Abstract
The aim of this study was to investigate the epidemiology of pyrazinamide (PZA) resistance and the associated risk factors as well as to evaluate the pncA gene loci as a marker for PZA resistance in China. A population-based multicenter study of pulmonary tuberculosis (TB) cases was carried out from 2011 to 2013 in four Chinese districts/counties with different geographic and socioeconomic features. Testing for multidrug-resistant tuberculosis (MDR-TB) and susceptibility to PZA was done by the proportion method on Lowenstein-Jensen medium and Bactec MGIT 960, respectively. Mutations in the pncA gene were identified by sequencing. Among 878 culture-positive cases, 147 (16.7%) were resistant to PZA, with a significantly higher proportion among MDR isolates than among the first-line drug-susceptible isolates (30.2% versus 7.7%; P < 0.001). In total, 136 isolates had a nonsynonymous pncA mutation, with a comparable diagnostic performance between Beijing family and non-Beijing family as well as between MDR-TB and first-line drug-susceptible TB. Furthermore, the mutations in isolates with high-level PZA resistance (MIC > 500 mg/liter) were observed mainly in three regions of the pncA gene (codons 51 to 76, codons 130 to 142, and codons 163 to 180). Patients with prior treatment history had a significantly higher risk for PZA monoresistance (odds ratio [OR], 2.86; 95% confidence interval [CI], 1.363 to 6.015) and MDR PZA resistance (OR, 6.47; 95% CI, 3.186 to 13.15), while the additional factors associated with MDR PZA resistance were the patient's age (OR, 1.02; 95% CI, 1.003 to 1.042), lung cavity (OR, 2.64; 95% CI, 1.296 to 5.391). These findings suggest that it is a priority to identify PZA resistance in MDR-TB and that a rapid molecular diagnostic test based on pncA mutations in the Chinese settings where MDR-TB prevalence is high should be developed.
INTRODUCTION
Drug-resistant Mycobacterium tuberculosis, which predominantly arises from the selection of naturally resistant bacterial strains through pressure from inappropriate chemotherapy, poses a serious challenge to global tuberculosis (TB) control (1). Because of its high bactericidal effect in an acidic environment, pyrazinamide (PZA) is usually recommended for the treatment of PZA-susceptible multidrug-resistant TB (MDR-TB) throughout the duration of the regimen (2). An increasing number of PZA-resistant strains has, however, been observed (1), and without precise knowledge of PZA susceptibility, its unstandardized utilization in treatment regimen could lead to resistance against other antibiotics as well (particularly against efficient oral drugs, such as fluoroquinolones [FQ], that are initially included in the regimen). The unavailability of low-cost drug susceptibility testing (DST) techniques, in addition to safety considerations related to the handling of MDR-TB strains, further inhibits our understanding of the scale of PZA resistance in resource-limited areas.
PZA is a prodrug that needs to be converted to an active form, pyrazinoic acid, by the bacterial pyrazinamidase (PZase) enzyme, which is encoded by pncA. Mutations or variations in pncA leading to the loss of PZase activity is the major mechanism leading to PZA resistance (3). However, while high numbers of PZA-resistant cases can be related to inactivation of the PZase, the genetic variants, including single nucleotide polymorphisms (SNPs) and small deletions, are highly diverse and scattered over the full length of the 561-bp pncA gene (4, 5). This complicates the development of molecular tests, as no “hot spot region” comprising the majority of mutations is observed in the pncA gene. Therefore, an in-depth knowledge of the variants found in PZA-resistant strains combined with evidence-based correlation with resistance phenotypes is needed to develop large-scale databases ensuring valid data interpretation especially in specific local settings.
China has one of the highest burdens of notified cases of Mycobacterium tuberculosis infection. In 2012, the Chinese Health Authorities reported a TB incidence of 39.5 cases per 100,000 people nationwide and that 2.3% of primary TB cases were resistant to both isoniazid (INH) and rifampin (RIF) or referred to as MDR-TB (6). Furthermore, according to the National Laboratory Surveillance System for Resistance in TB, 4,170 isolates from notified cases were MDR-TB cases. Despite the high burden of MDR-TB, the magnitude of PZA resistance in China is largely unknown. Since PZA is likely to remain a central component in the treatment of tuberculosis, it is critically important to understand the epidemiology of PZA resistance as well as its associated sociodemographic and clinical factors. The present study aimed to fill this gap by determining the epidemiology of PZA resistance in newly treated patients and previously treated patients as well as the factors associated with PZA resistance in a population-based multicenter study of tuberculosis in China.
MATERIALS AND METHODS
Settings.
Four districts/counties from Zhejiang (DQ), Jiangsu (GY), and Sichuan (GJ, RO) provinces were selected as study sites. The population of these regions ranged from 490,000 to 3,260,000. The TB incidence ranged from 38.2/100,000 to 78.8/100,000, and the case mortality rate ranged from 0.29% to 0.31% in 2011. The directly observed treatment, short-course (DOTS) strategy has been implemented since the early 1990s at all the study sites. All study sites were designated TB health facilities for TB diagnosis, treatment, and management. As recommended by the DOTS strategy, a standardized treatment regimen is routinely applied for drug-susceptible TB cases.
Study design.
