Abstract
Carbapenemase-producing Gram-negative bacilli have been a global concern over the past 2 decades because these organisms can cause severe infections with high mortality rates. Carbapenemase genes are often carried by mobile genetic elements, and resistance plasmids can be transferred through conjugation. We conducted whole-genome sequencing (WGS) to demonstrate that the same plasmid harboring a metallo-β-lactamase gene was detected in two different species isolated from a single patient. Metallo-β-lactamase-producing Achromobacter xylosoxidans (KUN4507), non-metallo-β-lactamase-producing Klebsiella pneumoniae (KUN4843), and metallo-β-lactamase-producing K. pneumoniae (KUN5033) were sequentially isolated from a single patient and then analyzed in this study. Antimicrobial susceptibility testing, molecular typing (pulsed-field gel electrophoresis and multilocus sequence typing), and conjugation analyses were performed by conventional methods. Phylogenetic and molecular clock analysis of K. pneumoniae isolates were performed with WGS, and the nucleotide sequences of plasmids detected from these isolates were determined using WGS. Conventional molecular typing revealed that KUN4843 and KUN5033 were identical, whereas the phylogenetic tree analysis revealed a slight difference. These two isolates were separated from the most recent common ancestor 0.74 years before they were isolated. The same resistance plasmid harboring blaIMP-19 was detected in metallo-β-lactamase-producing A. xylosoxidans and K. pneumoniae. Although this plasmid was not self-transferable, the conjugation of this plasmid from A. xylosoxidans to non-metallo-β-lactamase-producing K. pneumoniae was successfully performed. The susceptibility patterns for metallo-β-lactamase-producing K. pneumoniae and the transconjugant were similar. These findings supported the possibility of the horizontal transfer of plasmid-borne blaIMP-19 from A. xylosoxidans to K. pneumoniae in a single patient.
INTRODUCTION
Carbapenemase-producing Gram-negative bacilli, which include Enterobacteriaceae and nonfermenters, have been a global threat over the past 2 decades (1). These organisms can cause life-threatening infections, including bloodstream infections, nosocomial pneumonia, surgical site infections, peritonitis, endocarditis, and urinary tract infections. These organisms are usually resistant to all β-lactams and are often resistant to other classes of antibiotics. Therefore, infections caused by these organisms tend to have high mortality rates (2).
Carbapenemases related to resistance against carbapenems without any other permeability defects are so-called true carbapenemases belonging to Ambler molecular class A, B, or D (3). These carbapenemases include KPC- and GES-type β-lactamases (class A), metallo-β-lactamases (MBLs [class B]), and carbapenem-hydrolyzing class D β-lactamases (CHDLs) (e.g., OXA-48 and its derivatives). Among the MBL producers, IMP-, VIM-, and NDM-type MBLs are primarily identified. IMP-type MBLs, of which 52 variants have been found to date, are widespread in Asian and Australian regions (http://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/) (2).
The carbapenemase genes are known to be carried by mobile genetic elements (e.g., plasmids, transposons, integrons, and insertion sequences) that support the horizontal transfer of these resistance genes. Resistance plasmid (R plasmid) conjugation contributes to disseminating resistance genes between bacteria through conjugation (4). Two types of plasmids, conjugative and mobilizable plasmids, play important roles in the process of conjugative transfer. Conjugative plasmids contain a full set of conjugation genes, whereas mobilizable plasmids contain minimal gene sets that allow them to be transferred through conjugation when they cooccur with a conjugative plasmid in the same donor cell (4).
Although several studies have demonstrated the horizontal transfer of the carbapenemase gene in a single patient (5 – 9), data on the recipient strains are lacking. The main objective of this study was to clarify whether the interspecies dissemination of a plasmid harboring MBL genes can occur in a single patient.
MATERIALS AND METHODS
Clinical course and isolates used.
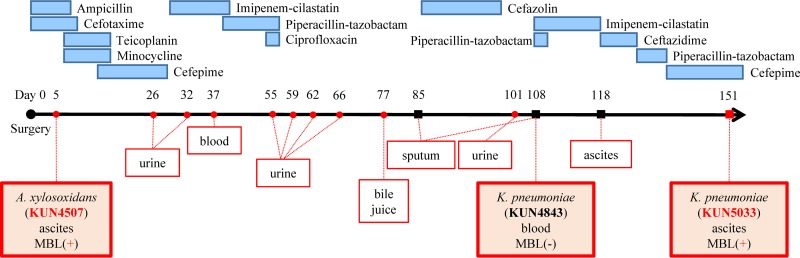
The studied patient was a 54-year-old male. He had liver cirrhosis caused by chronic hepatitis C virus infection and received a living-donor liver transplantation in 2009 at Kyoto University Hospital, Kyoto, Japan. Figure 1 shows the clinical course of this patient. First, MBL-producing Achromobacter xylosoxidans KUN4507 was isolated from ascites on postoperative day (POD) 5. Thereafter, MBL-producing A. xylosoxidans was sequentially detected in many specimens (e.g., urine, blood, bile juice, and sputum) until POD 101. On POD 37, MBL-producing A. xylosoxidans caused bacteremia, and piperacillin-tazobactam was used as a treatment. This bacteremic episode was the only infection caused by MBL-producing A. xylosoxidans, and isolates from other sites were considered to be colonizations because antimicrobial treatment against these isolates was not needed. Klebsiella pneumoniae was first detected on POD 85 and caused pneumonia, which was treated with cefazolin. Subsequently, bacteremia caused by K. pneumoniae KUN4843 occurred on POD 108. Although these K. pneumoniae isolates did not produce MBL, MBL-producing K. pneumoniae KUN5033 was detected in the ascites on POD 151. We used A. xylosoxidans KUN4507 and K. pneumoniae KUN4843 and KUN5033 to analyze the horizontal transfer of the MBL gene from A. xylosoxidans to K. pneumoniae isolated from a single patient. The Institutional Review Board of Kyoto University Hospital approved this study protocol. No informed consent was needed because the data were analyzed anonymously.
