estimated to reach 30.7 cases per million/year (16) and is believed to have clinical differences and prognostic dissimilarities to its non-endemic form, but no differences in immunopathology (15,17,18). As the difference in endemicity may be due to environmental factors, we do not distinguish in this study individuals of EPF from those with the sporadic form.
Among the genes previously reported to be differentially expressed in PF, several are known for their central role in immune responses. In this study, we selected, among these, six genes accounting for different types of immune responses (i.e., innate or adaptive, cellular or humoral). CD1D encodes the D member of the CD1 family of major histocompatibility complex (MHC)-like glycoproteins, responsible for capturing and presenting lipid and glycolipid antigens to T cells (19). The cytotoxic T lymphocyte antigen-4 (CTLA4) gene is well known for encoding a negative regulator of T cell responses, contributing to the control of autoreactivity (20). Killer cell lectin-like receptor subfamily D member 1 (KLRD1 or CD94) and subfamily G member 1 (KLRG1) genes are both in the natural killer (NK) cell complex (NKC) in the chromosomal region 12p13. The former codes for a receptor establishing disulphide-bonded heterodimers with natural killer cell group 2 (NKG2) family members, acting together as activating or inhibitory NK cell receptors. The latter, KLRG1, codes for an inhibitory receptor expressed on the surface of mainly NK and CD4+ and CD8+ αβ T cells, where it potentially raises their activation thresholds, ultimately preventing autoreactivity (21–23). The natural killer cell granule protein 7 (NKG7) gene is expressed in activated NK cells and possibly in some T lymphocyte subsets, where it may play a role in T-cell effector function (24). Finally, the tumor necrosis factor superfamily member 13b (TNFSF13B) gene, commonly also referred as B cell activating factor (BAFF) or B-lymphocyte stimulator (BLYS), codes for a cytokine important for the survival and proliferation of B and T cells (25).
In this context and considering pemphigus an interesting immune-related disease model, we investigated whether 3’UTR polymorphisms overlapping with predicted miRNA binding sites in the aforementioned PF-differentially expressed genes could be contributing to the previously observed mRNA variable levels. Through this approach, we describe an association between a KLRG1 3’UTR polymorphism and PF, and unveil a functional mechanism for the association that may also elucidate the KLRG1 mRNA differential levels previously observed in the disease.
2. Material and Methods
2.1. Samples
Informed consent was obtained from all participants. This study was performed according to Brazilian federal laws and approved by the Human Research Ethics Committee of the Federal University of Paraná. Peripheral blood was collected for DNA extraction from 333 PF patients and 427 clinically healthy controls from the endemic area. Individuals were mostly contacted at Hospital Adventista do Pênfigo, in Campo Grande, Mato Grosso do Sul state. Some individuals were from: Curitiba (Hospital de Clínicas da UFPR, Hospital de Dermatologia Sanitária São Roque and Hospital Santa Casa de Misericórdia), in Paraná state; Ribeirão Preto (Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto da USP), in São Paulo state; and Uberaba (Lar da Caridade Hospital do Fogo Selvagem de Uberaba), in Minas Gerais state. As the Brazilian population has a mixed ethnic origin, to avoid effects of population structure, individuals were classified in two different groups for further sample adjustment: EU, of predominantly European origin; and M, of mixed European and African origin.
An additional sample of 146 healthy donors, contacted at the blood bank of the Hospital de Clínicas da UFPR, was enrolled in this study for peripheral blood mononuclear cell (PBMC) isolation from venous blood by density gradient centrifugation with Histopaque® 1077 (Sigma-Aldrich Inc., Saint Louis, MO) according to manufacturer’s manual.
2.2. Genotyping
Three different online-available bioinformatics tools were consulted to verify SNP ability to disrupt/create miRNA ligation: MirSNP, mirsnpscore, and PolymiRTS. Nine SNPs in the 3’UTR (Table 1) of six candidate genes previously shown to be differentially expressed in PF (Table S1) (8) were selected given their putative ability to disrupt/create a miRNA binding site (Table S2): rs116898958, rs185198828, and rs4145212 (TNFSF13B), rs3009 (NKG7), rs1805672 (KLRG1), rs2537752 (KLRD1), rs139105990 (CTLA4), rs16839951, and rs422236 (CD1D). All the aforementioned SNPs were predicted to fall within seed sequences and to disrupt/create a miRNA binding site according to at least one of the referred tools.
Table 1.
Genes and their 3’UTR SNPs analyzed in this association study.
| Gene |
Analyzed SNPs |
Reference variationb |
||
|---|---|---|---|---|
| Official Symbol | Alias | Locationa | ||
| CD1D | CD1A, R3 | 1q23.1 | rs422236 | G>T |
| rs16839951 | A>G | |||
| CTLA4 | ALPS5, CD152 | 2q33.2 | rs139105990 | G>A |
| KLRD1 | CD94 | 12p13.2 | rs2537752 | T>A |
| KLRG1 | 2F1, CLEC15A | 12p13.31 | rs1805672 | A>G |
| NKG7 | GIG1, GMP-17 | 19q13.41 | rs3009 | G>A |
| TNFSF13B | BAFF, BLYS | 13q33.3 | rs4145212 | T>A |
| rs116898958 | C>T | |||
| rs185198828 | G>T | |||
Location is according to Ensembl genome browser.
forward to the genome according to dbSNP.
The NKG7, KLRG1, and KLRD1 SNPs were genotyped by the SNPlex™ System (Applied Biosystems, Foster City, CA), which consists of an oligonucleotide ligation assay in a multiplex PCR, followed by capillary electrophoresis (26). The TNFSF13B (BAFF), CTLA4, and CD1D SNPs were genotyped by iPLEX® MassARRAY® (Sequenom® Inc, San Diego, CA), which uses time of flight (TOF) mass spectrometry (27). Further genotyping of the PF-associated rs1805672 SNP in PBMC of healthy donors was performed through TaqMan® SNP Genotyping Assay according to the manufacturer’s protocol (Applied Biosystems).
2.3. Gene and miRNA expression analysis
KLRG1 and the high confidence miR-584-5p, predicted to bind to the 3’UTR of KLRG1 mRNA in the presence of rs1805672 A allele, as well as the miR-584-5p-harboring gene, SH3TC2, were quantified in PBMC of healthy donors through reverse transcription followed by real-time polymerase chain reaction (RT-qPCR). For this end, cDNA was produced from the same aliquot of total RNA isolated and purified through the phenol-guanidinium thiocyanate method: 40 ng for total cDNA production and detection of KLRG1 and SH3TC2, and 1 ng for miR-584-5p-specific cDNA. TaqMan® Gene Expression Assay and TaqMan® MicroRNA Assay (Applied Biosystems) were used for the detection of KLRG1 mRNA and miR-584-5p levels, respectively. For detection of SH3TC2 mRNA with FastStart Essential DNA Green Master (Roche Applied Science, Penzberg, Germany), the following primers were designed to achieve best coverage: 5’ ATCTGGAAGTTCTCCACCTA 3’ (forward) and 5’ GTACTTGTGGTCAAAGAGGAG 3’ (reverse). All qPCR assays were carried out in duplicates.