A population-based, STROBE-ME (strengthening reporting of observational studies in epidemiology-molecular epidemiology)-compliant cross-sectional study was conducted from January 2011 to December 2013. Patients were enrolled if they (i) were diagnosed as pulmonary tuberculosis patients at the local TB designated health facilities during the study period, (ii) were above 18 years old, (iii) had TB culture-positive sputum, and (iv) gave written consent for their clinical data and specimens to be used for further research. The sputum was collected in a sterile screw-top plastic container and sent to the local TB dispensaries for microbiological culture and then sent to the up-level prefectural TB laboratory for first-line drug susceptibility testing, including PZA. The strains were cultivated on Lowenstein-Jensen medium (L-J medium) and forwarded to the School of Public Health, Fudan University, for the molecular analysis. Data regarding demographic characteristics (age, gender, and geographic origin of the patient) and sputum smear microscopy were obtained by questionnaire interviews and reviews of the medical records. The patients' identification information was replaced by a unique code in data input and analysis. The questionnaires, informed consent forms, and study protocols were approved by the ethics committee of the School of Public Health, Fudan University.
Identification of strains and drug susceptibility testing.
Identification of the M. tuberculosis isolates was performed using standard microbiological tests (niacin accumulation test and nitrate reduction test). Susceptibility to first-line anti-tuberculosis drugs, including isoniazid (INH), rifampin (RIF), ethambutol (EMB), and streptomycin (STR), was tested using the proportion method on L-J medium (INH, 0.2 mg/liter; RIF, 40.0 mg/liter; EMB, 2.0 mg/liter; and STR, 4.0 mg/liter) according to the standard procedure recommended by World Health Organization (WHO) guidelines (7, 8). The strains were examined for their susceptibility to 100 mg/liter of PZA using a mycobacterial growth indicator tube (MGIT) PZA (Nippon Becton-Dickinson Co., Ltd., Tokyo, Japan) antimicrobial susceptibility testing assay, as previously described (9). For quality assurance, the DST was repeated for 10% of the isolates by an external technician from Shanghai Municipal Center for Disease Control and Prevention who had passed the WHO's external quality control assurance with a consistency of over 95%. No discrepancies were recorded. MDR was defined as resistance to at least INH and RIF. Other drug resistance (ODR) was defined as resistance to at least one of the four first-line anti-TB drugs (INH, RIF, EMB, and STR) but not MDR. New cases were defined as cases with no more than a 1-month history of previous anti-TB treatment. Previously treated cases were defined as cases with more than 1-month history of previous anti-TB treatment (10).
MIC testing of PZA-resistant isolates.
MIC tests for PZA resistant isolates were performed using the MGIT 960 platform according to the manufacturer's standard protocol (11). Briefly, bacterial suspensions were transferred to MGIT tubes containing serial PZA dilutions ranging from 64 to 1,024 mg/liter. Control tubes without any antibiotics were inoculated with undiluted and 1:10-diluted bacterial suspensions. The MIC was defined as the first antibiotic concentration that showed less growth than the 1:10-diluted control of the corresponding strain, i.e., the lowest concentration of the drug that inhibited more than 90% of the bacterial population. Duplicates of the M. tuberculosis H37Rv reference strain were included in each run as inter- and intrareplication quality controls. A MIC of more than 500 mg/liter was defined as high-level resistance to PZA (4, 12).
Molecular characterization of PZA resistance.
Using the method of Jureen et al. (13), the 561-bp pncA gene, along with additional regions of approximately 200 bp up- and downstream of the gene, was sequenced using the pncA_F3 (AAGGCCGCGATGACACCTCT) and pncA_R4 (GTGTCGTAGAAGCGGCCGAT) primers. These primers were used in a standard PCR to give a template for the subsequent sequencing reactions. The pncA_F3 and pncA_R4 primers, as well as the P3-F (ATCAGCGACTACCTGGCCGA) and P4-R (GATTGCCGACGTGTCCAGAC) primers, were used to subdivide the PCR fragment into two overlapping bidirectional sequencing reaction fragments. The sequencing reactions were performed using a BigDye Terminator cycle sequencing kit and were later run on a model 3100 genetic analyzer (Applied Biosystems, Inc., Foster City, CA). Retrieved sequences were analyzed with the (ClustalW) Vector NTI Advance (version 9) software (InfoMax, Inc.) using the wild-type H37Rv strain's pncA gene (Rv2043c) as the master reference.
Beijing family determination.
Spoligotyping was performed by using commercial kits from Isogen Bioscience BV (Maarssen, The Netherlands). Comparison of spoligotyping patterns with the SpolDB4 database identified M. tuberculosis isolates belonging to the Beijing family, i.e., defined as strains that only hybridized to the last nine spacer oligonucleotides (spacers 35 to 43) (14).
Statistical analysis.
Sensitivity was defined as the proportion of PZA-resistant isolates harboring a nonsynonymous pncA mutation, while the specificity was determined as the proportion of PZA-susceptible isolates harboring a wild-type pncA gene. An uncorrected chi-square test or Fisher exact test was used to compare the distributions of low-level and high-level MICs among mutations in different pncA regions. The area under the receiver operating characteristic curve (ROC-AUC) was calculated to assess the performance of pncA mutations in predicting PZA resistance, yielding values from 0.5 (no predictive power) to 1.0 (perfect prediction). Sensitivity, specificity, and ROC-AUC were calculated by OPENEPI (http://www.openepi.com/DiagnosticTest/DiagnosticTest.htm). Statistical analysis of risk factors was performed using SPSS for Windows, version 19.0 (SPSS Inc., Chicago, IL, USA). A P value of ≤0.05 was considered statistically significant. Association between studied variables and PZA resistance was calculated using odds ratios (ORs) with 95% confidence intervals (CIs), respectively, by univariate and multivariate analysis in a binary logistic regression model.