FIG 1.
Clinical course and bacterial isolates. The red circles indicate clinical isolates of metallo-β-lactamase (MBL)-producing Achromobacter xylosoxidans. The black squares indicate clinical isolates of Klebsiella pneumoniae (non-MBL producer). The red square indicates a clinical isolate of MBL-producing K. pneumoniae. The numbers above each isolate indicate the day from surgery (living-donor liver transplantation). The red boxes without shading indicate the sites from which each isolate was isolated. The red boxes with shading indicate the isolates used in this study, the isolation sites of these isolates, and MBL production. The blue boxes with shading indicate the antimicrobial agents, and the length of the boxes indicates the duration of antimicrobial therapy for each agent. There was no treatment between POD 57 and POD 85 because this patient showed no sign of sepsis after isolation of MBL-producing A. xylosoxidans.
Antimicrobial susceptibility testing and metallo-β-lactamase-production.
Antimicrobial susceptibility testing was performed using the broth microdilution method with dry plates (Eiken Chemical, Tokyo, Japan) according to CLSI guidelines (10). MBL detection was phenotypically performed using a double-disk synergy test with sodium mercaptoacetate (metallo-β-lactamase SMA Eiken; Eiken Chemical) and three commercial Kirby-Bauer (KB) disks (Eiken Chemical) containing 30 μg of ceftazidime (CAZ), 10 μg of imipenem (IPM), or 10 μg of meropenem (MEPM) as previously described (11).
Molecular typing was performed using pulsed-field gel electrophoresis (PFGE) with a GenePath system (Bio-Rad, Tokyo, Japan) and the restriction enzyme XbaI (TaKaRa Bio, Tokyo, Japan) as previously described (12). A multilocus sequence typing (MLST) analysis was performed for the K. pneumoniae isolates according to the Institut Pasteur MLST and whole-genome MLST databases (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html) (13).
Whole-genome sequencing (WGS).
The total genomic DNA of each bacterial isolate was extracted using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Samples were prepared using a Nextera XT DNA sample preparation kit according to the manufacturer's instructions and sequenced (2- by 300-mer reads) on a MiSeq benchtop sequencer (Illumina, Inc., San Diego, CA, USA). To filter out low-quality regions, raw Illumina paired-end reads were trimmed using ERNE-FILTER (14), with the following parameters: min-mean-phred-quality, 20; min-phred-value-mott, 20; and min-size, 50. De novo assembly was performed using SPAdes with default parameters (15), and the assembly quality was analyzed using QUAST (16).
Phylogenetic tree and SNP analysis.
In addition to the Illumina reads of K. pneumoniae KUN4843 and KUN5033, those of 35 K. pneumoniae isolates were downloaded from the Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra/) (see Table S1 in the supplemental material) to identify homologous recombination sites and for phylogenetic tree analysis. Trimmed reads were mapped to a reference genome using Burrows-Wheeler Aligner (BWA) (17). The 5,438,894-bp K. pneumoniae Kp52.145 genome (GenBank accession number FO834906) was used for reference because this strain was the first isolate with complete genome sequence data available and an isolation date. Single nucleotide polymorphism (SNP) calling was performed using the Genome Analysis Toolkit (18) and SAMtools (19). High-quality SNPs were based on the following parameters: depth of coverage of >10 and Phred quality score of >20. The core genome sequence was defined as the sequences that were shared by all strains. The consensus genome sequence for each strain was generated using VCFtools (https://vcftools.github.io/man_latest.html). To identify the homologous recombination sites, BRATNextGen was used as previously described (20). The core genome sequences of 37 K. pneumoniae isolates without recombination sites were used for the phylogenetic tree analysis. The maximum-likelihood tree was generated using RAxML with the general time-reversible (GTR) gamma substitution model and 100 rapid bootstrap replicates (21).
Molecular clock analysis.
To estimate the time at which the two K. pneumoniae isolates KUN4843 and KUN5033 separated from their most recent common ancestor (MRCA), a molecular clock analysis was performed using the core genome sequences of 17 K. pneumoniae isolates with known isolation dates (see Table S1 in the supplemental material) using the program Bayesian Analysis of Population Structure (BEAST) (22). The parameters were as follows: Hasegawa-Kishino-Yano substitution model, coalescent constant size tree prior, and Markov chain Monte Carlo chain length of 100,000,000. Tracer was used to evaluate convergence to ensure that Bayesian runs reached an effective sample size greater than 200 (22).
Plasmid analysis.
