Abstract
DNA gyrase mutations are a major cause of quinolone resistance in Mycobacterium tuberculosis. We therefore conducted the first comprehensive study to determine the diversity of gyrase mutations in pre-extensively drug-resistant (pre-XDR) (n = 71) and extensively drug-resistant (XDR) (n = 30) Thai clinical tuberculosis (TB) isolates. All pre-XDR-TB and XDR-TB isolates carried at least one mutation within the quinolone resistance-determining region of GyrA (G88A [1.1%], A90V [17.4%], S91P [1.1%], or D94A/G/H/N/V/Y [72.7%]) or GyrB (D533A [1.1%], N538D [1.1%], or E540D [2.2%]). MIC and DNA gyrase supercoiling inhibition assays were performed to determine the role of gyrase mutations in quinolone resistance. Compared to the MICs against M. tuberculosis H37Rv, the levels of resistance to all quinolones tested in the isolates that carried GyrA-D94G or GyrB-N538D (8- to 32-fold increase) were significantly higher than those in isolates bearing GyrA-D94A or GyrA-A90V (2- to 8-fold increase) (P < 0.01). Intriguingly, GyrB-E540D led to a dramatic resistance to later-generation quinolones, including moxifloxacin, gatifloxacin, and sparfloxacin (8- to 16-fold increases in MICs and 8.3- to 11.2-fold increases in 50% inhibitory concentrations [IC50s]). However, GyrB-E540D caused low-level resistance to early-generation quinolones, including ofloxacin, levofloxacin, and ciprofloxacin (2- to 4-fold increases in MICs and 1.5- to 2.0-fold increases in IC50s). In the present study, DC-159a was the most active antituberculosis agent and was little affected by the gyrase mutations described above. Our findings suggest that although they are rare, gyrB mutations have a notable role in quinolone resistance, which may provide clues to the molecular basis of estimating quinolone resistance levels for drug and dose selection.
INTRODUCTION
The global control of tuberculosis is becoming a challenge due to extensively drug-resistant tuberculosis (XDR-TB), a potentially life-threatening form of tuberculosis that is estimated to have caused approximately 9.7% of multidrug-resistant tuberculosis (MDR-TB) cases in 2015 (1). The mainstay treatment regimen for MDR-TB consists of quinolones due to their bactericidal activity (2). The later-generation quinolones, e.g., moxifloxacin (MXF) and gatifloxacin (GAT), exhibit enhanced bactericidal activity and have shown an improvement in successfully treating the disease (3 – 5). Recently, DC-159a, a novel 8-methoxyfluoroquinolone, has shown lower MICs against quinolone-resistant Mycobacterium tuberculosis than the later-generation quinolones and promising in vivo activity for combination therapy of drug-susceptible and quinolone-resistant tuberculosis (6, 7).
Hence, quinolones are considered to be the cornerstone of the current treatment for MDR-TB; however, the use of quinolones in treating MDR-TB is debatable due to concerns about emerging resistance in M. tuberculosis (8, 9). Resistance to quinolones can pave the way for the emergence of XDR-TB. In Thailand, the quinolone resistance rates among MDR-TB cases increased from 9% in 2005 to 15% in 2011 (10, 11). Early detection of quinolone-resistant tuberculosis will help in choosing the proper regimen for the treatment of MDR-TB and in preventing the development of XDR-TB. Genotypic methods have been proposed as a rapid means to shorten the time required for quinolone resistance detection (12).
DNA gyrase is the sole target of quinolones in M. tuberculosis because DNA topoisomerase IV is absent in the M. tuberculosis genome (13). Quinolone binds to the quinolone binding pocket (QBP) in the DNA gyrase-DNA complex. Resistance to quinolones in M. tuberculosis results primarily from mutations in gyrA and gyrB, particularly in the quinolone resistance-determining regions (QRDRs) of GyrA (QRDR-A, residues 74 to 113) and GyrB (QRDR-B, residues 500 to 540), which cause alterations in the amino acids of the QBP (14, 15). A wide variety of mutations within the QRDRs have been reported; however, different amino acid substitutions in gyrase may cause different levels of resistance to quinolones (16, 17).