2.4. Reporter gene assays and oligonucleotide transfection assay
The 3’UTR of KLRG1, a possible miR-584-5p target, was PCR amplified using human genomic DNA as template and LongAmp® Taq DNA Polymerase (New England BioLabs, Ipswich, MA). The PCR amplicons containing rs1805672 A allele overlapping with a putative miR-584-5p seed-sequence were subsequently gel purified, and ligated into the pmirGLO vector (Promega, Fitchburg, WI). The reporter construct harboring miR-584-5p-disrupting rs1805672 G allele was constructed using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The Dual-Luciferase® Reporter Assay System (Promega), with Firefly and Renilla luciferase activities reporting and normalizing the assay, respectively, was used according to the manufacturer’s instructions to measure the silencing effect of co-transfected miR-584-5p. In order to verify the potential of this miRNA to downregulate KLRG1 full messages, miR-584-5p mirVana™ miRNA Mimic or Negative Control #1 oligonucleotides (Applied Biosystems) were transfected using Lipofectamine 2000 and Opti-MEM (Invitrogen Carlsbad, CA) following the manufacturer’s protocol. KLRG1 mRNA levels were then measured through RT-qPCR, with GAPDH as the reference gene for assay normalization. For detection of KLRG1 and GAPDH mRNAs with FastStart Essential DNA Green Master (Roche Applied Science, Penzberg, Germany), the following primers were designed to achieve best coverage: 5’ CTCACACCTCCTTGTGATAAC 3’ (forward) and 5’ TTGTTCCTCAGACCAATCCA 3’ (reverse); and 5’ GAAGGTGAAGGTCGGAGTC 3’ (forward) and 5’ GAAGATGGTGATGGGATTTC 3’ (reverse), respectively. All assays were carried out in PBMC (Cellular Technology, Shaker Heights, OH) and in duplicates or triplicates.
2.5. Statistical analyses
Prior to genetic association analysis, the proportion of EU and M individuals was adjusted, excluding some individuals, to be equal between patients and controls in the total sample, therefore avoiding effects of population structure. Considering the limited sizes of EU and M subgroups, we did not perform genetic association analysis of these subgroups alone.
For the genetic association analysis, allelic, genotypic, haplotypic and allele carrier frequencies were compared between patients and controls of the adjusted total sample by Fisher’s exact test, available through VassarStats (VassarStats: Website for Statistical Computation). Haplotypes were inferred through PLINK v1.07 for SNPs residing in the same gene (rs16839951 and rs422236 of CD1D; rs116898958 and rs4145212 of TNFSF13B), or for SNPs from genes closely linked in the same chromosome (rs2537752 of KLRD1 and rs1805672 of KLRG1). Odds ratios (OR) with confidence intervals (95% CI) were also estimated through VassarStats. When a class had value of zero in patients or in controls, the P value was calculated by Fisher’s exact test, as described above, and then 0.5 was added in each class to estimate the OR and CI. Hardy-Weinberg equilibrium was tested for all loci, using chi-square to compare observed and expected genotypic frequencies. Yates correction was applied whenever the expected value of any class was below 5 and the P value under 0.1.
Correlation analyses between SH3TC2 mRNA and miR-584-5p, SH3TC2 and KLRG1 mRNAs, and KLRG1 mRNA and miR-584-5p levels in PBMC, regardless of or based on genotype, were performed through VassarStats considering a nonparametric distribution (Spearman correlation). Reporter gene assays for both rs1805672 constructs as well as the oligonucleotide transfection assay were compared to their respective negative conditions (mirVana™ miRNA Negative Control #1) through the t-test on VassarStats. For the reporter gene assays, rs1805672 constructs were also compared to each other. KLRG1 mRNA levels were compared based on rs1805672 genotype (G+ vs. A/A) through the Mann-Whitney test also on VassarStats. For all statistical analyses, the significance level was set at P<0.05, considering a two-tailed test.
3. Results
3.1. The A/G genotype of KLRG1 rs1805672 is associated with increased predisposition to PF
Among all nine variations, the following positions were found to be monomorphic in both patient and control groups: rs139105990 of CTLA4 (monoallelic) and rs185198828 of TNFSF13B (MAF: 0.2% and 0.3%, respectively). For polymorphic positions, comparisons of allelic, genotypic and allele carrier frequencies between PF patients and endemic controls, considering the total or ethnically-adjusted samples, revealed that the KLRG1 rs1805672 A/G genotype is associated with increased susceptibility to the disease (Table 2; A/G vs. A/A: P=0.038; OR=1.60). Moreover, haplotypic analyses for pairs of SNPs residing in the same gene (CD1D, TNFSF13B) or chromosome (12p13, i.e., SNPs of KLRD1 and KLRG1) did not reveal any significant difference between patients and controls (Table 3).
Table 2.
Allelic, genotypic and allele carrier frequencies of seven SNPs compared between the PF patient and the endemic control groups.