RESULTS
Demographic and clinical characteristics of patients.
From 2011 to 2013, a total of 912 pulmonary TB patients were diagnosed and registered at the 4 study sites. Of the 912 sputum samples collected from these patients, 878 (96.3%) were included in the present study, while 34 (3.7%) were excluded due to culture negativity or lack of drug resistance profiles for the first-line anti-TB drugs. Among the included 878 patients, 608 (69.2%) were male, with an average age of 47.3 (±19.2) years. Additionally, 185 (21.1%) were previously treated with first-line drugs and 227 (25.9%) had cavities present in their lungs. There were no statistically significant differences between the subjects from the four counties in terms of demographic (gender and age) or clinical characteristics (previously treated, sputum smear positive, and cavity) (Table 1).
TABLE 1.
Demographic and clinical characteristics of patients and drug resistance patterns at diagnosis in the four studied sites
| Variablea | Value for each study site |
Total (n = 878) | χ2 value | P value | |||
|---|---|---|---|---|---|---|---|
| GY (n = 254) | DQ (n = 216) | GJ (n = 237) | RO (n = 171) | ||||
| Male, no. (%) | 184 (72.4) | 152 (70.4) | 157 (66.2) | 115 (67.3) | 608 (69.2) | 2.668 | 0.446 |
| Age, yrs (mean ± SD) | 45.3 ± 19.5 | 48.2 ± 18.0 | 47.5 ± 19.5 | 49.1 ± 19.9 | 47.3 ± 19.2 | 1.626b | 0.182 |
| Previously treated, no. (%) | 56 (22.0) | 41 (19.0) | 52 (21.9) | 36 (21.1) | 185 (21.1) | 0.820 | 0.845 |
| Sputum smear positive, no. (%) | 159 (62.6) | 140 (64.8) | 143 (60.3) | 91 (53.2) | 533 (60.7) | 5.945 | 0.114 |
| Cavity, no. (%) | 70 (27.6) | 47 (21.8) | 62 (26.2) | 48 (28.1) | 227 (25.9) | 2.724 | 0.436 |
| First-line drug susceptibility, no. (%) | |||||||
| DS | 116 (45.7) | 134 (62.0) | 128 (54.0) | 100 (58.5) | 478 (54.4) | 22.47 | 0.001c |
| MDR | 34 (13.4) | 25 (11.6) | 40 (16.9) | 30 (17.5) | 129 (14.7) | ||
| ODR | 104 (40.9) | 57 (26.4) | 69 (29.1) | 41 (24.0) | 271 (30.9) | ||
| PZA susceptibility, no. (%) | |||||||
| PZA resistance | 44 (17.3) | 37 (17.1) | 38 (16.0) | 28 (16.4) | 147 (16.7) | 0.187 | 0.980 |
| PZA monoresistance | 12 (4.7) | 13 (6.0) | 6 (2.5) | 6 (3.5) | 37 (4.2) | 3.779 | 0.286 |
| PZA resistance/MDR | 8 (3.1) | 9 (4.2) | 11 (4.6) | 11 (6.4) | 39 (4.4) | 2.657 | 0.448 |
DS, drug susceptible to isoniazid, rifampin, ethambutol, and streptomycin; MDR, multidrug resistant; ODR, other first-line drug resistant; PZA, pyrazinamide.
F value in ANOVA test.
P < 0.05.
Drug susceptibility profile.
In total, 400 (45.6%) isolates were resistant to at least one of the four first-line anti-TB drugs, with 129 (14.7%) being simultaneously resistant to INH and RIF (MDR). Using the MGIT 960 system, PZA resistance was observed in 147 (16.7%) of the 878 isolates and ranged from 16.0% to 17.3% at the four study sites. By comparison, the MDR isolates showed a significantly higher proportion of PZA resistance than did the first-line drug-susceptible isolates (30.2% versus 7.7%; P < 0.001), while higher PZA resistance were also observed in ODR isolates than in the drug-susceptible isolates (26.2% versus 7.7%; P < 0.001).
Molecular characterization of the pncA gene.
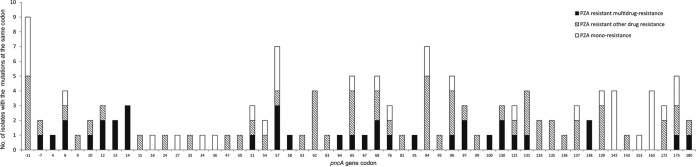
Sequencing of the pncA gene revealed that 136 isolates (15.5%) harbored sequence alterations, including 133 nonsynonymous substitutions and 3 large fragment insertions or deletions. Additionally, one isolate carried a synonymous mutation (Ala36Ala). All substitutions occurred between nucleotide −11 and codon 180. The distribution of the substitutions is shown in Fig. 1. Nucleotide −11, located in the putative promoter region, harbored the most frequent substitutions (9/133 [6.8%]), while codon 57 and codon 94 had the second most frequent mutation (7/133 [5.3%]). Overall, the distributions of mutations were quite comparable between isolates with PZA monoresistance and PZA-resistant MDR (Fig. 1).
FIG 1.
Number of mutations at each codon in the pncA gene among different resistance patterns.