Plasmids of each strain were extracted using a PureLink HiPure Plasmid Midiprep kit (Thermo Fisher Scientific, MA, USA) according to the manufacturer's instructions. Plasmid sequence data were extracted from whole-genome sequencing data sets generated by Illumina MiSeq. To identify the contigs derived from the plasmids, a Basic Local Alignment Search Tool (BLAST) analysis was performed for all generated contigs (23). We defined the contig containing the plasmid components as the plasmid-derived contig. Furthermore, to circularize the plasmids extracted from A. xylosoxidans KUN4507 and K. pneumoniae KUN4843, WGS with a PacBio RS II system (Pacific Biosciences [PacBio], Menlo Park, CA USA) was also performed by Macrogen Japan. PacBio long reads were analyzed using SMRT analysis software (24). A hierarchical genome assembly process (HGAP) was used for de novo assembly, and Quiver was used to polish the data. For one plasmid extracted from A. xylosoxidans KUN4507, PCR and Sanger sequencing were performed to close the gap with the primers listed in Table S2 in the supplemental material. The PCR conditions were as follows: 2 min at 95°C, followed by 35 cycles of 10 s at 98°C, 30 s at 55°C, and 5 min at 72°C. The Rapid Annotation using Subsystem Technology (RAST) annotation pipeline (25), the Microbial Genome Annotation Pipeline (MiGAP) (www.migap.org), and BLAST were used to annotate these plasmids. To compare the harbored plasmids, Illumina reads from each isolate were mapped to circularized plasmids (pKUN4507_1, pKUN4843_1, and pKUN4843_2) and contigs derived from other plasmids using BWA.
Plasmid conjugation analysis.
An in vitro conjugation assay was performed using the liquid mating method as previously described with certain modifications (26). We used A. xylosoxidans KUN4507 as the donor. Azide-resistant Escherichia coli ME8067, streptomycin-resistant E. coli ME8568 derived from HB101, and K. pneumoniae KUN4843 were used as the recipients (see Table S3 in the supplemental material). For the conjugation experiment between A. xylosoxidans KUN4507 and E. coli ME8067, deoxycholate hydrogen sulfide lactose (DHL) agar with azide (100 μg/ml) and ceftazidime (2 μg/ml) was used as the selective agar. For the other conjugation experiments, a suitable selective agar was not found. Therefore, we selected transconjugants by the following method. First, we used the culture conditions to select transconjugants. A. xylosoxidans is an obligate aerobe, whereas E. coli and K. pneumoniae are facultative anaerobes. After liquid mating, the donor and recipient mixture was cultured on DHL agar with ceftazidime (2 μg/ml) under anaerobic conditions for 24 h. Then, red-purple colonies identified as Enterobacteriaceae were selected and used for subculturing. Finally, a colony PCR was performed to confirm the species and MBL gene. The PCR primers were manually designed to target specific regions of the 16S rRNA for each species as well as blaIMP-19 (see Table S2 in the supplemental material). The PCR reagents were as follows: 1× Ex Taq buffer, 1.5 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphates, 0.3 μM (each) species-specific primers, 1 μM primers for blaIMP-19 genes, and 1 U of Ex Taq polymerase (TaKaRa Bio, Otsu, Japan) to a volume of 30 μl achieved by the addition of ultrapure water. A toothpick was used to collect individual colonies, and it was then dipped into the reaction tube. Amplification was conducted in a TaKaRa PCR Thermal Cycler Dice Touch (TaKaRa Bio). After an initial denaturation for 2 min at 95°C, 30 cycles were completed, each consisting of 10 s at 98°C, 30 s at 55°C, and 60 s at 72°C. A final extension of 5 min at 72°C was applied with an infinite hold at 4°C. The species and MBL genes were confirmed by electrophoresis of the amplified PCR products using QIAxcel (Qiagen). Each conjugation experiment was performed three times.
Accession numbers.
The genome data have been deposited in the GenBank/DDBJ database under accession numbers DRX052506, DRX052507, DRX052508, LC155906, LC155907, LC155908, and LC155909.
RESULTS
Antimicrobial susceptibility testing and molecular typing.
The antimicrobial susceptibility testing results are shown in Table 1. K. pneumoniae KUN5033 and the transconjugant derived from K. pneumoniae KUN4843 (recipient) that acquired blaIMP-19 revealed nearly the same MICs for the test antimicrobial agents. This result suggests that blaIMP-19 is related to β-lactam resistance, including resistance to carbapenems. The MICs of cephalosporins and carbapenems for the transconjugant were higher than those for the recipient strain (K. pneumoniae KUN4843). All of the isolates analyzed in this study were susceptible to piperacillin-tazobactam and colistin (with the exception of A. xylosoxidans KUN4507). The PFGE analysis revealed that K. pneumoniae KUN4843 and KUN5033 showed the same PFGE patterns (see Fig. S1 in the supplemental material). Furthermore, these isolates belonged to the same MLST (sequence type [ST] 29).
TABLE 1.
