Abstract
Cellular retinoic acid binding proteins (CRABPs) bind all-trans-retinoic acid (atRA) tightly. This study aimed to determine whether atRA is channeled directly to cytochrome P450 (CYP) CYP26B1 by CRABPs, and whether CRABPs interact directly with CYP26B1. atRA bound to CRABPs (holo-CRABP) was efficiently metabolized by CYP26B1. Isotope dilution experiments showed that delivery of atRA to CYP26B1 in solution was similar with or without CRABP. Holo-CRABPs had higher affinity for CYP26B1 than free atRA, but both apo-CRABPs inhibited the formation of 4-OH-RA by CYP26B1. Similar protein-protein interactions between soluble binding proteins and CYPs may be important for other lipophilic CYP substrates.
Keywords: Cytochrome P450, substrate channeling, retinoic acid, binding proteins
Introduction
All-trans-retinoic acid (atRA), is the main active metabolite of vitamin A and is responsible for many of the biological effects of vitamin A [1–3]. The cellular activities of atRA are believed to be mediated by a network of proteins including the enzymes synthesizing atRA, the CYP26 enzymes that are the major atRA clearing enzymes, retinoic acid receptors (RARs) and cellular retinoic acid binding proteins (CRABPs) [1–4]. Inside cells atRA is predominantly bound to its cellular binding proteins CRABP-I and CRABP-II [5,6] while in plasma, atRA is believed to be bound to albumin. atRA acts as a transcriptional activator by binding to specific transcription factors such as RARs, and the transcriptional activity of atRA has been shown to be increased by CRABP-II expression [7–9]. The effect of CRABP-II on RAR activation is likely due to the fact that atRA is channeled by CRABP-II but not CRABP-I to RARα, thereby facilitating transcriptional activity [8]. It has also been shown that in response to atRA binding, CRABP-II localizes to the nucleus whereas CRABP-I does not [7]. However, in the absence of atRA, CRABP-II co-localized with the endoplasmic reticulum (ER) [10]. In contrast, increased expression of CRABP-I was shown to decrease the differentiation-specific gene expression in F9 teratocarcinoma cells [11]. Together these studies show that CRABPs play an important role in mediating atRA actions.
atRA is eliminated mainly by oxidative metabolism [1,12,13]. Cytochrome P450 enzymes of the 26 family (CYP26s) are believed to be mainly responsible for metabolism of atRA during development and adult life [12–16]. Although CYP26A1 and CYP26B1 do metabolize atRA efficiently in the absence of CRABPs [17,18], it has also been shown that atRA bound to CRABP-I is metabolized[19,20], and CRABP-I increases the efficiency of atRA catabolism [11]. Since atRA binds both CRABP-I and CRABP-II tightly (Kd 0.06 and 0.13 nM, respectively) [8], it is expected that when CRABPs are expressed, no free atRA exists within the cell. Hence, the free-drug hypothesis, which states that drug binding to pharmacological receptors and metabolic enzymes is driven by free drug in solution and drug bound to other proteins is unavailable for receptor binding, suggests that atRA is either not available for metabolism or metabolism is greatly reduced in the presence of CRABPs. Despite this, in F9 teratocarcinoma cells, increased metabolism of atRA was observed when CRABP-I expression increased [11]. Similarly, in rat testes microsomes, that likely express CYP26B1, the depletion of atRA in the presence and absence of CRABP was identical, but the overall metabolite formation was decreased with increasing CRABP-I [19,20]. Taken together this data suggests that the free drug hypothesis is not applicable for atRA metabolism, and the CRABP(s) deliver atRA directly to the CYP enzymes responsible for atRA metabolism without a requirement of free atRA in cytosol. However, direct interaction between CRABPs and CYP26 enzymes has not been demonstrated, and the identity of the CRABPs and/or CYPs involved in the targeted metabolism of atRA have not been characterized. In addition, direct channeling of a substrate from a cytosolic carrier protein to any specific membrane bound mammalian CYP enzyme has not been previously demonstrated. The aim of this study was to determine whether CRABPs directly channel atRA to CYP26B1 and whether the CRABPs and CYP26B1 interact with each other.
Materials and Methods
Chemicals and Reagents
Ammonium acetate was obtained from Mallinckrodt Baker (Phillipsburg, NJ), and LC/MS-grade methanol, acetonitrile (ACN) and ethyl acetate from Fisher Scientific (Fairlawn, NJ). CYP3A4 and CYP2C8 Supersomes™ coexpressed with cytochrome P450 reductase and cytochrome b5 were purchased from BD Biosciences (Woburn, MO). 4-OH-RA was synthesized as previously described [21]. atRA and NADPH were purchased from Sigma Aldrich (St. Louis, MO). atRA-d5 and 4-oxo-13-cis-RA-d3 were obtained from Toronto Research Chemicals (North York, ON).