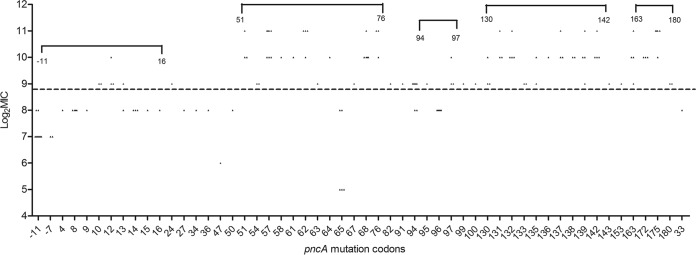
In order to investigate the role of specific pncA mutations in PZA resistance, MICs were determined for each pncA substitution (Fig. 2). For the 133 isolates with nonsynonymous substitutions in pncA, 72.9% (97/133) had a high-level MIC (>500 mg/liter) (see Table S1 in the supplemental material). Different MICs were seen for the same amino acid change in the 7 typical substitutions, including the promoter (A-11G), Trp68Cys, Thr76Pro, Phe94Leu, Val130Ala, Thr135Pro, and Val163Gly. However, among them, only genetic mutations in codon 94 were simultaneously associated with both low and high levels of PZA resistance. In addition, 71 of 97 highly PZA-resistant (MIC > 500 mg/liter) isolates contained a substitution in one of the following regions: codon 51 to 76, codon 130 to 142, and codon 163 to 180; 58 of 97 demonstrated an extremely high-level MIC (MIC ≥ 1,024 mg/liter). The pncA mutations also concentrated in two other regions (codon 94 to 97 and nucleotide −11 to codon 16) for which the corresponding MICs demonstrated a relatively low level of PZA resistance (Fig. 2). A comparison of MIC distributions (low-level and high-level) in the five regions mentioned above demonstrated statistical difference (P < 0.001) (Table 2).
FIG 2.
Pyrazinamide MIC distribution among the isolates with mutations in the different codons of the pncA gene. The MIC is presented for each isolate and the symbols above the dotted line indicate high-level pyrazinamide resistance (MIC > 500 mg/liter).
TABLE 2.
Regions of the pncA gene correlated with low- or high-level pyrazinamide resistance
| Region | No. of isolates with mutation | No. (%) correlated with: |
Pa | |
|---|---|---|---|---|
| Low-level PZA resistance (MIC ≤ 500 mg/liter) | High-level PZA resistance (MIC > 500 mg/liter) | |||
| Codons 51–76 | 34 | 5 (14.7) | 29 (85.3) | |
| Codons 130–142 | 28 | 0 (0.0) | 28 (100.0) | |
| Codons 163–180 | 14 | 0 (0.0) | 14 (100.0) | <0.001 |
| Codons 94–97 | 16 | 6 (37.5) | 10 (62.5) | |
| Nucleotide −11 to codon 16 | 29 | 21 (72.4) | 8 (27.6) | |
Calculated by comparing all the codon regions of the pncA gene by using the Fisher exact test.
The performance of pncA sequencing data to predict PZA resistance was evaluated by comparison with the result obtained from the phenotypic MGIT 960 system (Table 3). Overall, 131 isolates containing nonsynonymous mutations/deletions were found among PZA-resistant isolates and 5 were found among PZA-susceptible isolates. Molecular diagnosis identified 89.1% of PZA-resistant isolates. For MDR isolates and Beijing family isolates, the sensitivities reached 94.9% and 89.7%, respectively. Performance was comparable between Beijing family isolates (sensitivity, 89.7%; 95% CI, 83.46% to 93.77%) and non-Beijing family isolates (sensitivity, 81.8%; 95% CI, 52.30% to 94.86%), as well as between MDR (sensitivity, 94.9%, 95% CI, 83.11% to 98.58%) and first-line drug-susceptible isolates (sensitivity, 91.9%; 95% CI, 78.70% to 97.20%). Furthermore, the sensitivity of pncA gene mutations/deletions to predict PZA resistance ranged from 81.8% to 94.7% at the study sites, showing no statistically difference based on the 95% CI of the sensitivity and specificity.
TABLE 3.
Comparison of nonsynonymous pncA gene mutations with pyrazinamide resistance results based on Bactec MGIT 960
| PZA susceptibilitya | No. of isolates with: |
% sensitivity (95% CI) | % specificity (95% CI) | ROC-AUC (95% CI) | |
|---|---|---|---|---|---|
| pncA nonsynonymous mutations/deletions | pncA wild typeb or synonymous mutations | ||||
| Overall, n = 878 | |||||
| Resistant | 131 | 16 | 89.1 (83.05, 93.19) | 99.3 (98.41, 99.71) | 0.94 (0.925, 0.957) |
| Susceptible | 5 | 726 | |||
| MDR, n = 129 | |||||
| Resistant | 37 | 2 | 94.9 (83.11, 98.58) | 96.7 (90.65, 98.86) | 0.96 (0.907, 0.985) |
| Susceptible | 3 | 87 | |||
| ODR, n = 271 | |||||
| Resistant | 60 | 11 | 84.5 (74.35, 91.12) | 99.0 (96.43, 99.73) | 0.92 (0.878, 0.947) |
| Susceptible | 2 | 198 | |||
| DS, n = 478 | |||||
| Resistant | 34 | 3 | 91.9 (78.70, 97.20) | 100.0 (99.14, 100.00) | 0.96 (0.938, 0.975) |
| Susceptible | 0 | 441 | |||
| Beijing family, n = 739 | |||||
| Resistant | 122 | 14 | 89.7 (83.46, 93.77) | 99.2 (98.07, 99.65) | 0.94 (0.925, 0.960) |
| Susceptible | 5 | 598 | |||
| Non-Beijing family, n = 139 | |||||
| Resistant | 9 | 2 | 81.8 (52.30, 94.86) | 100.0 (97.09, 100.00) | 0.91 (0.849, 0.951) |
| Susceptible | 0 | 128 | |||
| GY, n = 254 | |||||
| Resistant | 36 | 8 | 81.8 (68.04, 90.49) | 99.0 (96.59, 99.74) | 0.90 (0.861, 0.938) |
| Susceptible | 2 | 208 | |||
| DQ, n = 216 | |||||
| Resistant | 33 | 4 | 89.2 (75.29, 95.71) | 98.3 (95.19, 99.43) | 0.94 (0.897, 0.966) |
| Susceptible | 3 | 176 | |||
| GJ, n = 237 | |||||
| Resistant | 36 | 2 | 94.7 (82.71, 98.54) | 100.0 (98.11, 100.00) | 0.97 (0.944, 0.990) |
| Susceptible | 0 | 199 | |||
| RO, n = 171 | |||||
| Resistant | 26 | 2 | 92.9 (77.35, 98.02) | 100.0 (97.38, 100.00) | 0.96 (0.924, 0.987) |
| Susceptible | 0 | 143 | |||
DS, drug susceptible to isoniazid, rifampin, ethambutol, and streptomycin; MDR, multidrug resistant; ODR, other first-line drug resistant; PZA, pyrazinamide.