| Gene/SNP | PF (%) | Control (%) | OR | 95% CI | P |
|---|---|---|---|---|---|
| CD1D | |||||
| rs422236 | |||||
| T | 234 (49.8) | 190 (50.5) | Ref. | ||
| G | 236 (50.2) | 186 (49.5) | 1.03 | 0.79 – 1.35 | 0.836 |
| TT | 54 (23.0) | 52 (27.7) | Ref. | ||
| TG | 126 (53.6) | 86 (45.7) | 1.41 | 0.88 – 2.26 | 0.187 |
| GG | 55 (23.4) | 50 (26.6) | 1.06 | 0.62 – 1.82 | 0.891 |
| T+ | 180 (76.6) | 138 (73.4) | 1.19 | 0.76 – 1.85 | 0.497 |
| G+ | 181 (77.0) | 136 (72.3) | 1.26 | 0.81 – 1.95 | 0.310 |
| rs16839951 | |||||
| A | 435 (92.6) | 352 (91.7) | Ref. | ||
| G | 35 (7.4) | 32 (8.3) | 0.89 | 0.54 – 1.46 | 0.702 |
| AA | 201 (85.5) | 163 (84.9) | Ref. | ||
| AG | 33 (14.0) | 26 (13.6) | 1.03 | 0.59 – 1.79 | 1.000 |
| GG | 1 (0.4) | 3 (1.6) | 0.27 | 0.03 – 2.62 | 0.331 |
| A+ | 234 (99.6) | 213 (97.7) | 3.71 | 0.38 – 35.99 | 0.331 |
| G+ | 34 (14.5) | 35 (16.1) | 1.05 | 0.62 – 1.79 | 0.891 |
| KLRD1 | |||||
| rs2537752 | |||||
| T | 218 (50.9) | 177 (53.6) | Ref. | ||
| A | 210 (49.1) | 153 (46.4) | 1.11 | 0.84 – 1.49 | 0.465 |
| TT | 55 (25.7) | 47 (28.5) | Ref. | ||
| TA | 108 (50.5) | 83 (50.3) | 1.11 | 0.69 – 1.80 | 0.712 |
| AA | 51 (23.8) | 35 (21.2) | 1.25 | 0.70 – 2.22 | 0.465 |
| T+ | 163 (76.2) | 130 (78.8) | 0.86 | 0.53 – 1.40 | 0.621 |
| A+ | 159 (74.3) | 118 (71.5) | 1.15 | 0.73 – 1.81 | 0.561 |
| KLRG1 | |||||
| rs1805672 | |||||
| A | 323 (75.1) | 260 (78.3) | Ref. | ||
| G | 107 (24.9) | 72 (21.7) | 1.20 | 0.85 – 1.68 | 0.343 |
| AA | 117 (54.4) | 105 (63.3) | Ref. | ||
| AG | 89 (41.4) | 50 (30.1) | 1.60 | 1.03 – 2.47 | 0.038 |
| GG | 9 (4.2) | 11 (6.6) | 0.73 | 0.29 – 1.84 | 0.641 |
| A+ | 206 (95.8) | 155 (93.4) | 1.62 | 0.66 – 4.02 | 0.356 |
| G+ | 98 (45.6) | 61 (36.7) | 1.44 | 0.95 – 2.18 | 0.094 |
| NKG7 | |||||
| rs3009 | |||||
| A | 279 (65.2) | 225 (67.4) | Ref. | ||
| G | 149 (34.8) | 109 (32.6) | 1.10 | 0.81 – 1.49 | 0.538 |
| AA | 92 (43.0) | 83 (49.7) | Ref. | ||
| AG | 95 (44.4) | 59 (35.3) | 1.45 | 0.94 – 2.26 | 0.118 |
| GG | 27 (12.6) | 25 (15.0) | 0.97 | 0.52 – 1.81 | 1.000 |
| A+ | 187 (87.4) | 142 (85.0) | 1.22 | 0.68 – 2.19 | 0.549 |
| G+ | 122 (57.0) | 84 (50.3) | 1.31 | 0.87 – 1.97 | 0.214 |
| TNFSF13B | |||||
| rs4145212 | |||||
| T | 447 (78.1) | 304 (74.5) | Ref. | ||
| A | 125 (21.9) | 104 (25.5) | 0.82 | 0.61 –1.10 | 0.194 |
| TT | 171 (59.8) | 114 (55.9) | Ref. | ||
| TA | 105 (36.7) | 76 (37.3) | 0.92 | 0.63 – 1.34 | 0.700 |
| AA | 10 (3.5) | 14 (6.9) | 0.48 | 0.20 – 1.11 | 0.088 |
| T+ | 276 (96.5) | 190 (93.1) | 2.03 | 0.88 – 4.67 | 0.094 |
| A+ | 115 (40.2) | 90 (44.1) | 0.85 | 0.59 – 1.23 | 0.404 |
| rs116898958 | |||||
| C | 650 (97.6) | 831 (97.3) | Ref. | ||
| T | 16 (2.4) | 23 (2.7) | 0.89 | 0.47 – 1.70 | 0.718 |
| CC | 317 (95.2) | 404 (94.6) | Ref. | ||
| CT | 16 (4.8) | 23 (5.4) | 0.89 | 0.46 – 1.71 | 0.744 |
| TT | 0 (0.0) | 0 (0.0) | NC | NC | NC |
| C+ | 333 (100) | 427 (100) | NC | NC | NC |
| T+ | 16 (4.8) | 23 (5.4) | 0.89 | 0.46 – 1.71 | 0.744 |
OR: odds ratio. CI: confidence interval. P: two-tailed P value. Ref.: reference allele or genotype. NC: not calculated. Both rs139105990 (CTLA4) and rs185198828 (TNFSF13B) were monomorphic in the patient and control groups. Genotypic frequencies were in agreement with those expected by Hardy-Weinberg equilibrium for all loci, except for rs3009 in the control group (P=0.01).
Table 3.
Haplotypic frequencies of three SNP pairs compared between the PF patient and the endemic control groups.
| Haplotype | PF (%) | Control (%) | OR | 95% CI | P |
|---|---|---|---|---|---|
| CD1D | |||||
| rs16839951, rs422236 | |||||
| AG | 232 (49.8) | 186 (49.5) | 1.01 | 0.77 – 1.33 | 0.945 |
| GT | 32 (6.9) | 30 (8.0) | 0.85 | 0.51 – 1.43 | 0.596 |
| AT | 202 (43.3) | 160 (42.5) | 1.03 | 0.78 – 1.36 | 0.834 |
| KLRG1, KLRD1 | |||||
| rs1805672, rs2537752 | |||||
| GA | 74 (14.6) | 45 (12.0) | 1.25 | 0.84 – 1.86 | 0.275 |
| AA | 175 (34.6) | 130 (34.8) | 0.99 | 0.75 – 1.31 | 1.000 |
| GT | 71 (14.0) | 51 (13.6) | 1.03 | 0.70 – 1.52 | 0.921 |
| AT | 186 (36.7) | 148 (39.6) | 0.89 | 0.67 – 1.17 | 0.400 |
| TNFSF13B | |||||
| rs116898958, rs4145212 | |||||
| TA | 10 (2.1) | 7 (1.8) | 1.16 | 0.44 – 3.07 | 0.810 |
| CA | 101 (21.5) | 86 (22.6) | 0.94 | 0.67 – 1.29 | 0.739 |
| CT | 359 (76.4) | 286 (75.3) | 1.16 | 0.43 – 3.10 | 0.809 |
OR: odds ratio. CI: confidence interval. P: two-tailed P value.
3.2. KLRG1 mRNA and miR-584-5p levels positively correlate in healthy donors regardless of genotype and possibly in those carrying the rs1805672 G allele
Considering the aforesaid association of the KLRG1 rs1805672 A/G genotype with A/A set as reference and the tendency of association of G+ with PF (Table 2; P=0.094; OR=1.44), we hypothesized that the G allele of rs1805672 could be disrupting a miRNA binding site and, therefore, contributing to the observed higher levels of KLRG1 in untreated patients – KLRG1 was previously reported by our group to be 2.2-fold overexpressed in untreated patients with the generalized form of PF compared to healthy individuals (Table S1). To test this hypothesis, we focused on a single high confidence miRNA, which binding to KLRG1 is supposed to be lost in the presence of the G allele, as predicted by two out of three online bioinformatics tools (Table S2, underlined).