Enzyme expression
Recombinant CYP26B1 was expressed in Sf-9 cells using the baculovirus system and microsomes were prepared as described previously [18]. P450 reductase was expressed in E. Coli and purified as described previously [22]. For incubations P450 reductase was added to the membrane preparations as previously described [18]. The CRABP expression vectors were provided by Dr. Noa Noy (Case Western University) and the CRABP-I and CRABP-II were expressed in E. Coli and purified as described previously [3] and the CRABP concentration was determined by BCA assay, UV absorbance and fluorescence titration. The binding of atRA was confirmed by fluorescence titration as described [8]. Holo-CRABPs were generated by mixing equal concentrations of atRA and CRABPs and the complete 1:1 binding was verified by fluorescence titration and UV spectroscopy.
Formation of 4-OH-RA by P450s in the presence and absence of CRABP-I and CRABP-II
Five pmol/mL of CYP3A4 and CYP2C8 were pre-incubated in 0.5 mL for 3 min with 1 mM NADPH and the reactions were initiated by addition of 1 µM holo-CRABP-I, holo-CRABP-II or atRA. The reaction was quenched after 10 minutes by the addition of an equal volume of acetonitrile followed by 5 mL ethyl acetate containing 30 pmol 4-oxo-RA-d3 as internal standard. For CYP26B1, 5 pmol/mL P450 and 10 pmol/mL P450 reductase were preincubated with 1 mM NADPH for 3 minutes and the reaction was initiated by adding 50 nM atRA alone or 50 nM holo-CRABP-I or holo-CRABP-II. The reactions were quenched after 5 min with an equal volume of acetonitrile followed by the addition of 5 mL ethyl acetate and 30 pmol 4-oxo-RA-d3. Following a liquid-liquid extraction, samples were dried under nitrogen and resuspended in 100 µL acetonitrile. 4-OH-RA formation was analyzed by LC-MS/MS as previously described [21]. All incubations were conducted in triplicate.
Effect of CRABP-I and CRABP-II on CYP26B1 mediated 4-OH-RA formation
The effect of CRABP-I and CRABP-II on the Km and kcat of 4-OH-RA formation by CYP26B1 was determined using 5 pmol/mL CYP26B1 with 10 pmol/mL P450 reductase in 0.5 mL incubations. The incubations were conducted with eight different concentrations (3.9 – 500 nM) of holo-CRABP-I, holo-CRABP-II or free atRA or atRA in the presence of albumin (20 mg/mL) and 4-OH-RA formation was measured. atRA was prebound to CRABP in a 1:1 ratio. CYP26B1 was preincubated with 1 mM NADPH for 3 min at 37°C and reactions were initiated with substrate. Following a 5 min incubation at 37°C, the reaction was quenched with an equal volume of acetonitrile, internal standard added (100 pmol 4-oxo-RA-d3) and 4-OH-RA was extracted with 5 mL ethyl acetate. The concentration of 4-OH-RA was determined by LC-MS/MS analysis as previously described (21).
The inhibition of CYP26B1 by excess apo-CRABP was determined using 5 pmol/mL CYP26B1 and 10 pmol/mL reductase in 0.5 mL incubations. CYP26B1 and reductase were preincubated 3 min at 37°C with 1 mM NADPH and CRABP-I or CRABP-II at 6 different concentrations (0 – 500 nM). The reaction was initiated with 50 nM atRA and incubated for 5 min at 37°C. The reaction was quenched with equal volume of acetonitrile, 100 pmol internal standard (4-oxo-RA-d3) was added and 4-OH-RA was extracted with 5 mL ethyl acetate. The concentration of 4-OH-RA was determined by LC-MS/MS as previously described [21].
Isotope dilution experiments with holo-CRABP-I, holo-CRABP-II and free atRA
For isotope dilution, 10 pmol CYP26B1 with 100 pmol P450 reductase were incubated in 0.5 mL with 100 nM holo-CRABP-I or holo-CRABP-II (bound with either atRA or atRA-d5) and 100 nM free atRA or atRA-d5 for 2 minutes. The reaction was initiated by adding simultaneously holo-CRABP and free atRA. The isotope dilution was conducted both using holo-CRABP-I and holo-CRABP-II with atRA and adding atRA-d5 free in solution and using holo-CRABP-I and holo-CRABP-II with atRA-d5 and adding atRA free in solution. Formation of 4-OH-RA and 4-OH-RA-d4 was monitored by LC-MS/MS as previously described [18], and the ratio between labeled and unlabeled products calculated. To control for intrinsic isotope effect of 4-hydroxylation, atRA and atRA-d5 (100 nM each) were co-incubated with CYP26B1 as described above in the absence of CRABP-I and CRABP-II and the formation of 4-OH-RA and 4-OH-RA-d4 measured.