Wild type refers to the isolates without the identified mutation in the pncA gene.
Sociodemographic and clinical characteristics associated with PZA resistance.
In primary univariate analysis (Table 4), age, previous treatment history and Beijing family genotype were found to be associated with PZA resistance; previous treatment history predicted a higher hazard of PZA monoresistance. Age, previous treatment history, cavity, and Beijing family genotype were positively associated with PZA resistance in MDR-TB patients.
TABLE 4.
Sociodemographic and clinical characteristics associated with pyrazinamide resistance
| Risk factor | No. (%) or mean age ± SD |
PZA monoresistance vs DS |
PZA resistance/MDR vs DS |
PZA resistance vs DS |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PZA monoresistancea (n = 37) | PZA resistance/MDRb (n = 39) | PZA resistance (n = 147) | DSc (n = 441) | Pd | OR (95% CI)d | Pd | OR (95% CI)d | Pd | OR (95% CI)d | |
| Male | 30 (81.1) | 26 (66.7) | 106 (72.1) | 301 (68.3) | 0.110 | 1.99 (0.855–4.649) | 0.838 | 0.93 (0.464–1.864) | 0.381 | 1.20 (0.796–1.816) |
| Age, yrs | 51.1 ± 21.3 | 54.4 ± 20.3 | 54.4 ± 20.3 | 46.8 ± 18.8 | 0.189 | 1.01 (0.994–1.030) | 0.018e | 1.02 (1.004–1.039) | 0.003e | 1.02 (1.005–1.025) |
| Previously treated, no. (%) | 12 (32.4) | 20 (51.3) | 53 (36.1) | 62 (14.1) | 0.004e | 2.93 (1.402–6.143) | <0.001e | 6.44 (3.250–12.74) | <0.001e | 3.45 (2.241–5.302) |
| Sputum smear positive, no. (%) | 23 (62.2) | 25 (64.1) | 91 (61.9) | 260 (59.0) | 0.703 | 1.14 (0.573–2.282) | 0.531 | 1.24 (0.629–2.457) | 0.528 | 1.13 (0.771–1.659) |
| Cavity | 10 (27.0) | 18 (46.2) | 38 (25.9) | 112 (25.4) | 0.827 | 1.09 (0.511–2.318) | 0.007e | 2.52 (1.295–4.896) | 0.913 | 1.02 (0.668–1.570) |
| Beijing family | 35 (94.6) | 38 (97.4) | 136 (92.5) | 369 (83.7) | 0.096 | 3.42 (0.803–13.52) | 0.050e | 7.42 (1.002–44.87) | 0.009e | 2.41 (1.242–4.287) |
PZA monoresistance, isolates resistant to pyrazinamide but susceptible to isoniazid, rifampin, ethambutol, and streptomycin.
PZA resistance/MDR, isolates resistant to pyrazinamide as well as resistant to at least isoniazid and rifampin.
DS, isolates susceptible to all the 5 tested drugs (isoniazid, rifampin, ethambutol, streptomycin, and pyrazinamide).
P and OR (95% CI) were derived from the binary logistic regression model.
P < 0.05.
In the multivariate analysis (Table 5), age, previous treatment history, and Beijing family were retained in the model associated with PZA resistance, while in addition to the above variables, presence of cavities was also retained in the model of PZA resistance/MDR-TB and age was removed from the model of PZA monoresistance. Previous treatment history was a significant predictor of high risk for PZA resistance (P < 0.001; OR, 3.30; 95% CI, 2.131 to 5.101) both for PZA monoresistance (P = 0.005, OR, 2.86; 95% CI, 1.363 to 6.015) and for PZA resistance/MDR (P < 0.001, OR, 6.47; 95% CI, 3.186 to 13.15). Age was found to be associated with PZA resistance/MDR (P = 0.024; OR, 1.02; 95% CI, 1.003 to 1.042) and PZA resistance (P = 0.012; OR, 1.01; 95% CI, 1.003 to 1.023), while Beijing family genotype was associated with PZA resistance (P = 0.019; OR, 2.25; 95% CI, 1.140 to 4.451). Additionally, MDR-TB patients with cavitation were more likely to be infected with PZA-resistant strains (P = 0.008; OR, 2.64; 95% CI, 1.296 to 5.391).