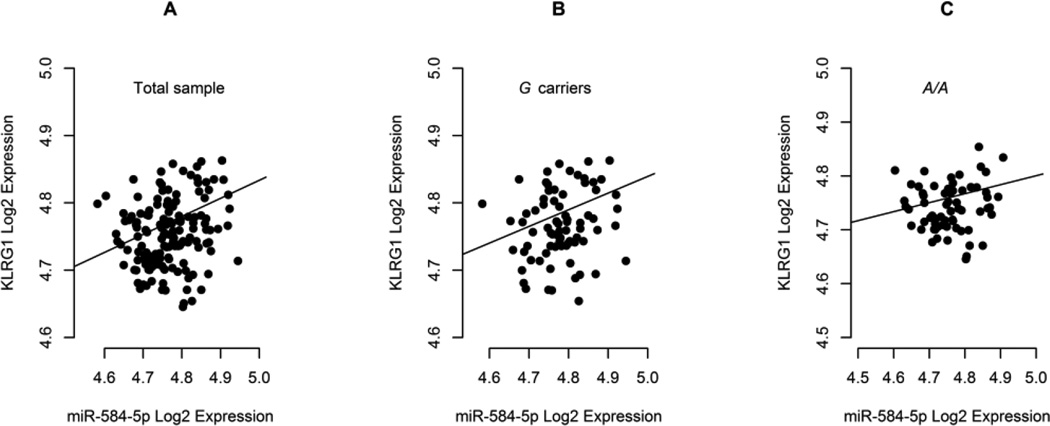
Both KLRG1 mRNA and miR-584-5p levels were quantified in PBMC of healthy donors and tested for correlation in the total sample, i.e., independently of genotype distribution. In this scenario, we found a positive correlation between KLRG1 and miR-584-5p levels in PBMC of 146 healthy donors (Figure 1A; P=0.030; rS=0.180). This was also verified in individuals carrying the miR-584-5p-disrupting G allele as a trend (Figure 1B; P=0.100; rS=0.187; n=75), but not in A/A donors (Figure 1C; P=0.495; rS=0.086; n=65), where miR-584-5p binding site is believed to be fully preserved, therefore allowing for KLRG1 mRNA decay or inhibition of protein synthesis.
Figure 1.
Correlation plots between miR-584-5p and KLRG1 mRNA levels in PBMC of healthy donors after Log2 transformation of Cq values.
Spearman correlation analysis indicated (A) significant positive correlation for the total sample (P=0.030; rS=0.180; n=146), (B) a trend for positive correlation in carriers of the miR-584-5p-disrupting G allele (P=0.100; rS=0.187; n=75) and (C) absence of correlation in individuals with the A/A genotype, where the miR-584-5p seed site is supposed to be fully available (P=0.495; rS=0.086; n=65).
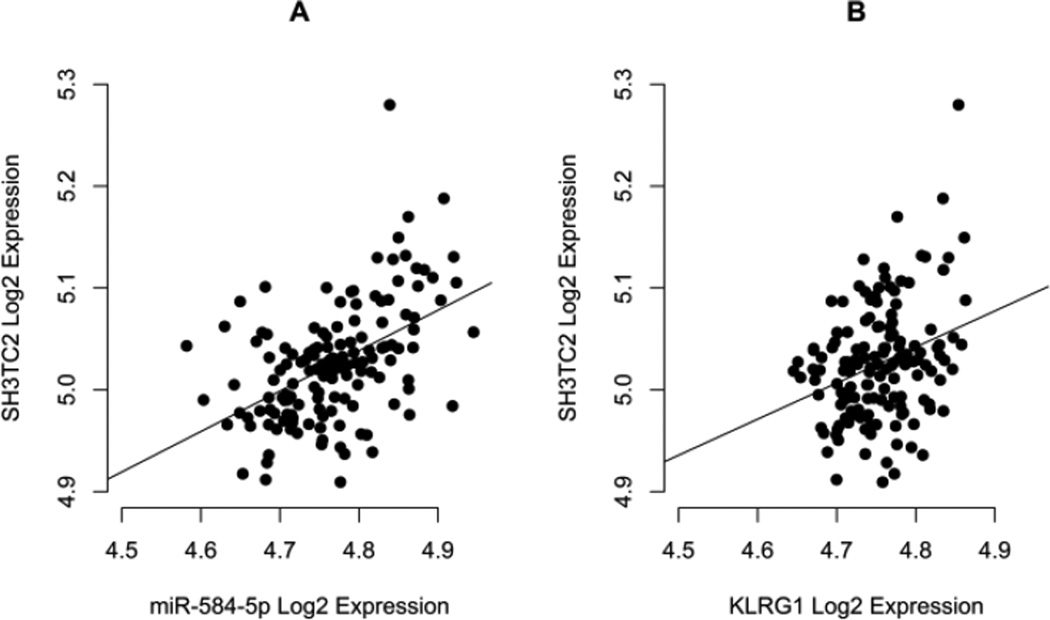
In order to validate our correlation approach and corroborate the hypothesis of direct proportional accumulation of KLRG1 mRNA and mature miR-584-5p as suggested by the aforementioned positive correlation, we also investigated miR-584-5p host gene, SH3TC2. As expected, SH3TC2 mRNA and miR-584-5p levels were positively correlated in PBMC (Figure 2A; P<0.0001; rS=0.474; n=146), as was also the SH3TC2 and KLRG1 mRNA levels (Figure 2B; P=0.002; rS=0.250; n=146). Interestingly, when considering the rs1805672 genotype, positive significant correlation was observed between SH3TC2 and KLRG1 mRNA levels only for carriers of the G allele (data not shown; P=0.007; rS=0.312; n=75. For A/A individuals: P=0.075; rS=0.222; n=65).
Figure 2.
Correlation plots between miR-584-5p and SH3TC2, and KLRG1 and SH3TC2 mRNA levels in PBMC of healthy donors after Log2 transformation of Cq values.
Spearman correlation analysis confirmed (A) the expected significant positive correlation between miR-584-5p and its host gene, SH3TC2, mRNA levels (P<0.0001; rS=0.474; n=146) and revealed (B) a significant positive correlation between KLRG1 and SH3TC2 mRNA levels (P=0.002; rS=0.250; n=146), corroborating our hypothesis that KLRG1 and SH3TC2 accumulate at directly proportional levels in PBMC, which in turn explains positive correlation between KLRG1 mRNA and miR-584-5p in the case where individuals are carriers of miR-584-5p-disrupting G allele.