Determination of kinetic constants
The velocity (v) of 4-OH-RA formation at increasing concentrations of atRA or holo-CRABP as a substrate ([RA]T) was fit to the equation:
| (1) |
where ET, is the total CYP26B1 concentration, kcat is the catalytic rate constant and Km substrate affinity constant. The velocity of 4-OH-RA formation both as a function of increasing holo-CRABP (1:1 CRABP:atRA ratio) and increasing total CRABP (constant atRA concentration) was globally fit to the equation:
| (2) |
where Kd is the affinity constant for atRA with apo-CRABP and Ki is the affinity constant for apo-CRABP with CYP26B1. The Kd values used for CRABP-I and CRABP-II were 0.062 nM and 0.14 nM, respectively as reported [8]. The values of free atRA in solution ([RA]u) and free CRABP-I and CRABP-II ([BP]u) in solution were estimated from total atRA and CRABP concentrations ([RA]T and [BP]T, respectively) and the binding constants using the quadratic formula:
| (3) |
| (4) |
For comparison the formation kinetics of 4-OH-RA from atRA by CYP26B1 in the presence of CRABP-I and CRABP-II and in the absence of any protein-protein interaction between CYP26B1 and CRABPs (the free drug hypothesis model) was simulated using equation (5) and the same ET, kcat and Km values as described above.
| (5) |
The [RA]u was calculated by Equation 3.
All parameters were estimated via nonlinear regression using Solver for Microsoft Excel (Frontline Systems, Incline Village, NV, USA). All statistical analyses were conducted using GraphPad v.5 (La Jolla, CA, USA).
Results
CRABP-I and CRABP-II deliver atRA to CYP26B1 but not to CYP2C8 and CYP3A4
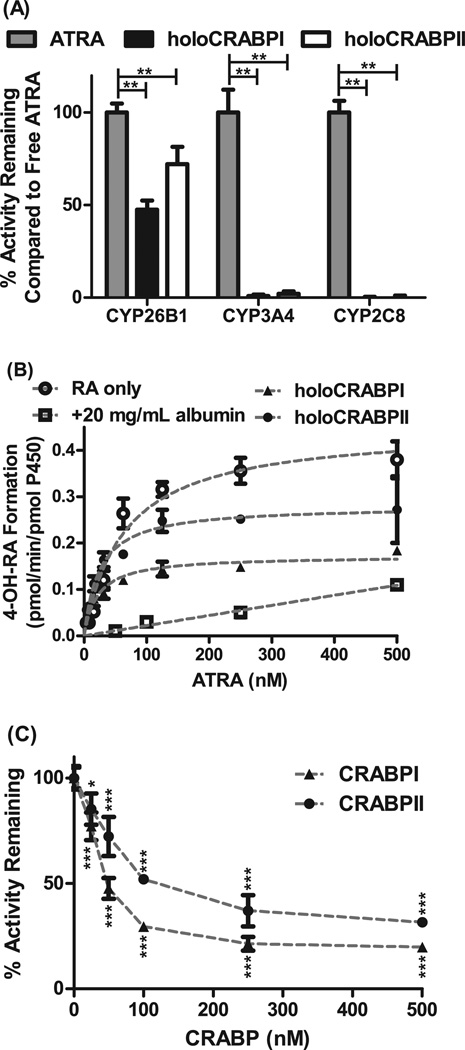
To determine whether CRABP-I or CRABP-II affect the metabolism of atRA by CYP enzymes, atRA was incubated with CYP26B1, CYP2C8 and CYP3A4 in the presence and absence of CRABP-I and CRABP-II. CYP26B1, CYP2C8 and CYP3A4 have all been shown previously to metabolize atRA to 4-OH-RA with Km values of 19 nM, 13 µM and 19 µM, respectively [18,23], and formation of 4-OH-RA was detected with each CYP in this study as well (Figure 1A). In the presence of CRABP-I, the formation rate of 4-OH-RA by CYP26B1 was decreased by 52% (p<0.001) whereas formation of 4-OH-RA by CYP3A4 and CYP2C8 was decreased by 98% (Figure 1A). In the presence of CRABP-II the formation rate of 4-OH-RA by CYP26B1 was decreased by 28% (p<0.001). The formation of 4-OH-RA by CYP3A4 and CYP2C8 was reduced by 99% in the presence of CRABP-II compared to free atRA (Figure 1A). These data suggest that CRABP-I and CRABP-II target atRA selectively for metabolism by CYP26B1 and prevent the metabolism of atRA by other CYP enzymes. The fact that metabolism by CYP3A4 and CYP2C8 was effectively abolished in the presence of CRABP-I and CRABP-II demonstrates that atRA is extensively bound to CRABPs (effective sink in the absence of a receiving enzyme) in the incubations. Based on this data CRABPs minimize the presence of free atRA in solution and prevent the availability of free atRA for metabolism via partitioning to lipid and membrane access to the CYP. As such, these observations indicate that CRABP-I and CRABP-II deliver atRA directly to CYP26B1.