TABLE 5.
Multivariate analysis of demographic and clinical characteristics associated with pyrazinamide resistance
| Variablec | β | P | OR (95% CI)a |
|---|---|---|---|
| PZA monoresistance vs DS | |||
| Previously treated | 1.052 | 0.005b | 2.86 (1.363–6.015) |
| Beijing family | 1.186 | 0.109 | 3.28 (0.766–14.00) |
| PZA resistance/MDR vs DS | |||
| Age | 0.022 | 0.024b | 1.02 (1.003–1.042) |
| Previously treated | 1.867 | <0.001b | 6.47 (3.186–13.15) |
| Cavity | 0.972 | 0.008b | 2.64 (1.296–5.391) |
| Beijing family | 1.849 | 0.074 | 6.35 (0.838–48.14) |
| PZA resistance vs DS | |||
| Age | 0.013 | 0.012b | 1.01 (1.003–1.023) |
| Previously treated | 1.193 | <0.001b | 3.30 (2.131–5.101) |
| Beijing family | 0.812 | 0.019b | 2.25 (1.140–4.451) |
OR and 95% CI were calculated in the binary logistic regression model by backward selection.
P < 0.05.
DS, first-line drug susceptible; MDR, multidrug resistant; PZA, pyrazinamide.
DISCUSSION
It has been widely accepted that PZA resistance could compromise anti-TB treatment, particularly MDR-TB treatment (15 – 17); however, there are few epidemiological data on PZA resistance, especially in high MDR-TB burden areas. In contrast to earlier hospital-based studies or those conducted in a single region in China (18), the present study was multicenter, to better understand the epidemiological situation of PZA resistance across China.
This is the first reported population-based study of PZA resistance among TB cases in China. PZA susceptibility testing by Bactec MGIT 960 revealed that 16.7% (147/878) of the studied patients were PZA resistant. Simultaneously, a higher level of PZA resistance (30.2% [39/129]) was found in the MDR-TB isolates, which is consistent with previously reported studies with PZA resistance proportions ranging from 36% to 54% among MDR-TB patients (18 – 20). A significantly higher proportion of PZA resistance was observed in MDR cases than in first-line drug-susceptible cases (30.2% versus7.7%; P < 0.001). Unfortunately, the overall PZA monoresistance proportion was 7.7%, higher than that reported in most areas with low TB incidence. This might have two explanations. First, the PZA monoresistance proportion could be explained by the high proportion of previously treated patients included in the present study. Second, this could be due to an endemic spread of PZA monoresistant strains, which has been observed elsewhere (21), or a combination of these factors. This suggests that PZA resistance has been established throughout China and that PZA susceptibility testing should be prioritized, especially among MDR-TB patients (even in areas with a low prevalence of MDR-TB).
For the 147 strains phenotypically resistant to PZA, mutations/deletions in the pncA gene were detected in 131 cases, indicating a sensitivity of 89.1%. In addition, as previous studies demonstrated, mutations were scattered throughout the pncA gene. Among the 731 phenotypically PZA-susceptible isolates, we found 726 isolates with a wild-type pncA gene, resulting in a specificity of 99.3%. Most importantly, the diagnostic performances of pncA mutations were similar irrespective of genotype (Beijing family versus non-Beijing family), MDR, and geographic area of origin. The most frequent alteration was observed in the putative promoter (nucleotide position −11), which might influence the initiation of protein synthesis and subsequent binding of the ribosome to the pncA mRNA (22). Additionally, the frequent mutation found at codon 57 (7/130 [5.4%]) was reported to be involved in the suppression of metal iron (Mg2+/Fe2+) binding, leading to a drastic decrease of PZase activity (23), while the mutation at codon 94 (Phe94Leu/Cys/Ser) has been shown to be engaged in the hydrophobic core, which would decrease the stability of the PZase. To our knowledge, 4 SNPs (7 isolates) are reported here for the first time (Gly16Ser, Asp33Ala, Thr153Ile, and Val163Gly), indicating the highly diverse patterns of pncA mutations. The isolates carrying these novel mutations (n = 5) also had a high-level MIC, and the role of these mutations needs to be studied further.
Additionally, due to the clinical significance of MIC in TB treatment (24), we also related MICs to the pncA mutations, in an attempt to search for certain regions or substitutions that led to different levels of PZA resistance. In total, 97 of 133 PZA-resistant isolates demonstrated a relatively high MIC (>500 mg/liter), suggesting high-level PZA resistance. Most interestingly, the majority of those cases were associated with mutations in three specific regions of the pncA gene (codon 51 to 76, codon 130 to 142, and codon 163 to 180). As previously reported (25), these three pncA regions related to high-level PZA resistance might be associated with the activity of PZase. As structural models of the PZase have revealed, these three specific regions are located in loop 4, loop 7, and loop 10, which have been proven to be the part of PZase active-site region (loop 7) and the metal coordination site (loop 4) (26). Our results were consistent with those studies and suggested that substitutions influencing the function of the PZase catalytic site could result in a high level of PZA resistance. Additionally, we also observed that for 7 codons, the same substitution led to different MICs. This may suggest that other mechanism apart from pncA mutation could also influence the level of PZA resistance. Given the genetic profile of pncA in the PZA-resistant isolates, a method based on these specific pncA regions might be of particular interest for a rapid molecular test of PZA resistance.