3.3. The rs1805672 G allele disrupts a miR-584-5p binding site on the 3’UTR of KLRG1
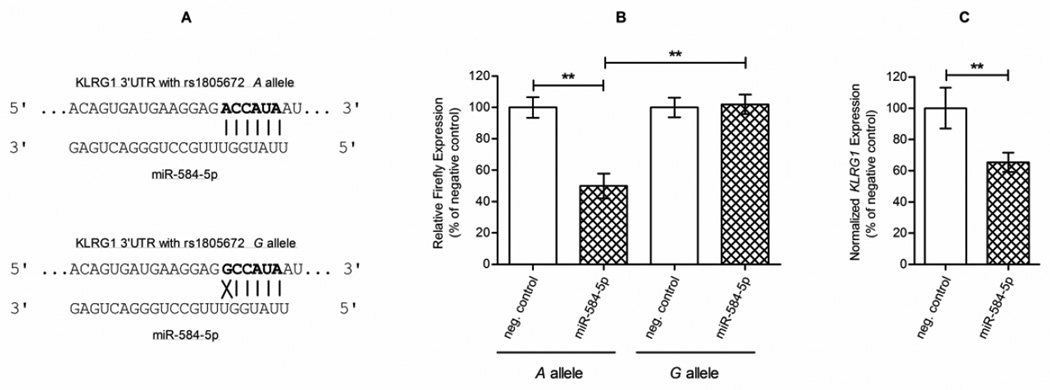
The putative binding site of miR-584-5p on KLRG1 3’UTR was confirmed through TargetScan Release 7.0. Interaction of miR-584-5p with KLRG1 3’UTR is predicted to take place in only one site, where it overlaps with rs1805672 polymorphism (Figure 3A). Dual-luciferase reporter assays were performed to verify the interaction between miR-584-5p and KLRG1 mRNA 3’UTR in the presence of rs1805672 A allele. Luciferase activity was significantly decreased by approximately 50% in the presence of miR-584-5p when compared to the miRNA-negative transfection condition (Figure 3B; P=0.006). Using the mutated vector, i.e., harboring rs1805672 G allele, no significant modulation of luciferase activity was observed when cells were co-transfected with miR-584-5p and compared to the miRNA-negative control (P=0.391). The ability of rs1805672 G allele to disrupt miR-584-5p-mediated luciferase downregulation through the 3’UTR of KLRG1 was confirmed when comparing both miRNA-transfected conditions (Figure 3B; P=0.005). Finally, our observation that miR-584-5p downregulates KLRG1 mRNA, as detected by RT-qPCR of PBMC transfected with the miRNA mimic (Figure 3C; P=0.007), confirms the silencing effect of miR-584-5p on KLRG1 full messages.
Figure 3.
Putative and validated interaction between miR-584-5p and the 3’UTR of KLRG1 in the context of rs1805672 polymorphism.
According to TargetScan algorithm, (A) the predicted interaction of miR-584-5p with KLRG1 mRNA takes place from position 1044 to 1050 of the 3’UTR (seed sequence in bold), when an A is present at position 1044 (top), being such interaction lost when a G is present instead (bottom). Our dual-luciferase reporter assays, with Renilla as a normalizer for Firefly expression, confirmed such interaction by (B) showing that, in the presence of the A allele, luciferase activity was significantly decreased by approximately 50% in the presence of miR-584-5p (vs. “neg. control”, miRNA-negative transfection condition, left columns; P=0.006; 49.9% decreased). The mutated vector harboring the G allele showed no significant modulation of luciferase activity when co-transfected with miR-584-5p (vs. “neg. control”, miRNA-negative transfection condition, right columns; P=0.391). Comparison of both miRNA-transfected conditions confirmed the ability of the G allele to disrupt, through the 3’UTR of KLRG1, miR-584-5p-mediated luciferase downregulation (cross-hatched columns; P=0.005). Transfection of miR-584-5p mimic in PBMC resulted in (C) a decrease of approximately 65% of KLRG1 mRNA levels (vs. “neg. control”, miRNA-negative transfection condition; P=0.007; 65.3% decreased), extending the reported silencing effect of this miRNA on KLRG1 3’UTR to its full messages as well.
3.4. The miR-584-5p-disrupting G/G genotype marks the highest KLRG1 mRNA levels in the fibroblast cell GTEx data
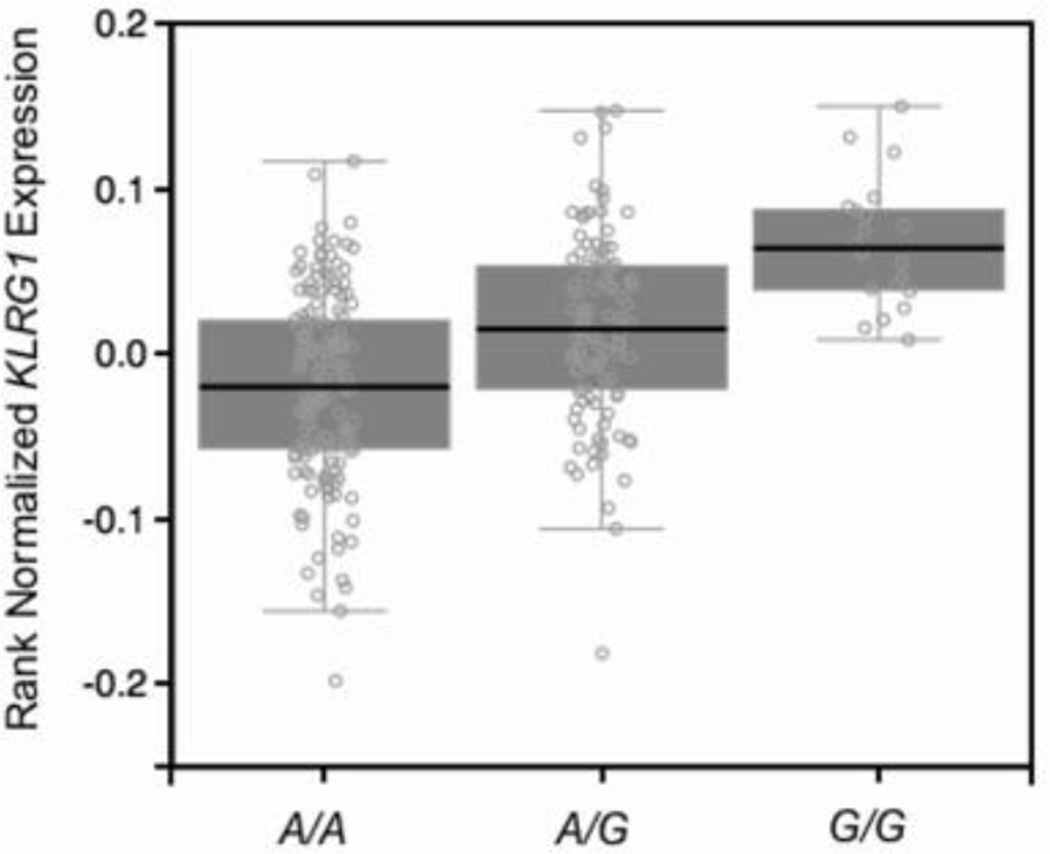
Our observations that the rs1805672 G allele disrupts a miR-584-5p binding site on KLRG1 3’UTR, raised the question whether the levels of KLRG1 mRNA were significantly higher in PBMC of G+ healthy individuals when compared to the A/A genotype. Indeed, KLRG1 mRNA accumulated at significantly higher levels in carriers of the G allele (data not shown; P=0.004). This effect would also be expected in G/G individuals, though the size of our PBMC sample would not allow comparison with this genotype. We explored the RNA-Seq data from fibroblasts present in the GTEx database and checked if rs1805672 could display an expression Quantitative Trait Locus (eQTL) effect on KLRG1 in an independent and larger sample. We found a strong cis-eQTL effect of this SNP on KRLG1 mRNA levels (Figure 4; P=3.4×10−13; effect size=0.48; n=272), with the G/G genotype marking the highest KLRG1 mean levels. In summary, our findings are consistent with a miR-584-5p–dependent posttranscriptional modulation of KLRG1 mRNA levels through the rs1805672 eQTL.