Figure 1. Effect of CRABP-I and CRABP-II on atRA metabolism by CYP26B1.
Panel A shows the 4-OH-RA formation from atRA by CYP26B1, CYP3A4 and CYP2C8 in the presence and absence of CRABP-I and CRABP-II. While atRA is metabolized efficiently by CYP26B1 in the presence of CRABP-I and CRABP-II, metabolism by CYP3A4 and CYP2C8 is completely prevented in the presence of CRABPs. (**p<0.001, two-way ANOVA with Bonferroni post-test) Panel B shows the determination of Km and kcat values for 4-OH-RA formation by CYP26B1 when holo-CRABP-I, holo-CRABP-II, free atRA or atRA bound with albumin is used as a substrate. The apparent Km and kcat values were 64.6 ± 10.3 nM and 0.45 ± 0.02 pmol/min/pmol CYP for free atRA, 21.7 ± 2.9 nM and 0.17 ± 0.006 pmol/min/pmol P450 for holoCRABPI, and 24.3 ± 4.2 nM and 0.28 ± 0.01 pmol/min/pmol CYP for holoCRABPII. Panel C shows the inhibition of CYP26B1 mediated 4-OH-RA formation by excess apo-CRABP. atRA concentration was 50 nM in this experiment. (*p<0.05, ***p<0.0001, one-way ANOVA with Dunnett post-hoc test).
CYP26B1 accepts holo-CRABPs and free atRA as substrates
Previous research has shown that CYP26B1 can obtain free atRA in solution as a substrate and presence of CRABP-I or CRABP-II is not required for atRA turnover or delivery to CYP26B1 in vitro [18]. In addition, since atRA is a high affinity ligand of CYP26B1 it is expected that any atRA that dissociates from CRABPs during the incubation will bind to CYP26B1. Hence, isotope dilution experiments were conducted to determine whether CYP26B1 obtains atRA as a substrate directly from CRABP-I and CRABP-II or whether ligand release to solution is required prior to binding to CYP26B1. When atRA and atRA-d5 were incubated with CYP26B1, an isotope effect was observed and metabolism of the atRA-d5 was 50% slower than that of atRA (4-OH-RA/4-OH-RA-d4 ratio was 1.5) likely due to the fact that the deuterium atoms in the labeled compound are located at C-4 and C-18 of atRA, the sites of metabolism. When atRA was bound with CRABP-I or CRABP-II and coincubated with atRA-d5 free in solution, 4-OH-RA/4-OH-RA-d4 ratios of 1.69 ± 0.24 and 2.58 ± 0.51 were observed with CRABP-I and CRABP-II respectively. The 4-OH-RA/4-OH-RA-d4 ratios were 1.37 ± 0.14 and 0.81 ± 0.02 when atRA-d5 was bound with the CRABP-I and CRABP-II, respectively, and atRA was free in solution. When the isotope dilution values were corrected for the isotope effect of faster metabolism of unlabeled than labeled atRA, the ratio between the products arising from bound and free atRA was 1.1 for CRABP-I and 1.8 for CRABP-II. In the absence of channeling, CYP26B1 should obtain the substrate from free in solution and the ratio of products arising from bound atRA over free atRA should approach zero. A ratio of 1 indicates that channeling does occur but the CYP does not differentiate between the holo-CRABP and atRA free in solution as substrates. A ratio of 1.8 obtained with CRABP-II suggests that the channeling of atRA from CRABP-II to CYP26B1 is preferred over delivery of atRA to CYP26B1 from solution or the lipid bilayer. Taken together the isotope dilution data suggests that holo-CRABPs interact directly with CYP26B1 via protein-protein interaction that results in atRA delivery to CYP26B1.