Nonetheless, the mutations that have been found in the gyrase genes do not necessarily correlate with resistance to quinolones. A particular mutation may be a genotypic variation rather than a cause of quinolone resistance. Based on previous studies, there are many methods for assessing the effects of DNA gyrase mutations, e.g., consideration of the presence of these mutations in quinolone-susceptible isolates (18, 19), analysis of the inhibitory effect of quinolones on DNA gyrase supercoiling activity (20), and transduction of these mutations into quinolone-susceptible strains with subsequent determination of susceptibility to quinolones (21).
Due to a lack of comprehensive data on gyrase mutations conferring quinolone resistance in clinical isolates of M. tuberculosis in Thailand, we determined the mutations in the entire sequence of the gyrase genes to distinguish between mutations associated with quinolone resistance in quinolone-resistant isolates, i.e., pre-XDR-TB and XDR-TB isolates, and those unrelated to quinolone resistance in quinolone-susceptible isolates, i.e., drug-sensitive TB and MDR-TB isolates. Additionally, the MICs for the M. tuberculosis clinical isolates harboring specific gyrase mutations were measured to determine the effect of different mutations on quinolone resistance levels. Supercoiling-inhibitory activities of quinolones against mycobacterial DNA gyrase were examined to reveal the impact of an uncharacterized mutation on quinolone resistance in M. tuberculosis.
MATERIALS AND METHODS
The study protocol was approved by the IRB of the Faculty of Medicine Siriraj Hospital, Mahidol University EC no. 029/2557.
Mycobacterial isolates and culture conditions.
Among 17,619 culture-positive samples from 288 hospitals in 46 provinces of Thailand, a total of 1,493 MDR-TB isolates, 206 pre-XDR-TB isolates, and 80 XDR-TB isolates were obtained in the Drug-Resistant Tuberculosis Research Laboratory, Faculty of Medicine Siriraj Hospital, Thailand, between December 2003 and March 2013. In the present study, 27 quinolone-susceptible isolates, i.e., drug-sensitive TB (n = 18) and MDR-TB (n = 9) isolates, and 101 quinolone-resistant isolates, i.e., pre-XDR-TB (n = 71) and XDR-TB (n = 30) isolates, from 92 patients were randomly selected from available stock cultures to obtain approximately 10 quinolone-resistant isolates from each year. Phenotypic resistance to four quinolones, i.e., ofloxacin (OFX), levofloxacin (LVX), MXF, and GAT, at 2 μg/ml was previously screened by the agar proportion method (22). At the time the screening began in 2003, there were no CLSI-recommended critical concentrations for later-generation quinolones (22). Therefore, the critical concentration of 2 μg/ml for OFX was also applied for later-generation quinolones in our laboratory since that time. At the time the isolates were collected, CLSI's revised critical concentration had not yet been adopted in our laboratory. A concentration of 2 μg/ml had been continuously used for LVX based on the WHO guideline from 2008 (23) and for MXF based on the WHO guideline from 2012 (24). The mycobacterial cells were kept in Middlebrook 7H9 broth medium supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment and 15% glycerol and were stored at −20°C until use. For preparation of genomic DNA, mycobacterial cells were recovered on Middlebrook 7H10 agar with or without 2 μg/ml of OFX for susceptible isolates and resistant isolates, respectively, for 4 weeks at 37°C.
Antimicrobial agents.
The following quinolones were obtained as pure substances from their manufacturers: OFX (Bio Basic Inc., Amherst, NY), ciprofloxacin (CIP) (Sigma-Aldrich Co., St. Louis, MO), LVX and sparfloxacin (SPX) (Sigma-Aldrich Co., Buchs, Switzerland), MXF (Sigma-Aldrich Laborchemikalien GmbH, Seelze, Germany), GAT (U.S. Pharmacopeia, Rockville, MD), and DC-159a (Daiichi-Sankyo Pharmaceuticals Co., Ltd., Tokyo, Japan).
Genomic DNA preparation.