Figure 4.
Expression Quantitative Trait Locus (eQTL) effect of rs1805672 on KLRG1.
Based on RNA-Seq data from fibroblasts present in the GTEx database, we show a strong cis-eQTL effect of rs1805672 on KLRG1 mRNA levels (P=3.4×10−13; effect size=0.48). The higher KLRG1 mRNA levels in individuals with miR-584-5p-disrupting G/G genotype is noteworthy.
4. Discussion
By means of a case-control study involving nine 3’UTR SNPs distributed among six immune-related genes previously found to be differentially expressed in PF, we describe an association between the disease and the A/G genotype of KLRG1 rs1805672, in addition to a trend of increased PF susceptibility for rs1805672 G carriers. These findings lead us to suggest that the lack of association with the rs1805672 G/G genotype is likely due to its low frequency in both patients and controls. All SNPs were chosen based on their putative ability to disrupt/create a miRNA binding site. Therefore, we focused our efforts on the miRNA-mRNA interface, hypothesizing that the genetic association could account, at least in part, for the mRNA differential levels.
Among the approaches for validating in silico-predicted miRNA-mRNA interactions, the correlation analyses between their levels is among the most commonly used strategies, especially when focusing on the interactome of a given trait (28–30). We used this same approach, but at a single candidate miRNA-mRNA interaction, and provide the first biological evidence that rs1805672 of KLRG1 overlaps with a miRNA targeting site. Interestingly, we found a positive correlation between KLRG1 mRNA and miR-584-5p levels in the total sample (i.e., regardless of genotypes) and a same-direction trend in carriers of the miR-584-5p-disrupting G allele. Although positive correlation is not intuitively expected between bona fide miRNA-mRNA pairs given the canonical mode of action of miRNAs, this relationship has been observed in several studies (31–35).
It has been verified that positive correlation between miRNA-mRNA pairs regardless of targeting relationships occurs between intronic miRNAs and their host genes, as expression of the former is dependent on transcription of the latter (36). In fact, miR-584-5p is an intronic miRNA, however not located in the same chromosome of KLRG1. Instead, miR-584-5p is located in chromosome 5 as an intron of the SH3 domain and tetratricopeptide repeats 2 gene (SH3TC2). Along with our results, this leads us to hypothesize that, although residing in different chromosomes, KLRG1 and miR-584-5p host gene share some degree of transcriptional regulation in PBMC. This hypothetical gene expression co-regulation will allow both RNAs to accumulate at proportional levels when miR-584-5p binding site in KLRG1 3’UTR is not present in two copies, i.e., when individuals are carriers of the rs1805672 G allele, as showed by our correlation analyses. In fact, quantification of the mRNA of SH3TC2 and correlation evaluation between it and miR-584-5p and KLRG1 in the same PBMC sample showed positive correlation, validating the correlation approach – as positive correlation is expected between intronic miRNAs and their host genes – and corroborating our hypothesis of KLRG1 and SH3TC2 co-expression. Such co-expression would be indicative of a common signaling pathway regulating transcription of both genes. In fact, it has been reported that miR-584 expression is upregulated by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappa B) signaling (37). The MHC class I, a positive regulator of KLRG1 (38), has been known to be upregulated by NF-kappa B (39,40). Therefore, differential activity status of NF-kappa B between PBMCs can accompany low or high KLRG1/SH3TC2 expressions. Finally, since (1) miR-584-5p targets KLRG1, but rs1805672 G allele disrupts binding of this microRNA, and (2) transcription of both KLRG1 and SH3TC2 can be coordinately regulated by NF-kappa B, there is a possibility that protein synthesis of KLRG1 harboring rs1805672 G allele is not affected by the miR-584-5p once NF-kappa B is activated.
Gene reporter assay is a gold-standard technique to validate miRNA-mRNA direct interactions (41). Through this technique, we deliver another layer of evidence for the predicted miR-584-5p-mediated KLRG1 posttranscriptional regulation. The magnitude of rs1805672 A allele posttranscriptional silencing effect through miR-584-5p (~50% downregulation of firefly’s activity) is in agreement with previously observed aggregated effects of translational repression and mRNA degradation of certain miRNAs on their targets (42). In addition, our finding that the rs1805672 G allele dramatically abrogates miR-584-5p-mediated downregulation of luciferase’s activity allows us to suggest that this SNP may play a major role in modulating KLRG1 mRNA levels. In fact, these levels significantly differed among PBMCs of A/A and G+ individuals, being higher in the latter. Moreover, an additive effect of the G allele on KLRG1 mRNA levels can be deduced when considering our eQTL analysis of the GTEx database, as A/G and G/G individuals showed the intermediate and highest KLRG1 mean levels for the rs1805672 eQTL, respectively. Clearly, several other SNPs in KLRG1 3’UTR may contribute to its posttranscriptional regulation through miRNAs and potentially associate with susceptibility to autoimmune diseases. Likewise, nucleotide variation within a miRNA seed sequence and its flanking regions could contribute to differential levels of targeted mRNA. However, analysis of miR-584-5p sequence available in genome browsers does not report SNPs located within the seed sequence or its flanking regions. Interestingly, there are multiple reported SNPs in the miR-584-5p precursor sequence. Since SNPs of miRNA precursors can affect maturation processing (43,44), it can be expected that the expression of mature miR-584-5p may be modulated by SNPs, ultimately affecting cleavage or translation of its targets, e.g., KLRG1. Taken together, the aforesaid findings allow us to conclude that rs1805672 marks differential KLRG1 mRNA levels – very likely and with considerable extent – through disrupting a miR-584-5p binding site.