Holo-CRABPs have lower apparent Km and kcat than atRA with CYP26B1
To further evaluate the metabolism of atRA from holo-CRABP-I and holo-CRABP-II, the apparent Michaelis-Menten kinetics for holo-CRABP-I and holo-CRABP-II were determined in comparison to atRA free in solution (Figure 1B). The apparent Km and kcat values (Figure 1B) were both decreased for holo-CRABP-I and holo-CRABP-II (p<0.0001) when compared to free atRA in solution. Formation of 4-OH-RA from free atRA had a Km of 64.6 ± 10.3 nM and a kcat of 0.45 ± 0.02 pmol/min/pmol P450. Formation of 4-OH-RA from holo-CRABP-I had a kcat of 0.17 ± 0.006 pmol/min/pmol and an apparent Km of 21.7 ± 2.9 nM. With holo-CRABP-II the kcat value was 0.28 ± 0.01 pmol/min/pmol and apparent Km 24.3 ± 4.2 nM. If the unbound fractions for atRA were accounted for in the incubations the unbound apparent Km was decreased 27-fold in the presence of CRABP-I and 18-fold in the presence of CRABP-II in comparison to atRA. As a control of the effect of a nonspecific protein sink to atRA hydroxylation by CYP26B1, albumin was added into the incubations at a concentration of 20 mg/mL. At this albumin concentration the unbound fraction of atRA was 0.01. As expected based on the free drug hypothesis, presence of albumin decreased atRA 4-hydroxylation rate significantly (p<0.05 for 250 nM and 500 nM atRA, paired t-test) in comparison to atRA free in solution (Figure 1B) demonstrating that free drug hypothesis accurately describes atRA metabolism in the presence of albumin. The higher product formation velocity from holo-CRABPs in comparison to albumin bound atRA further supports a direct protein-protein interaction between CRABPs and CYP26B1. The fact that the apparent Km was decreased for holo-CRABPs in comparison to free atRA is in contrast to the free drug hypothesis and can only be explained via direct delivery of atRA from CRABPs to CYP26B1.
Apo-CRABPs inhibit CYP26B1 mediated metabolism of atRA
The decreased Kcat value of 4-OH-RA formation from holo-CRABP-I and holo-CRABP-II suggested that CRABPs act as noncompetitive inhibitors of CYP26B1. To test whether CRABP-I and CRABP-II are inhibitors of CYP26B1, increasing concentrations of apo-CRABPs were added to incubations of holo-CRABPs. Increasing concentrations of apo-CRABPs inhibited the formation of 4-OH-RA in a concentration dependent manner (Figure 1C). holo-CRABP-I and holo-CRABP-II also inhibited the metabolism of 9-cisRA by CYP26B1 (data not shown). Together these data showt that apo-CRABP-I and apo-CRABP-II interact directly with CYP26B1 and inhibit atRA metabolism. Such direct inhibitory interaction between apo-CRABPs and CYP26B1 may be important in the control of cellular concentrations of atRA by allowing fine tuning CYP26B1 activity based on atRA to CRABP concentration ratio.
Kinetic Model of CRABP-CYP26B1 interactions
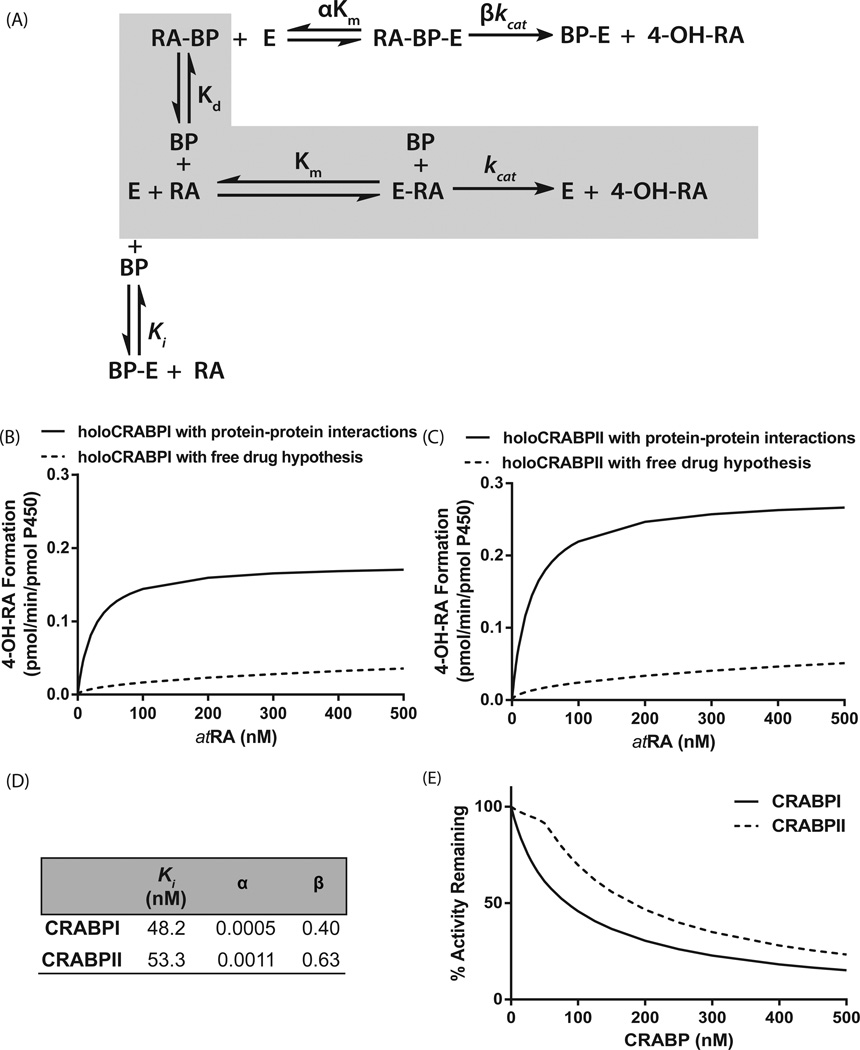
To further establish the kinetics of the interaction between CYP26B1 and CRABP-I and CRABP-II, a kinetic scheme and a velocity equation of the interactions was generated (Figure 2A), The model was globally fitted to the product formation data obtained in the presence and absence of CRABPs to obtain kinetic parameters (Figure 2).. Based on the kinetic parameter estimates, apo-CRABP-I and apo-CRABP-II inhibit CYP26B1 within biologically relevant concentrations (~50 nM) and the affinity of holo-CRABP-I (αKm=0.024 nM) and holo-CRABP-II (αKm=0.059 nM) with CYP26B1 is higher than that of atRA free in solution. However, the kinetic analysis also suggests that the catalytic rates of atRA hydroxylation are decreased by the presence of CRABP-I and CRABP-II. The formation of 4OH-RA by CYP26B1 in the presence of CRABPs was then simulated using the traditional free drug hypothesis model with sequestration of atRA by CRABPs and using the complete protein-protein interaction model (Figure 2B and C).