Risk factors associated with PZA resistance among cases of M. tuberculosis infection are less well understood, especially in countries with a high TB burden. In the present study, we did find that PZA monoresistance was exclusively associated with a history of previous treatment. Additionally, given the important role of PZA in MDR-TB treatment, we also investigated the factors associated with PZA resistance among MDR-TB patients. In a multivariate analysis, factors significantly associated with MDR-PZA resistance included age and lung cavitation. Meanwhile the association between Beijing family isolates and drug resistance has remained controversial, with discrepant findings in China (27 – 29); in the present study, the association between Beijing family and PZA resistance was observed in strains resistant to other first-line drugs rather than in isolates with PZA monoresistance and MDR. This may rule out the possibility that the Beijing family in China has a propensity toward development of resistance to PZA. Alternatively, a secondary explanation could be that the risk of PZA resistance might be further increased within Beijing family strains due to other host characteristics relevant to resistance to other first-line drugs (age and previous treatment). According to these observations and also for the development of a novel method for rapid identification of Beijing family strains (30), we would recommend testing for PZA susceptibility in previously treated patients and especially those infected with strains of the Beijing family if possible.
Our study was subject to several limitations. Since PZA susceptibility testing has not being routinely conducted in China, we decided to focus on four study sites with the ability to perform these tests. For the other areas that do not routinely test for PZA resistance, the prevalence may have been underestimated due to selection bias. Considering this limitation, we selected four areas with different geographic and socioeconomic characteristics in China. We therefore think that the findings can give a reliable picture of the real situation. In addition, the phenotypic testing of PZA resistance was performed with the Bactec MGIT 960 system, which is the reference method recommended by the WHO. The risk of false results using this methodology is well known (31), but because we randomly chose 10% of the sputum culture samples and repeated the phenotypic DST with over 95% consistency (compared to the previous testing), we estimate that our Bactec MGIT 960 method is reliable.
Conclusion.
The significantly higher prevalence of PZA resistance in MDR-TB isolates (30%) highlights the need for PZA susceptibility testing in patients infected with MDR-TB, and since PZA is an essential part of anti-MDR-TB treatment, PZA resistance is undoubtedly an important public health issue which needs to be addressed. Additionally, the reliable performance of pncA mutations in detecting PZA resistance and the association between specific regions of pncA and high-level PZA resistance show the value of a rapid molecular test for PZA resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the grants from the Sweden-China (VR-NSFC) joint project (principal investigator, Biao Xu and Sven Hoffner; no. 81361138019), the National Natural Science Foundation of China (NSFC) (principal investigator, Yi Hu; no. 81373063), as well as a TDR small grant (principal investigator, Yi Hu; no. PO201377850).
The content of the paper is solely the responsibility of the authors and does not necessarily represent the official view of the NSFC.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02687-15.
REFERENCES
- 1.Zumla A, George A, Sharma V, Herbert RH, Baroness Masham of, Oxley IA, Oliver M. 2015. The WHO 2014 global tuberculosis report–further to go. Lancet Global Health 3:e10–e12. doi: 10.1016/S2214-109X(14)70361-4. [DOI] [PubMed] [Google Scholar]
- 2.Caminero JA, Sotgiu G, Zumla A, Migliori GB. 2010. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis 10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 3.Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 4.Tan Y, Hu Z, Zhang T, Cai X, Kuang H, Liu Y, Chen J, Yang F, Zhang K, Tan S, Zhao Y. 2014. Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol 52:291–297. doi: 10.1128/JCM.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajendran V, Sethumadhavan R. 2014. Drug resistance mechanism of PncA in Mycobacterium tuberculosis. J Biomol Struct Dyn 32:209–221. doi: 10.1080/07391102.2012.759885. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Zhang H, Ruan Y, Chin DP, Xia Y, Cheng S, Chen M, Zhao Y, Jiang S, Du X, He G, Li J, Wang S, Chen W, Xu C, Huang F, Liu X, Wang Y. 2014. Tuberculosis prevalence in China, 1990–2010; a longitudinal analysis of national survey data. Lancet 383:2057–2064. doi: 10.1016/S0140-6736(13)62639-2. [DOI] [PubMed] [Google Scholar]
- 7.Chinese Anti-tuberculosis Association. 2006. The laboratory science procedure of diagnostic bacteriology in tuberculosis, 1st ed Chinese Education and Culture Press, Beijing, China. [Google Scholar]
- 8.Canetti G, Froman S, Grosset J, Hauduroy P, Langerova M, Mahler HT, Meissner G, Mitchison DA, Sula L. 1963. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ 29:565–578. [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2012. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed Document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 10.WHO/IUATLD. 2009. Guidelines for surveillance of drug resistance in tuberculosis. WHO/HTM/TB/2009.422. WHO, Geneva, Switzerland. [Google Scholar]