Two to three loopfuls of the mycobacterial culture were suspended in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and the suspension was heated at 80°C for 20 min to kill the mycobacterial cells. The heat-killed mycobacterial suspension was used for DNA extraction via an enzymatic method using lysozyme, and the DNA was then purified with cetyltrimethylammonium bromide-NaCl and chloroform-isoamyl alcohol (25). The DNA purities and concentrations were measured using a NanoDrop1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Amplification and sequencing of the gyrB and gyrA genes.
The entire nucleotide sequences of the gyrB and gyrA genes were amplified in 50-μl PCR mixtures consisting of 2.5 mM MgSO4, 0.25 mM concentrations of each of the four deoxynucleoside triphosphates (Thermo Fisher Scientific, Vilnius, Lithuania), 2 units of Platinum Taq DNA high-fidelity polymerase (Invitrogen, Carlsbad, CA), 20 pmol of a pair of amplification primers (Table 1), and 200 ng of template DNA under the following conditions: 2 min of initial denaturation at 94°C; 35 amplification cycles, with each cycle consists of 1 min at 94°C for denaturation, 1 min at 56°C for annealing, and 4 min at 68°C for extension; and a final extension at 68°C for 10 min. The 4,784-bp amplified products, which contained the 39-bp upstream region of gyrB, 2,145-bp gyrB, 34-bp intergenic region, 2,517-bp gyrA, and 49-bp downstream region of gyrA, were run on a 1% agarose gel and visualized with ethidium bromide staining. The amplified products were sequenced in both directions using an ABI Prism 3730XL Analyzer (Applied Biosystems, Foster City, CA) by Macrogen (Seoul, South Korea) for the determination of the nucleotide sequences of the entire gyrB and -A genes.
TABLE 1.
Nucleotide sequences of the primers used in this study
| Primer | Nucleotide sequencea | Comment |
|---|---|---|
| gyrB_Fw | 5′-GCACCAGGAAGAAAGATGTCC-3′ | Amplification (nucleotide positions 5084–5104)b |
| gyrA_Rv | 5′-TTCCTCCTCAGATCGCTACG-3′ | Amplification (nucleotide positions 9867–9848)b |
| gyrA_cln_Fw | 5′-ATGACAGACACGACGTTGCCGCCTG-3′ | Cloning, bp 1–25 of gyrA |
| gyrA_cln_Rv | 5′-CATCGTCGTCGCTCGAGCCTGATTAA-3′ | Cloning, inserted XhoI site |
| gyrB_cln_Fw | 5′-ATGGGTAAAAACGAGGCCAGAAGATC-3′ | Cloning, bp 1–25 of gyrB |
| gyrB_cln_Rv | 5′-TGCATCTCCTGCAGGATGTCAACCG-3′ | Cloning, inserted SbfI site |
| Mut_GB538_Fw | 5′-GGTGCTAAAGGACACCGAAGTTCAG-3′ | Mutagenesis |
| Mut_GB538_Rv | 5′-CGGTCGATGCGCGCTTTC-3′ | Mutagenesis |
| Mut_GB540_Fw | 5′-AGAACACCGACGTTCAGGCGATC-3′ | Mutagenesis |
| Mut_GB540_rv | 5′-TTAGCACCCGGTCGATGC-3′ | Mutagenesis |
The restriction enzyme site or mutated nucleotide is underlined.
Nucleotide position on chromosome of M. tuberculosis H37Rv (accession no. NC_000962.3).
Analysis of gyrase polymorphisms.
Short DNA sequences of 300 to 750 bp in length with quality values of ≥20 were assembled using the DNA Baser Sequence Assembler v4.3.0 software (Heracle BioSoft SRL Romania, 2012). The consensus sequences of each isolate were compared to that of M. tuberculosis H37Rv (GenBank accession number NC_000962.3) using the ClustalW Multiple Alignment in BioEdit v7.2.0 software (26).
Spoligotyping.
Spoligotyping was performed as a part of our prior study (27) using a spoligotyping kit (Ocimum Biosolution, Hyderabad, India) (28). Gyrase mutation data and spoligotyping information were analyzed together to rule out mutations that may have represented lineage-specific mutations that were not involved in quinolone resistance.