Figure 2.
Comparison of the models of the metabolism of atRA by CYP26B1 according to the free drug hypothesis and incorporating the protein–protein interactions between CRABPs and CYP26B1. Panel (A) shows the kinetic scheme of the interactions between CRABPs (BP), CYP26B1 (E), and atRA. The shaded gray area indicates the kinetic scheme for the free drug hypothesis, while the complete scheme shows the effect of substrate channeling and protein–protein interactions. Panels (B) and (C) show the simulated product formation curves in the presence of CRABP-I (B) and CRABP-II (C) using either the kinetic parameters obtained from a global fit to the data in Fig. 1B,C (solid lines, protein–protein interaction model) or using the independently measured kinetic constants in the absence of protein–protein interactions (dotted line, free drug hypothesis). The fitted kinetic constants for CRABP interactions with CYP26B1 according to the substrate channeling model are shown in panel (D). Panel (E) shows the simulated product formation curves in the presence of increasing concentrations of apo-CRABP using the kinetic constants from the global fit of the experimental data to the protein–protein interaction model. Using the fitted parameters shown in panel (D), velocity was calculated using Eqn (2) for a range of atRA and CRABP concentrations (0–500 nM) and plotted using GRAPHPAD PRISM v5. The simulations were conducted as described in Materials and methods section.
DISCUSSION
For several decades the role of the individual CRABPs in mediating atRA metabolism has been unclear. The CRABPs are highly conserved across species and the expression patterns of CRABP-I and CRABP-II are distinctly different both during embryonic development and adult life [6]. It has been well established that CRABP-II facilitates transcriptional activation of RARs by atRA [7]. However, the role of CRABP-I has not been as well determined in various tissues with different expression of atRA metabolizing enzymes, and the overall effects of both CRABPs on metabolism of atRA have not been established. In fact, the effect of CRABP-II on atRA metabolism has not been previously reported in any system. In F9 cells CRABP-I expression has been shown to result in lower sensitivity of the cells to atRA [21], and CRABP-I silencing experiments in F9 cells showed that increasing expression of CRABP-I increased the metabolism of atRA and decreased the cellular half-life of atRA [11]. In rat testes microsomes, atRA bound to CRABP-I was shown to be metabolized with greater efficiency than free atRA indicating a direct role of CRABP-I in atRA metabolism [19]. It is important to note that in all of these systems the enzymes responsible for atRA metabolism are not known. This study shows that both CRABP-I and CRABP-II interact specifically with CYP26B1. In addition, the data shows that while apo-CRABP-I and apo-CRABP-II inhibit the metabolism of atRA by CYP26B1, holo-CRABPs specifically deliver atRA to CYP26B1 and the affinity of holo-CRABPs to CYP26B1 is significantly greater than that of free atRA. This increase in the affinity of holo-CRABPs to CYP26B1 in comparison to the decreased kcat leads to an increased intrinsic clearance (kcat/Km) of holo-CRABPs, potentially explaining the observed faster metabolic rates of atRA in cells with higher CRABP expression and the higher metabolic efficiency in rat testes microsomes in the presence of CRABP-I.