- 11.Werngren J, Sturegard E, Jureen P, Angeby K, Hoffner S, Schon T. 2012. Reevaluation of the critical concentration for drug susceptibility testing of Mycobacterium tuberculosis against pyrazinamide using wild-type MIC distributions and pncA gene sequencing. Antimicrob Agents Chemother 56:1253–1257. doi: 10.1128/AAC.05894-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin A, Takiff H, Vandamme P, Swings J, Palomino JC, Portaels F. 2006. A new rapid and simple colorimetric method to detect pyrazinamide resistance in Mycobacterium tuberculosis using nicotinamide. J Antimicrob Chemother 58:327–331. doi: 10.1093/jac/dkl231. [DOI] [PubMed] [Google Scholar]
- 13.Jureen P, Werngren J, Toro JC, Hoffner S. 2008. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob Agents Chemother 52:1852–1854. doi: 10.1128/AAC.00110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Qing HZ, Enkhsaikan D, Nymadawa P, van Embden JD. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol 33:3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang KC, Leung CC, Yew WW, Leung EC, Leung WM, Tam CM, Zhang Y. 2012. Pyrazinamide may improve fluoroquinolone-based treatment of multidrug-resistant tuberculosis. Antimicrob Agents Chemother 56:5465–5475. doi: 10.1128/AAC.01300-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helbling P, Altpeter E, Egger JM, Zellweger JP. 2014. Treatment outcomes of multidrug-resistant tuberculosis in Switzerland. Swiss Med Wkly 144:w14053. [DOI] [PubMed] [Google Scholar]
- 17.Bastos ML, Hussain H, Weyer K, Garcia-Garcia L, Leimane V, Leung CC, Narita M, Pena JM, Ponce-de-Leon A, Seung KJ, Shean K, Sifuentes-Osornio J, Van der Walt M, Van der Werf TS, Yew WW, Menzies D, Collaborative Group for Meta-analysis of Individual Patient Data in MDR-TB. 2014. Treatment outcomes of patients with multidrug-resistant and extensively drug-resistant tuberculosis according to drug susceptibility testing to first- and second-line drugs: an individual patient data meta-analysis. Clin Infect Dis 59:1364–1374. doi: 10.1093/cid/ciu619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Q, Zhao LL, Li F, Fan YM, Chen YY, Wu BB, Liu ZW, Pan AZ, Zhu M. 2015. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob Agents Chemother 59:1690–1695. doi: 10.1128/AAC.04541-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons SO, van der Laan T, Mulder A, van Ingen J, Rigouts L, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2014. Rapid diagnosis of pyrazinamide-resistant multidrug-resistant tuberculosis using a molecular-based diagnostic algorithm. Clin Microbiol Infect 20:1015–1020. doi: 10.1111/1469-0691.12696. [DOI] [PubMed] [Google Scholar]
- 20.Pierre-Audigier C, Surcouf C, Cadet-Daniel V, Namouchi A, Heng S, Murray A, Guillard B, Gicquel B. 2012. Fluoroquinolone and pyrazinamide resistance in multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 16:221–223, i–ii. doi: 10.5588/ijtld.11.0266. [DOI] [PubMed] [Google Scholar]
- 21.Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y. 2000. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother 44:528–532. doi: 10.1128/AAC.44.3.528-532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KW, Lee JM, Jung KS. 2001. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Korea. J Korean Med Sci 16:537–543. doi: 10.3346/jkms.2001.16.5.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Deng JY, Bi LJ, Zhou YF, Zhang ZP, Zhang CG, Zhang Y, Zhang XE. 2008. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. FEBS J 275:753–762. doi: 10.1111/j.1742-4658.2007.06241.x. [DOI] [PubMed] [Google Scholar]
- 24.Gumbo T, Chigutsa E, Pasipanodya J, Visser M, van Helden PD, Sirgel FA, McIlleron H. 2014. The pyrazinamide susceptibility breakpoint above which combination therapy fails. J Antimicrob Chemother 69:2420–2425. doi: 10.1093/jac/dku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheen P, Lozano K, Gilman RH, Valencia HJ, Loli S, Fuentes P, Grandjean L, Zimic M. 2013. pncA gene expression and prediction factors on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis 93:515–522. doi: 10.1016/j.tube.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du X, Wang W, Kim R, Yakota H, Nguyen H, Kim SH. 2001. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry 40:14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 27.Gao GJ, Lian L, Sun Y, Wei J, Xiao J, Wang X, Zhang L, Zhao X, Yang D, Zhao HX, Zhao H, Wang HZ, Wan KL, Li XW. 2015. Drug resistance characteristics of Mycobacterium tuberculosis isolates to four first-line antituberculous drugs from tuberculosis patients with AIDS in Beijing, China. Int J Antimicrob Agents 45:124–129. doi: 10.1016/j.ijantimicag.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Yuan L, Huang Y, Mi LG, Li YX, Liu PZ, Zhang J, Liang HY, Li F, Li H, Zhang SQ, Li WJ. 2015. There is no correlation between sublineages and drug resistance of Mycobacterium tuberculosis Beijing/W lineage clinical isolates in Xinjiang, China. Epidemiol Infect 143:141–149. doi: 10.1017/S0950268814000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Pang Y, Wang Y, Liu C, Zhao Y. 2014. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int J Antimicrob Agents 43:231–235. doi: 10.1016/j.ijantimicag.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Alonso M, Navarro Y, Barletta F, Martinez Lirola M, Gotuzzo E, Bouza E, Garcia de Viedma D. 2011. A novel method for the rapid and prospective identification of Beijing Mycobacterium tuberculosis strains by high-resolution melting analysis. Clin Microbiol Infect 17:349–357. doi: 10.1111/j.1469-0691.2010.03234.x. [DOI] [PubMed] [Google Scholar]
- 31.Simons SO, van Ingen J, van der Laan T, Mulder A, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2012. Validation of pncA gene sequencing in combination with the mycobacterial growth indicator tube method to test susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol 50:428–434. doi: 10.1128/JCM.05435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.