Antimicrobial susceptibility testing of each isolate and transconjugant
| Antibiotic agent | MICs (μg/ml) for: |
|||
|---|---|---|---|---|
| A. xylosoxidans KUN4507 | K. pneumoniae KUN4843 | K. pneumoniae KUN5033 | Transconjuganta | |
| Piperacillin | ≤4 | >64 | >64 | >64 |
| Piperacillin-tazobactamb | 4/4 | 4/4 | 8/4 | 8/4 |
| Cefazolin | >8 | >8 | >8 | >8 |
| Cefotaxime | >16 | ≤1 | >16 | >16 |
| Ceftazidime | >16 | ≤1 | >16 | >16 |
| Cefepime | >16 | ≤1 | >16 | >16 |
| Imipenem | >8 | ≤0.25 | >8 | 8 |
| Meropenem | >8 | ≤0.25 | 4 | 2 |
| Aztreonam | >16 | ≤1 | ≤1 | ≤1 |
| Gentamicin | >8 | ≤1 | ≤1 | ≤1 |
| Tobramycin | >8 | ≤1 | ≤1 | ≤1 |
| Amikacin | >32 | ≤1 | ≤1 | ≤1 |
| Ciprofloxacin | >4 | >4 | >4 | >4 |
| Levofloxacin | >4 | >4 | >4 | >4 |
| Minocycline | >16 | >16 | >16 | >16 |
| Colistin | 4 | ≤2 | ≤2 | ≤2 |
Transconjugant derived from K. pneumoniae KUN4843 that acquired blaIPM-19.
The MIC of piperacillin-tazobactam is indicated by the concentration of piperacillin-tazobactam.
Whole-genome sequencing statistics.
The sequencing statistics are shown in Tables S1 and S4 in the supplemental material. The average sequencing coverage was approximately 45×. The numbers of contigs and N50 (bp) of each isolate (A. xylosoxidans KUN4507 and K. pneumoniae KUN4843 and KUN5033) were 148/82,254 bp, 98/361,546 bp, and 135/330,754 bp, respectively (see Table S4). The breadth of coverage compared to the K. pneumoniae Kp52.145 reference sequence was 87.2% (KUN4843) and 83.3% (KUN5033) (see Table S1).
Phylogenetic tree and SNP analysis.
Although conventional molecular typing analyses revealed that the two K. pneumoniae isolates (KUN4843 and KUN5033) were identical, we conducted phylogenetic tree analysis using SNPs to compare these isolates in more detail. Initially, the total number of SNP sites compared to the K. pneumoniae Kp52.145 reference sequence including homologous recombination sites was 112,689 (average 26,636 ± 589). BRATNextGen detected 346 recombination sites (see Fig. S2 in the supplemental material). The total base pair length of the recombination sites was 193,842 bp (average, 560 ± 2,643 bp). The total number of SNP sites compared to the K. pneumoniae Kp52.145 reference sequence excluding recombination sites was 103,517 (average, 24,739 ± 564). The total base pair of the core genome sequence from 37 K. pneumoniae isolates was 4,258,806 bp (78.3% of the reference genome). Although differences between K. pneumoniae KUN4843 and KUN5033 were not observed by conventional molecular typing, a phylogenetic tree and SNP analysis revealed that these isolates were slightly different from each other (see Fig. S3). K. pneumoniae KUN4843 had 24,132 SNPs, and KUN5033 had 24,131 SNPs compared to the K. pneumoniae Kp52.145 reference genome. Of these SNPs, 24,128 SNPs were common among these isolates. Compared with the sequence of the MRCA, K. pneumoniae KUN4843 had four SNPs, and K. pneumoniae KUN5033 had three SNPs (Table 2).
TABLE 2.
Summary of SNPs that differ between Klebsiella pneumoniae isolates KUN4843 and KUN5033
| Isolate | SNP position (nt)a | Reference base | Alternate baseb | Mutation type | Region | Functional description of the region (GenBank accession no.) |
|---|---|---|---|---|---|---|
| KUN4843 | 526319 | G | A | Nonsynonymous | uxuA2 | Similar to mannonate dehydratase from Lactococcus lactis subsp. cremoris (KGH33940) |
| 1329788 | G | A | Synonymous | Unnamed protein | Highly similar to phospholipid-glycerol acyltransferase from Enterobacter cloacae (EPR31355) | |
| 1350124 | C | T | Synonymous | sbcC | Similar to ATP-dependent double-stranded DNA exonuclease SbcC from Escherichia coli O103:H2 strain 12009; enterohemorrhagic Escherichia coli (BAI29239) | |
| 2885006 | C | G | Nonsynonymous | hipA | Similar to protein HipA from Escherichia coli strain K-12 (LN832404) | |
| KUN5033 | 2704847 | G | T | Noncoding | ||
| 3607720 | G | A | Nonsynonymous | edd | Highly similar to phosphogluconate dehydratase from Escherichia coli O6 (EZA74923) | |
| 4062427 | C | T | Nonsynonymous | ompC | Highly similar to outer membrane protein C from Klebsiella pneumoniae (KHF63615) |
Position in the reference strain K. pneumoniae Kp52.145 (GenBank accession number FO834906). nt, nucleotide.
Alternate nonreference allele.
Molecular clock analysis.