The observed product formation data with CRABPs and CYP26B1 can only be explained via substrate channeling and protein-protein interactions, and as shown in Figure 2, CRABPs simply sequestering free atRA from solution would result in distinctly different product formation kinetics with a higher apparent Km (unchanged unbound Km) but unchanged kcat. The observed protein-protein interactions appear to be substrate and CYP26 specific. Based on preliminary studies conducted by us (data not shown), CRABP-I and CRABP-II also interact with CYP26A1 and CYP26C1, the other members of the CYP26 family, in a similar manner. However, as shown here with CYP3A4 and CYP2C8, in the absence of CYP26s CRABPs sequester atRA from solution consistent with the free drug hypothesis. Similarly, in the presence of significant albumin binding, atRA is sequestered from metabolism despite its high affinity to CYP26B1 demonstrating the specificity of the CRABP-CYP26 interaction. Finally, the substrate channeling appears to be specific for atRA as a substrate as CRABP-I binding has been shown to prevent the metabolism of 4OH-atRA and 4oxo-atRA in rat testes microsomes [20] despite the fact that both metabolites bind CRABP-s with similar affinity as atRA[19] and are efficiently metabolized by CYP26B1 [18, 21].
The fact that both CRABPs deliver atRA to CYP26B1 is surprising, as only CRABP-II has been shown to deliver atRA to RAR and localize to the nucleus in the presence of atRA [7,8]. It was expected that only CRABP-I would deliver atRA to CYP26B1 in the ER for metabolism. However, all the data presented here suggest that both CRABP-I and CRABP-II interact with CYP26B1 and channel atRA to CYP26B1 although it is possible that post-translational modifications of CRABPs alter the interactions between CRABPs and CYP26B1. Our data is consistent with the reported localization data of CRABP-II [10] in which CRABP-II is associated with the ER and SUMOylation upon atRA binding in cells results in translocation of CRABP-II into the nucleus followed by export of CRABP-II from the nucleus and re-association of CRABP-II with the ER. Our data suggests that CRABP-II associates with the ER through tight interactions with CYP26B1.
The inhibition of CYP26B1 by CRABPs is in agreement with the previous data reported in rat testes microsomes in which the total amount of radioactive atRA metabolites decreased with increasing apo-CRABP-I concentrations [19]. Interestingly, the data shown here suggests that both CRABP-I and CRABP-II play an important role in regulating the metabolic rate of atRA and that the ratio of apo-CRABP to holo-CRABP for both CRABPs influences the metabolism of atRA. The inhibition of CYP26B1 by apo-CRABPs is biologically plausible as it allows the ratio of apo to holo-CRABP in the cell to mediate the metabolic rate of atRA. According to this model, if cellular atRA concentrations are low (lower than CRABP concentrations) apo-CRABP will inhibit the metabolism of atRA and aid in combatting atRA deficiency. Yet, in situations in which the cell is exposed to excess atRA (i.e. atRA concentrations exceed CRABP concentrations) holoCRABP will facilitate atRA metabolism. It is unlikely that the CRABPs fit within the active site of CYP26B1 and therefore the inhibition of CYP26B1 by CRABPs is expected to be due to CRABPs binding or interacting with a surface site of CYP26B1. It is possible that the CRABPs bind at an allosteric site decreasing catalytic activity but more studies are required to identify the interacting residues and subsequent possible conformational changes in the CYP26B1 protein.
At present there are no reports of a specific mammalian membrane bound CYP accepting a substrate directly from a soluble carrier protein. However, a crystal structure of a bacterial CYP107H1 in complex with the fatty acid linked to acyl carrier protein has been reported [24], providing structural insight how these carrier protein- CYP interactions may occur. Mammalian CYP enzymes are generally believed to gain access to their substrates via the lipid bilayer in which they are embedded, or through a substrate access channel facing the cytosol. It would be possible that CRABPs deliver atRA into the lipid membrane and not directly to the CYP. However, if this was the case, the delivery of the substrate would not be expected to be CYP-enzyme specific as shown here with the lack of holoCRABP metabolism by CYP3A4 and CYP2C8. Similarly, atRA can partition to the lipid bilayer in the presence of albumin but the metabolism of atRA in the presence of albumin was significantly less than in the presence of CRABPs supporting a specific substrate delivery by CRABPs to CYP26B1 via cytosolic access channel. Direct inhibition of CYP3A4 and CYP2C8 by CRABPs was overruled in an experiment using midazolam as a CYP3A4 substrate. CRABP-I and CRABP-II had no effect on CYP3A4 mediated hydroxylation of midazolam, a substrate that does not bind to CRABP (data not shown). Hence both the substrate delivery and the inhibitory effect of CRABPs was specific to the CYP enzyme considered as the endogenous atRA hydroxylase (CYP26B1).