Given these results, we believe to have unveiled a possible mechanistic explanation for the genetic association of rs1805672 with pemphigus foliaceus, where KLRG1 mRNA levels were found to be increased in untreated patients in relation to healthy controls (8). These higher levels of KLRG1 mRNA in the disease may be due, in part, to the higher frequency of the rs1805672 A/G genotype among patients when compared to controls, leading to an impaired miR-584-5p-mediated KLRG1 posttranscriptional downregulation. Independent of the other factors that may be contributing to this differential expression of KLRG1 in PF, the association between the KLRG1 genotype with both the disease and mRNA levels may allow us to postulate that KLRG1 variable levels have a causative effect towards the disease. Interestingly, KLRG1 protein has been shown to bind E-cadherin (CDH1) (45). More recently, autoantibodies against this classical cadherin have been detected in sera of about half of PF patients besides healthy individuals of an endemic area in Brazil. Importantly, anti-CDH1 autoantibodies were not detected in healthy subjects from a USA non-endemic location (11). Another striking observation is that the missing self recognition mediated by KLRG1 relies on its binding to a conserved site on E-cadherin that overlaps the site responsible for cell-cell adhesion (21). Therefore, above-normal KLRG1 levels could be impairing E-cadherin-dependent cell-cell adhesion, thus having a pathological role in pemphigus. Another possibility is that binding of KLRG1 to E-cadherin could be eliciting an intracellular response on keratinocytes expressing CDH1, leading to the activation of signaling pathways ultimately linked to “apoptolysis”, an interesting new concept for pemphigus’ pathogenesis (13). This hypothetical activation would not necessarily involve the recognized natural killer inhibitory response in which KLRG1 plays an important role (46,47). The supposed KLRG1-CDH1-dependent activation of intracellular signaling pathways in keratinocytes leading to “apoptolysis” would have an additive effect to those already described.
In this context, it remains to be verified whether a relationship between anti-CDH1 autoantibodies, PF-increased KLRG1 levels and KLRG1-CDH1 binding exists in pemphigus. Independent of the existence of this pathway in the disease, our results suggest that KLRG1 variable levels in immune-related conditions have the potential to be modulated by miR-584-5p for therapeutic purposes.
5. Conclusion
Here, we have shown that a 3’UTR polymorphism, rs1805672, marks differential KLRG1 levels by its verified potential to disrupt a miR-584-5p binding site. Although other factors are expected to contribute to the variable levels of KLRG1, the rs1805672 may be considered relevant as it was found to be associated with pemphigus foliaceus, an autoimmune disease, and therefore be clinically relevant for KLRG1-mediated immune responses. Besides proposing a functional elucidation for this association, our study adds value to the discovery of polymorphisms contributing to human variable gene expression, especially to those located in miRNA binding sites, as they may be considered for further therapeutic approaches, when designing alternative clinical trials based on RNA interference.
Supplementary Material
Highlights for.
Manuscript BBAGRM-16-127 titled “A 3’UTR polymorphism marks differential KLRG1 mRNA levels through disruption of a miR-584-5p binding site and associates with pemphigus foliaceus susceptibility” by Cipolla et al.
The KLRG1 rs1805672 G allele may increase susceptibility to pemphigus foliaceus
A molecular mechanism is proposed for G allele-dependent miR-584-5p binding loss
The G allele abrogates reduction of reporter levels with KLRG1 3’UTR and miR-584-5p
rs1805672 shows an expression Quantitative Trait Locus effect on KLRG1 expression
Higher levels of KLRG1 in the disease may relate to rs1805672 miR-disrupting effect
Acknowledgments
We would like to thank Bruno Zagonel Piovezan for performing the SNPlex® genotyping and Carolina Maciel Camargo for obtaining the PBMC sample from healthy donors.
Funding
This work was supported by the National Institutes of Health [grant number EY019463] to RML; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to MLPE; Fundação Araucária to MLPE; and Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES) [Bolsista CAPES – Processo n° 99999.006318/2015-00] to GAC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors of this study declare the absence of any conflict of interest.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang Z, Du R, Edwards A, Flemington EK, Zhang K. The sequence structures of human microRNA molecules and their implications. PLoS One. 2013;8:e54215. doi: 10.1371/journal.pone.0054215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cipolla GA. A non-canonical landscape of the microRNA system. Front. Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malheiros D, Panepucci RA, Roselino AM, Araújo AG, Zago MA, Petzl-Erler ML. Genome-wide gene expression profiling reveals unsuspected molecular alterations in pemphigus foliaceus. Immunology. 2014;143:381–395. doi: 10.1111/imm.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camargo CM, Augusto DG, Petzl-Erler ML. Differential gene expression levels might explain association of LAIR2 polymorphisms with pemphigus. Hum. Genet. 2016;135:233–244. doi: 10.1007/s00439-015-1626-6. [DOI] [PubMed] [Google Scholar]
- 10.Groves RW. Pemphigus: a brief review. Clin. Med. 2009;9:371–375. doi: 10.7861/clinmedicine.9-4-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores G, Culton DA, Prisayanh P, Qaqish BF, James K, Maldonado M, Aoki V, Hans-Filho G, Rivitti EA, Diaz LA. IgG autoantibody response against keratinocyte cadherins in endemic pemphigus foliaceus (fogo selvagem) J. Invest. Dermatol. 2012;132:2573–2580. doi: 10.1038/jid.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grando SA. Pemphigus autoimmunity: hypotheses and realities. Autoimmunity. 2012;45:7–35. doi: 10.3109/08916934.2011.606444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grando SA, Bystryn JC, Chernyavsky AI, Frusić-Zlotkin M, Gniadecki R, Lotti R, Milner Y, Pittelkow MR, Pincelli C. Apoptolysis: a novel mechanism of skin blistering in pemphigus vulgaris linking the apoptotic pathways to basal cell shrinkage and suprabasal acantholysis. Exp. Dermatol. 2009;18:764–770. doi: 10.1111/j.1600-0625.2009.00934.x. [DOI] [PubMed] [Google Scholar]
- 14.Vielmuth F, Waschke J, Spindler V. Loss of desmoglein binding is not sufficient for keratinocyte dissociation in pemphigus. J. Invest. Dermatol. 2015;135:3068–3077. doi: 10.1038/jid.2015.324. [DOI] [PubMed] [Google Scholar]
- 15.Aoki V, Sousa JX, Jr, Diaz LA. Pathogenesis of endemic pemphigus foliaceus. Dermatol. Clin. 2011;29:413–418. doi: 10.1016/j.det.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chagas AC, Ivo ML, Honer MR, Correa Filho R. Situation of endemic pemphigus foliaceus in Mato Grosso do Sul, Brazil, 1990–1999. Rev. Lat. Am. Enfermagem. 2005;13:274–276. doi: 10.1590/s0104-11692005000200022. [DOI] [PubMed] [Google Scholar]