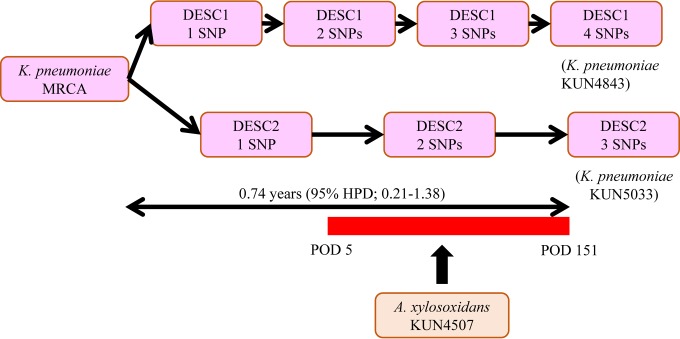
To assess when the two K. pneumoniae isolates (KUN4843 and KUN5033) diverged from the MRCA, we performed molecular clock analysis. All of the parameters had an effective sample size greater than 220. As a result of the molecular clock analysis, the MRCA was divided into two groups of descendants 0.74 years before K. pneumoniae KUN4843 and KUN5033 appeared (95% highest posterior density [HPD], 0.21 to 1.38) (see Fig. S4 in the supplemental material). These findings support the possibility of the occurrence of the MRCA and its two descendant variations in this patient and the likelihood of contact with A. xylosoxidans harboring the MBL gene during the clinical course.
Plasmid sequencing.
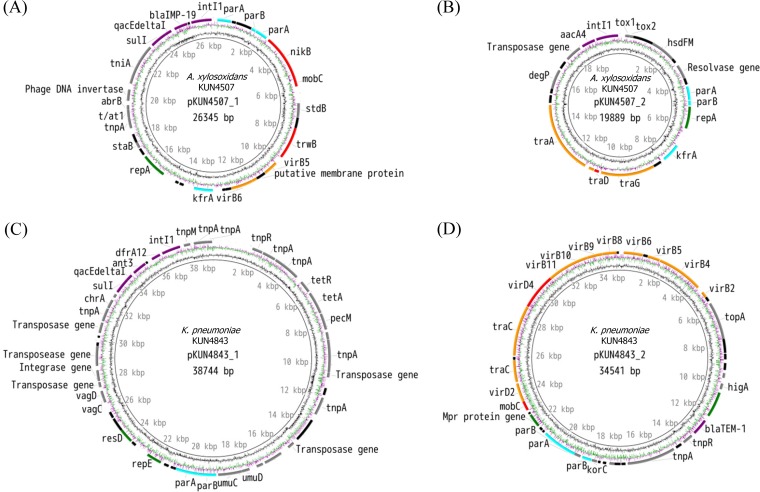
To analyze plasmid mobility, we revealed the sequence of plasmids using WGS. Two plasmids (pKUN4507_1 detected from A. xylosoxidans KUN4507 and K. pneumoniae KUN5033 and pKUN4843_2 detected from K. pneumoniae KUN4843 and KUN5033) could be circularized directly from contigs generated by SPAdes. One plasmid (pKUN4507_2) was circularized by PCR and Sanger sequencing, and another plasmid (pKUN4843_1) was circularized by PacBio RS II analysis. Two circularized plasmids (pKUN4507_1 with 26,345 bp and pKUN4507_2 with 19,889 bp) were detected in A. xylosoxidans KUN4507 (Fig. 2A and B). The plasmid incompatibility group was not determined in pKUN4507_1, and it was identified as a Col-like plasmid for pKUN4507_1 (88.5% similarity). One of the two plasmids, pKUN4507_1, harbored the MBL gene (blaIMP-19) located in the class 1 integron. In K. pneumoniae KUN4843, two circularized plasmids (pKUN4843_1 with 38,744 bp and pKUN4843_2 with 34,541 bp) were detected (Fig. 2C and D). Although pKUN4843_1 was revealed to be an IncR plasmid (100% identical), the plasmid incompatibility group was not determined in pKUN4843_2. Furthermore, as a result of the BLAST analysis, 6 of the 98 contigs belonging to KUN4843 were considered to be derived from plasmids other than the above two plasmids although they were not circularized. The total number of bases in these contigs was 103,616 bp. For convenience, we named these noncircularized contigs en masse “pKUN4843_others.” The pKUN4843_others contigs included contigs 22, 29, 34, 35, 39, and 53 (Table 3). The coding genes of plasmids related to conjugative transfer are shown in Table 3. Although pKUN4507_1 had all of the conjugative transfer gene components, many of the mating pair formation (MPF) genes were lacking. This result suggests that pKUN4507_1 is not a conjugative plasmid but a mobilizable plasmid. Because of the lack of conjugative transfer genes (relaxase genes and type IV secretion system coupling protein [T4CP] genes), another plasmid detected from A. xylosoxidans KUN4507, pKUN4507_2, was found to be nontransferable. These findings suggest that A. xylosoxidans KUN4507 cannot deliver the resistance genes (e.g., blaIMP-19) by itself to other bacteria through conjugative transfer. Of the plasmids detected in K. pneumoniae KUN4843, pKUN4843_1 had no transfer ability, whereas pKUN4843_2 had almost all of the sets of genes required for conjugative transfer except for virB7 and was considered to be a conjugative plasmid. The structure of this plasmid is similar to that of the conjugative plasmid pFSEC-01 described by Zhang et al. (27). Other conjugative transfer genes (finO, traA, traE, traI, traJ, traK, traL, traM, traX, and traY) were also found in pKUN4843_others. The pKUN4507_1 (100% identical) plasmid that harbored blaIMP-19 was detected by both MBL-producing isolates (A. xylosoxidans KUN4507 and K. pneumoniae KUN5033). Based on the mapping analysis using plasmids derived from K. pneumoniae KUN4843 as a reference and the K. pneumoniae KUN4843 and KUN5033 Illumina reads, K. pneumoniae KUN4843 and KUN5033 had the same plasmids, including pKUN4843_1, pKUN4843_2, and pKUN4843_others (Table 3).