In conclusion, this study shows that soluble cytosolic carrier proteins such as CRABPs can interact with ER membrane bound CYP enzymes and mediate oxidative metabolism of CYP substrates. This suggests that such interactions with other soluble carrier proteins and CYP enzymes may also occur and mediate the metabolism of steroids, fatty acids and other fat soluble vitamins. Similar to the CRABP-II- RAR interaction, the fatty acid binding proteins (FABPs) have been shown to interact with PPAR in the nucleus [9] and it is possible that FABPs also interact with CYP enzymes in the ER. This may have broad effects on the metabolism of fat soluble substrates by CYPs in the liver and other tissues.
Acknowledgments
This work was supported by NIH grants R01-GM081569 and R01-GM111772 and by a pilot grant from the Royalty Research Fund of University of Washington. The authors wish to thank Dr Sumit Goswami, Dr. Jayne E. Thatcher and Elijah Weber for their assistance in enzyme expression, preparation of microsomes and conduct of preliminary experiments, Dr Noa Noy for providing the CRABP expression vectors and Dr Wendel L Nelson for helpful discussions during this work.
REFERENCES
- 1.Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noy N. Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr. 2010;30:201–217. doi: 10.1146/annurev.nutr.28.061807.155509. [DOI] [PubMed] [Google Scholar]
- 3.Ross AC. Cellular metabolism and activation of retinoids: roles of cellular retinoid-binding proteins. Faseb J. 1993;7:317–327. doi: 10.1096/fasebj.7.2.8440409. [DOI] [PubMed] [Google Scholar]
- 4.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 5.Napoli JL, Posch KP, Fiorella PD, Boerman MH. Physiological occurrence, biosynthesis and metabolism of retinoic acid: evidence for roles of cellular retinol-binding protein (CRBP) and cellular retinoic acid-binding protein (CRABP) in the pathway of retinoic acid homeostasis. Biomed Pharmacother. 1991;45:131–143. doi: 10.1016/0753-3322(91)90101-x. [DOI] [PubMed] [Google Scholar]
- 6.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348(Pt 3):481–495. [PMC free article] [PubMed] [Google Scholar]
- 7.Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell Biol. 2002;22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 9.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majumdar A, Petrescu AD, Xiong Y, Noy N. Nuclear translocation of cellular retinoic acid-binding protein II is regulated by retinoic acid-controlled SUMOylation. J Biol Chem. 2011;286:42749–42757. doi: 10.1074/jbc.M111.293464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boylan JF, Gudas LJ. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem. 1992;267:21486–21491. [PubMed] [Google Scholar]
- 12.Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu Rev Nutr. 2011;31:65–87. doi: 10.1146/annurev-nutr-072610-145127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol. 2009;5:875–886. doi: 10.1517/17425250903032681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 16.White JA, Ramshaw H, Taimi M, Stangle W, Zhang A, Everingham S, Creighton S, Tam SP, Jones G, Petkovich M. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc. Natl. Acad. Sci. U S A. 2000;97:6403–6408. doi: 10.1073/pnas.120161397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem. Pharmacol. 2009;77:258–268. doi: 10.1016/j.bcp.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topletz AR, Thatcher JE, Zelter A, Lutz JD, Tay S, Nelson WL, Isoherranen N. Comparison of the function and expression of CYP26A1 and CYP26B1, the two retinoic acid hydroxylases. Biochem. Pharmacol. 2012;83:149–163. doi: 10.1016/j.bcp.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorella PD, Napoli JL. Expression of cellular retinoic acid binding protein (CRABP) in Escherichia coli. Characterization and evidence that holo-CRABP is a substrate in retinoic acid metabolism. J. Biol. Chem. 1991;266:16572–16579. [PubMed] [Google Scholar]
- 20.Fiorella PD, Napoli JL. Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J. Biol. Chem. 1994;269:10538–10544. [PubMed] [Google Scholar]
- 21.Shimshoni JA, Roberts AG, Scian M, Topletz AR, Blankert SA, Halpert JR, Nelson WL, Isoherranen N. Stereoselective formation and metabolism of 4-hydroxy-retinoic Acid enantiomers by cytochrome p450 enzymes. J Biol Chem. 2012;287:42223–42232. doi: 10.1074/jbc.M112.404475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woods CM, Fernandez C, Kunze KL, Atkins WM. Allosteric activation of cytochrome P450 3A4 by alpha-naphthoflavone: branch point regulation revealed by isotope dilution analysis. Biochemistry. 2011;50:10041–10051. doi: 10.1021/bi2013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thatcher JE, Zelter A, Isoherranen N. The relative importance of CYP26A1 in hepatic clearance of all-trans retinoic acid. Biochem. Pharmacol. 2010;80:903–912. doi: 10.1016/j.bcp.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cryle MJ, Schlichting I. Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc. Natl Acad Sci. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]