- 17.Castro RM, Proença NG. Similarities and differences between South American pemphigus foliaceus and Cazanave pemphigus foliaceus. An. Bras. Dermatol. 1983;53:137–139. [Google Scholar]
- 18.Stanley JR, Klaus-Kovtun V, Sampaio SA. Antigenic specificity of fogo selvagem autoantibodies is similar to North American pemphigus foliaceus and distinct from pemphigus vulgaris autoantibodies. J. Invest. Dermatol. 1986;87:197–201. doi: 10.1111/1523-1747.ep12695334. [DOI] [PubMed] [Google Scholar]
- 19.Park SH, Bendelac A. CD1-restricted T-cell responses and microbial infection. Nature. 2000;406:788–792. doi: 10.1038/35021233. [DOI] [PubMed] [Google Scholar]
- 20.Chikuma S, Bluestone JA. CTLA-4 and tolerance: the biochemical point of view. Immunol. Res. 2003;28:241–253. doi: 10.1385/IR:28:3:241. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Hofmann M, Wang Q, Teng L, Chlewicki LK, Pircher H, Mariuzza RA. Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition. Immunity. 2009;31:35–46. doi: 10.1016/j.immuni.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder MR, Weyand CM, Goronzy JJ. The double life of NK receptors: stimulation or co-stimulation? Trends Immunol. 2004;25:25–32. doi: 10.1016/j.it.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Vogler I, Steinle A. Vis-à-vis in the NKC: genetically linked natural killer cell receptor/ligand pairs in the natural killer gene complex (NKC) J. Innate. Immun. 2011;3:227–235. doi: 10.1159/000324112. [DOI] [PubMed] [Google Scholar]
- 24.Medley QG, Kedersha N, O’Brien S, Tian Q, Schlossman SF, Streuli M, Anderson P. Characterization of GMP-17, a granule membrane protein that moves to the plasma membrane of natural killer cells following target cell recognition. Proc. Natl. Acad. Sci. U. S. A. 1996;93:685–689. doi: 10.1073/pnas.93.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 2013;24:203–215. doi: 10.1016/j.cytogfr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, Wu PP, Wang Y, Spoonde AY, Koehler RT, et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J. Biomol. Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- 27.Jurinke C, van den Boom D, Cantor CR, Köster H. The use of MassARRAY technology for high throughput genotyping. Adv. Biochem. Eng. Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 28.Heiser D, Tan YS, Kaplan I, Godsey B, Morisot S, Cheng WC, Small D, Civin CI. Correlated miR-mRNA expression signatures of mouse hematopoietic stem and progenitor cell subsets predict “Stemness” and “Myeloid” interaction networks. PLoS One. 2014;9:e94852. doi: 10.1371/journal.pone.0094852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vishnubalaji R, Hamam R, Abdulla MH, Mohammed MA, Kassem M, Al-Obeed O, Aldahmash A, Alajez NM. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 2015;6:e1614. doi: 10.1038/cddis.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Zeng Y. Identification of miRNA-mRNA crosstalk in pancreatic cancer by integrating transcriptome analysis. Eur. Rev. Med. Pharmacol. Sci. 2015;19:825–834. [PubMed] [Google Scholar]
- 31.Hsieh WJ, Lin FM, Huang HD, Wang H. Investigating microRNA-target interaction-supported tissues in human cancer tissues based on miRNA and target gene expression profiling. PLoS One. 2014;9:e95697. doi: 10.1371/journal.pone.0095697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Baker S, Jiang H, Stuart G, Bai Y. Correlating bladder cancer risk genes with their targeting microRNAs using MMiRNA-Tar. Genomics Proteomics Bioinformatics. 2015;13:177–182. doi: 10.1016/j.gpb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res. Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, Mayfield RD. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics. 2013;14:725. doi: 10.1186/1471-2164-14-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YP, Li KB. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Jiang Q, Lou X, Ji X, Wen Z, Wu J, Tao H, Jiang T, He W, Wang C, et al. MicroRNAs up-regulated by CagA of Helicobacter pylori induce intestinal metaplasia of gastric epithelial cells. PLoS One. 2012;7:e35147. doi: 10.1371/journal.pone.0035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corral L, Hanke T, Vance RE, Cado D, Raulet DH. NK cell expression of the killer cell lectin-like receptor G1 (KLRG1), the mouse homolog of MAFA, is modulated by MHC class I molecules. Eur. J. Immunol. 2000;30:920–930. doi: 10.1002/1521-4141(200003)30:3<920::AID-IMMU920>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Schouten GJ, van der Eb AJ, Zantema A. Downregulation of MHC class I expression due to interference with p105-NF kappa B1 processing by Ad12E1A. EMBO J. 1995;14:1498–1507. doi: 10.1002/j.1460-2075.1995.tb07136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forloni M, Albini S, Limongi MZ, Cifaldi L, Boldrini R, Nicotra MR, Giannini G, Natali PG, Giacomini P, Fruci D. NF-kappaB, and not MYCN, regulates MHC class I and endoplasmic reticulum aminopeptidases in human neuroblastoma cells. Cancer Res. 2010;70:916–924. doi: 10.1158/0008-5472.CAN-09-2582. [DOI] [PubMed] [Google Scholar]
- 41.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han SJ, Marshall V, Barsov E, Quiñones O, Ray A, Labo N, Trivett M, Ott D, Renne R, Whitby D. Kaposi’s sarcoma-associated herpesvirus microRNA single-nucleotide polymorphisms identified in clinical samples can affect microRNA processing, level of expression, and silencing activity. J. Virol. 2013;87:12237–12248. doi: 10.1128/JVI.01202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tessmer MS, Fugere C, Stevenaert F, Naidenko OV, Chong HJ, Leclercq G, Brossay L. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int. Immunol. 2007;19:391–400. doi: 10.1093/intimm/dxm004. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura S, Kuroki K, Ohki I, Sasaki K, Kajikawa M, Maruyama T, Ito M, Kameda Y, Ikura M, Yamamoto K, et al. Molecular basis for E-cadherin recognition by killer cell lectin-like receptor G1 (KLRG1) J. Biol. Chem. 2009;284:27327–27335. doi: 10.1074/jbc.M109.038802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web References
This paper contains information obtained from the following URLs:
- dbSNP Home Page. http://www.ncbi.nlm.nih.gov/SNP/
- Ensembl Genome Browser. http://www.ensembl.org/index.html.
- GTEx Portal. http://www.gtexportal.org/
- 50.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MirSNP. http://bioinfo.bjmu.edu.cn/mirsnp/search/Search/
- 51.Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, Zhang D. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics. 2012;13:661. doi: 10.1186/1471-2164-13-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- mirsnpscore. http://www.bigr.medisin.ntnu.no/mirsnpscore/
- 52.Thomas LF, Saito T, Sætrom P. Inferrin g causative variants in microRNA target sites. Nucleic Acids Res. 2011;39:e109. doi: 10.1093/nar/gkr414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PolymiRTS Database 3.0. http://compbio.uthsc.edu/miRSNP/
- Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TargetScan Release 7.0. http://www.targetscan.org/vert_70/
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VassarStats: Website for Statistical Computation. http://vassarstats.net.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.