FIG 2.
Schema of circularized plasmids. The red arrows indicate mobility genes (relaxase gene and type IV secretion system coupling protein [T4CP] gene). The orange arrows indicate mating pair formation (MPF) genes. The aqua arrows indicate establishment genes. The green arrows indicate replication and stability genes. The gray arrows indicate other coding genes. The black arrows indicate hypothetical protein-encoding genes. The innermost ring shows GC content, and the middle ring shows GC skew. (A) pKUN4507_1 detected from Achromobacter xylosoxidans KUN4507 and Klebsiella pneumoniae KUN5033. The plasmid pKUN4507_1 harbors mobility genes and two MPF genes, which indicate a mobilizable plasmid. This plasmid harbored a class 1 integron including blaIMP-19. (B) pKUN4507_2, another plasmid detected in A. xylosoxidans KUN4507. The plasmid pKUN4507_2 lacks a relaxase gene and many MPF genes and is not considered to be transferable. (C) pKUN4843_1, including a class 1 integron and many transposase genes detected in Klebsiella pneumoniae KUN4843 and KUN5033. (D) pKUN4843_2, detected in K. pneumoniae KUN4843 and KUN5033. This plasmid has almost all of the genes required for conjugative transfer and was considered to be a conjugative plasmid.
TABLE 3.
Summary of the genes related to conjugative transfer
| Plasmid name (GenBank accession no.) | Length (bp) | Mobility gene(s) |
T4SS MPF gene(s)b | Plasmid transferability | Plasmid/contig profile of:d |
Similarity (% [WSS])c | |||
|---|---|---|---|---|---|---|---|---|---|
| Relaxase gene(s) | T4CPa | A. xylosoxidansKUN4507 | K. pneumoniae KUN4843 | K. pneumoniae KUN5033 | |||||
| pKUN4507_1 (LC155906) | 26,345 | nikB, mobC | trwB | virB5, virB6, traG | Mobilizable | + | − | + | 100 (100)e |
| pKUN4507_2 (LC155907) | 19,889 | traD | traA, traC, traG | Nontransferable | + | − | − | ||
| pKUN4843_1 (LC155908) | 38,744 | Nontransferable | − | + | + | 100 (99.99)f | |||
| pKUN4843_2 (LC155909) | 34,541 | mobC | virD4 | virB2, virB4, virB5, virB6, virB8, virB9, virB10, virB11, virD2, traC | Conjugative | − | + | + | 100 (100)f |
| pKUN4843_others | 103,616 | − | + | + | |||||
| Contig 22 | 57,649 | − | + | +h | 100f,g | ||||
| Contig 29 | 22,103 | − | + | +h | 100f,g | ||||
| Contig 34 | 8,638 | − | + | +h | 100f,g | ||||
| Contig 35 | 8,329 | finO, traA, traE, traI, traJ, traK traL, traM, traX, traY | − | + | +h | 100f,g | |||
| Contig 39 | 5,844 | − | + | +h | 100f,g | ||||
| Contig 53 | 1,053 | − | + | +h | 99.0f,g | ||||
Type IV secretion system coupling protein.
T4SS, type IV secretion system; MPF, mating pair formation.
Sequence similarity was determined using mapping analysis, and whole-sequence similarity (WSS) of circularized plasmids was calculated using ClustalW.
Presence (+) or absence (−) of the plasmid/contig is indicated.
Sequence similarity between A. xylosoxidans KUN4507 and K. pneumoniaeKUN5033.
Sequence similarity between K. pneumoniae KUN4843 and K. pneumoniae KUN5033.
Excluding the regions where depth of coverage is <10%.
The Illumina reads of K. pneumoniae KUN5053 could be mapped to this contig of KUN4843.
Plasmid conjugation.
To assess the mobility of the pKUN4507_1 plasmid, we performed plasmid conjugation analysis. Plasmid conjugation from A. xylosoxidans KUN4507 (donor) to E. coli (ME8067 and ME8568) (recipients) was not observed. However, the conjugation experiment between A. xylosoxidans KUN4507 (donor) and K. pneumoniae KUN4843 (recipient) was performed successfully, with an average conjugation frequency of 2.36 × 10−7 ± 0.13 × 10−7 transconjugants per recipient. These findings suggest that the MRCA and its two descendant variations harbored plasmids with the same structures (other than pKUN4507_1) and are capable of acquiring the plasmid pKUN4507_1 from A. xylosoxidans KUN4507.