Determination of MICs.
Due to limited resources, a subset of quinolone-susceptible isolates and quinolone-resistant isolates with a single mutation among the three most common GyrA mutations or GyrB mutations was randomly selected to determine whether the different gyrase mutations conferred different levels of resistance to quinolones. The MICs were determined according to the standard agar dilution method recommended by CLSI (29). Inocula of 105 CFU per spot of M. tuberculosis isolates were inoculated onto Middlebrook 7H10 agar supplemented with 10% oleic acid-albumin-dextrose catalase and 0.5% glycerol containing quinolones at concentrations ranging from 0.03 to 32 mg/liter in 2-fold dilutions. M. tuberculosis H37Rv was used as the reference strain. The inoculated agar plates were incubated at 37°C for 28 days. Each isolate was tested in triplicate. The MIC was defined as the lowest drug concentration that yielded no visible growth of M. tuberculosis.
Bacterial strains and plasmids.
The NEB 10-beta Escherichia coli strain (New England BioLabs, Ipswich, MA) was used as the cloning host for recombinant plasmid propagation. The NEB-express E. coli strain (New England BioLabs, Ipswich, MA) was used as the host for recombinant protein expression. The pMAL-c5x vector plasmid (New England BioLabs, Ipswich, MA) was used to create expression plasmids for M. tuberculosis DNA gyrase subunits A and B.
Construction of wild-type mycobacterial gyrase expression vectors.
Insert fragments were separately prepared by amplification of the gyrA and gyrB genes of M. tuberculosis H37Rv with two sets of primers (Table 1). The 3′-end overhangs of the PCR products were polished using the Quick Blunting kit (New England BioLabs, Ipswich, MA). The blunt-ended gyrA and gyrB fragments were digested with XhoI and SbfI, respectively, ligated into pMAL-c5X digested with XmnI/SalI and XmnI/SbfI, respectively, using the Quick Ligation kit (New England BioLabs, Ipswich, MA), and subsequently transformed into the chemically competent NEB 10-beta E. coli strain. Recombinant clones were selected from the colonies that grew on LB agar plates containing ampicillin (100 μg/ml). Successful insertions of gyrA and gyrB fragments in frame downstream of the tac promoter and malE were confirmed by digestion of the recombinant plasmid with PvuII (New England BioLabs, Ipswich, MA) and Sanger sequencing using an ABI Prism 3730XL Analyzer at Macrogen. Recombinant plasmids were propagated in the cloning host, extracted, and then transformed into the chemically competent NEB-express E. coli strain. The resulting recombinant gyrB plasmids were used as template DNAs for subsequent site-directed mutagenesis.
In vitro mutagenesis.
To investigate the effect of gyrB mutations on quinolone resistance, two pairs of mutagenesis primers were designed to introduce single-nucleotide substitutions A1612G or A1620C (Table 1) in wild-type (WT) gyrB, resulting in GyrB N538D or E540D mutations, respectively, by site-directed mutagenesis with the Q5 site-directed mutagenesis kit (New England BioLabs). The resulting mutant plasmids were sequenced to ensure the presence of mutations introduced into gyrB and the absence of undesired mutations using an ABI Prism 3730XL Analyzer at Macrogen.
Recombinant mycobacterial gyrase expression and purification.
Two milliliters of overnight cultures of NEB-express E. coli carrying recombinant WT or mutant plasmids was separately inoculated into 200 ml of rich medium containing 100 μg/ml ampicillin and grown at 28°C until the optical density at 600 nm reached 0.5 to 0.8. IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma-Aldrich Co., St. Louis, MO) was added to a final concentration of 0.3 mM and continuously incubated at 28°C for 8 h. E. coli cells were harvested by centrifugation at 3,300 × g for 20 min at 4°C (Heraeus Megafuge 1.0R; Kendro Laboratory International Sales, Ashville, NC). The supernatant was discarded, and the pellet was suspended in 5 ml of TGED buffer (50 mM Tris-HCl [pH 7.9], 10% glycerol, 1 mM EDTA, and 1 mM dithiothreitol [DTT]). The cell suspension was kept at −70°C overnight. Frozen bacterial cells were thawed at 4°C, 10 mg/ml of lysozyme was added to a final concentration of 1 mg/ml, and the cell suspension was further incubated at 4°C for 3 h and subsequently centrifuged at 11,400 × g for 60 min at 4°C (Allegra X-15R; Beckman Coulter, Inc., Indianapolis, IN). The cell lysate was collected for further purification.