DISCUSSION
The aim of this study was to demonstrate the horizontal transfer of the R plasmid harboring the MBL gene from A. xylosoxidans to K. pneumoniae in a single patient. Our data supported the possibility that the MRCA of K. pneumoniae or its descendant could acquire the plasmid harboring the MBL gene from A. xylosoxidans. Because two closely related descendant variants were detected in this patient's clinical specimens, it is reasonable to assume that the MRCA and its descendants occurred in this patient (Fig. 3). Lieberman et al. reported the similar detection of SNP differences in closely related clinical isolates around the same time (28). In their study of patients with cystic fibrosis, Burkholderia dolosa isolates accumulated SNPs over time, and closely related descendants were isolated at nearly the same time. Based on the molecular clock analysis performed in our study, it is possible that the horizontal gene transfer (HGT) occurred during this patient's clinical course. The evidence that no other MBL-producing K. pneumoniae isolates were detected in our hospital during this period (data not shown) also suggests that it is unlikely that this patient was infected by this organism as a nosocomial infection. To date, five studies on the horizontal transfer of the carbapenemase gene in a single patient have been published (5 – 9). These articles describe the same plasmid harboring the carbapenemase genes, such as KPC genes, MBL genes, and OXA-48 genes, among different bacterial species isolated from a single patient, and they mainly addressed in vitro conjugal transfer. Drieux et al. performed only in vivo conjugation experiments in an animal model, and four other studies performed conventional conjugation analyses. Although we did not perform conjugation using an animal model in our study, we used an actual clinical isolate as the recipient of the conjugation analysis. Therefore, the advantage of our study is that we not only demonstrated the transfer of the plasmid harboring the MBL gene but also supported the presence of the recipient strain in a single patient.
FIG 3.
SNPs accumulated from the most recent common ancestor (MRCA) of Klebsiella pneumoniae. Two variants of a K. pneumoniae descendant (DESC) occurred in the same patient. One of the two descendant (DESC2) variants was considered to be in close contact with Achromobacter xylosoxidans KUN4507, and the plasmid harboring the MBL gene (pKUN4507_1) was transferred from A. xylosoxidans KUN4507 to DESC2. The red bar indicates the duration of the contact of A. xylosoxidans KUN4507 with DESC2. HPD, highest posterior density; POD, postoperative day.
In addition, whole-genome sequencing was used to clarify the details of plasmid harboring genes related to conjugation. Conjugation is a DNA transfer process through the type IV secretion system (T4SS), and three groups of conjugative transfer genes (relaxase gene, T4CP gene, and MPF gene) perform central roles (29). To complete horizontal gene transfer through conjugation, all of the conjugative transfer genes are considered to be essential and must be located on the plasmids in a single donor bacterial cell. The plasmid harboring the MBL gene, pKUN4507_1 detected in A. xylosoxidans KUN4507, harbors relaxase genes and the T4CP gene but lacks many of the MPF genes. Therefore, this plasmid was not conjugative but mobilizable (not self-transferable). Moreover, because another plasmid detected in A. xylosoxidans KUN4507, pKUN4507_2, also lacked MPF genes, A. xylosoxidans KUN4507 alone could not transfer the plasmid harboring the MBL gene to other bacteria through so-called conjugation. The conjugation of plasmids harboring the MBL gene (pKUN4507_1) from A. xylosoxidans to E. coli was not successful. However, the conjugative transfer of this resistant plasmid between A. xylosoxidans and K. pneumoniae could be observed. One of the conjugation processes, triparental mating, can explain this horizontal transfer of a mobilizable plasmid. K. pneumoniae KUN4843 or other bacteria harboring a conjugative plasmid could act as helper strains during conjugation. In this process, it is conceivable that a conjugative plasmid was once transferred into A. xylosoxidans and thereafter, a mobilizable plasmid harboring blaIMP-19 (pKUN4507_1) was transferred into K. pneumoniae. As with other mechanisms of HGT, a DNA uptake system found in some bacteria (e.g., Neisseria gonorrhoeae) can also be considered (30).
In this evaluation of the clinical course of a single patient, K. pneumoniae was found to survive despite numerous exposures to antibiotic agents to which it is normally susceptible. Liao et al. reported that extracellular CHDL produced by Acinetobacter baumannii helped other susceptible bacteria to survive against the eradication of carbapenem and named this phenomenon the sheltering effect (31). This effect is strongly affected by a strong promoter of the CHDL gene. Regarding MBL production, certain promoter variants (e.g., a PcS variant) located on the class 1 integron integrase gene are known to be strong promoters (32). As we demonstrated in the previous study, the PcS strong promoter was found in pKUN4507_1, which harbors blaIMP-19 (33). Given the possibility of close contact between MBL-producing A. xylosoxidans and K. pneumoniae, the sheltering effect is an effective measure to promote the continued presence of this organism.
Our research also revealed the important findings that A. xylosoxidans could cause infection and become a reservoir of MBL genes. A. xylosoxidans is a nonfermentative, aerobic Gram-negative bacillus that can cause life-threatening infections, especially in immunocompromised patients (34). The clinical importance of this species has increased in recent decades. Several studies have reported that A. xylosoxidans harbors MBL genes, including blaIMP-1, blaIMP-19, blaVIM-1, blaVIM-2, and blaTMB-1 (33, 34). All of these MBL genes were located on a class 1 integron as one of the gene cassettes, whereas only blaIMP-1, blaIMP-19, and blaVIM-2 were revealed to be plasmid components. Although these three MBL genes were presumed to be transferred through conjugation, evidence supporting this hypothesis was lacking.
One limitation should be noted. Although we demonstrated the possibility of horizontal gene transfer in a single patient, we did not provide evidence of direct horizontal gene transfer of the plasmid harboring blaIMP-19. Therefore, we could not completely exclude other possibilities. First, the patient might have already carried the MBL-producing A. xylosoxidans and the K. pneumoniae strains when hospitalized. Second, the MBL-producing plasmid might have been transferred from K. pneumoniae to A. xylosoxidans or from a third, unknown donor to both A. xylosoxidans and K. pneumoniae. However, horizontal gene transfer of the plasmid harboring blaIMP-19 from MBL-producing A. xylosoxidans to K. pneumoniae strains isolated from a single patient was demonstrated by the conjugation analysis. Therefore, it is conceivable that the plasmid harboring blaIMP-19 could be transferred from A. xylosoxidans to K. pneumoniae.
In summary, our findings supported the possibility that the horizontal transfer of plasmid-borne blaIMP-19 occurs from A. xylosoxidans to K. pneumoniae in a single patient. Surprisingly, this plasmid was not self-transferable, and other conjugative plasmids were not found in the donor A. xylosoxidans. This result suggests that there is triparental mating or another mechanism associated with horizontal gene transfer, such as a DNA uptake system. Finally, clinicians should pay attention to the facts that MBL genes and other carbapenemase genes may be transferred and that the susceptibility patterns of clonal isolates can suddenly be changed in a single patient during his/her clinical course. The recent development of rapid identification tools, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), has greatly reduced the time required for species identification (35); therefore, the recognition of this type of change in antimicrobial susceptibility is more important than ever.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National BioResource Project (National Institute of Genetics, Japan) for providing the E. coli ME8067 and ME8568 recipient strains for the conjugation experiments.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00933-16.
REFERENCES
- 1.Akova M, Daikos GL, Tzouvelekis L, Carmeli Y. 2012. Interventional strategies and current clinical experience with carbapenemase-producing Gram-negative bacteria. Clin Microbiol Infect 18:439–448. doi: 10.1111/j.1469-0691.2012.03823.x. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcillan-Barcia MP, Alvarado A, de la Cruz F. 2011. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev 35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 5.Manageiro V, Ferreira E, Pinto M, Canica M. 2014. First description of OXA-48 carbapenemase harbored by Escherichia coli and Enterobacter cloacae from a single patient in Portugal. Antimicrob Agents Chemother 58:7613–7614. doi: 10.1128/AAC.02961-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocsis E, Lo Cascio G, Piccoli M, Cornaglia G, Mazzariol A. 2014. KPC-3 carbapenemase harbored in FIIk plasmid from Klebsiella pneumoniae ST512 and Escherichia coli ST43 in the same patient. Microb Drug Resist 20:377–382. doi: 10.1089/mdr.2013.0152. [DOI] [PubMed] [Google Scholar]
- 7.Drieux L, Bourgeois-Nicolaos N, Cremniter J, Lawrence C, Jarlier V, Doucet-Populaire F, Sougakoff W. 2011. Accumulation of carbapenemase-producing Gram-negative bacteria in a single patient linked to the acquisition of multiple carbapenemase producers and to the in vivo transfer of a plasmid encoding VIM-1. Int J Antimicrob Agents 38:179–180. doi: 10.1016/j.ijantimicag.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Sidjabat HE, Silveira FP, Potoski BA, Abu-Elmagd KM, Adams-Haduch JM, Paterson DL, Doi Y. 2009. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis 49:1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]
- 9.Luzzaro F, Docquier JD, Colinon C, Endimiani A, Lombardi G, Amicosante G, Rossolini GM, Toniolo A. 2004. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-beta-lactamase encoded by a conjugative plasmid. Antimicrob Agents Chemother 48:648–650. doi: 10.1128/AAC.48.2.648-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Notake S, Matsuda M, Tamai K, Yanagisawa H, Hiramatsu K, Kikuchi K. 2013. Detection of IMP metallo-beta-lactamase in carbapenem-nonsusceptible Enterobacteriaceae and non-glucose-fermenting Gram-negative rods by immunochromatography assay. J Clin Microbiol 51:1762–1768. doi: 10.1128/JCM.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 25.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lartigue MF, Poirel L, Aubert D, Nordmann P. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother 50:1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Sun B, Wang Y, Lei L, Schwarz S, Wu C. 2016. Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob Agents Chemother 60:658–661. doi: 10.1128/AAC.02114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. 2014. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet 46:82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 31.Liao YT, Kuo SC, Lee YT, Chen CP, Lin SW, Shen LJ, Fung CP, Cho WL, Chen TL. 2014. Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob Agents Chemother 58:3983–3990. doi: 10.1128/AAC.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papagiannitsis CC, Tzouvelekis LS, Miriagou V. 2009. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active p2 promoter. Antimicrob Agents Chemother 53:277–280. doi: 10.1128/AAC.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto M, Nagao M, Hotta G, Matsumura Y, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2012. Molecular characterization of IMP-type metallo-beta-lactamases among multidrug-resistant Achromobacter xylosoxidans. J Antimicrob Chemother 67:2110–2113. doi: 10.1093/jac/dks179. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Zhu Y, Ma Y, Liu F, Lu N, Yang X, Luan C, Yi Y, Zhu B. 2015. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob Agents Chemother 59:1152–1161. doi: 10.1128/AAC.04260-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singhal N, Kumar M, Kanaujia PK, Virdi JS. 2015. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.