Cell lysate was loaded onto a preequilibrated amylose resin column in TGED buffer. The column was washed with 15 column volumes of TGED buffer containing 1 M NaCl to remove contaminated genomic DNA and was continuously washed with 15 column volumes of TGED buffer. The maltose binding protein (MBP)-Gyr fusion proteins were eluted with TGED buffer containing 10 mM maltose, and 1-ml fractions of eluent were subsequently collected. Fractions containing the fusion protein were determined using the Bradford assay. The positive fractions were pooled and concentrated to 1 ml using Vivaspin15R (Sartorius Stedim Biotech GmbH, Göttingen, Germany).
Factor Xa was added to the MBP-Gyr fusion protein solution to a final concentration of 1%, wt/vol. The reaction mixture was incubated at 4°C for 8 h. After factor Xa cleavage, the 93-kDa GyrA and 78-kDa GyrB subunits were separated from the 42-kDa MBP (Fig. 1). The cleavage mixture was dialyzed twice against 50 mM Tris-HCl (pH 7.9), 30% glycerol, and 5 mM dithiothreitol at 4°C for 4 h and was then incubated overnight. The dialyzed enzyme was stored at −20°C. Protein concentrations and compositions were determined using a NanoDrop1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and SDS-PAGE (Mini-Protean tetra cell; Bio-Rad Laboratories, Inc., Hercules, CA), respectively. Concentrations of purified DNA gyrase ranged from 1.89 to 4.25 mg/ml from a 1,600-ml culture.
FIG 1.
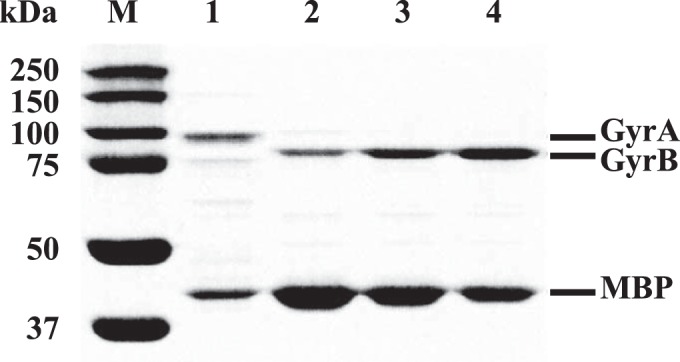
SDS-PAGE analysis of recombinant M. tuberculosis DNA gyrase stained with Coomassie blue. Lane M, protein standard marker; lane 1, WT GyrA (93 kDa); lane 2, WT GyrB (78 kDa); lane 3, GyrB-N538D (78 kDa); lane 4, GyrB-E540D (78 kDa). MBP, maltose binding protein (42 kDa).
Supercoiling inhibition assay.
Inhibitory effects of quinolones against DNA gyrase supercoiling activity were determined by the ATP-dependent supercoiling assay as described previously (30) with minor modifications. The purified WT GyrA was preincubated with WT GyrB, GyrB-N538D, or GyrB-E540D at room temperature for 15 min to reconstitute the active heterotetrameric GyrA2GyrB2 complex. One unit of the active gyrase complex was added to the reaction mixture (total volume, 20 μl) containing supercoiling assay buffer (20 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 1 mM ATP, 1 mM spermidine, 20 mg/liter tRNA, 20 mg/liter bovine serum albumin) and 0.2 μg of relaxed pBR322 plasmids (Inspiralis Ltd., Norwich, United Kingdom) as a substrate. The reaction was performed at 37°C for 1 h and terminated by addition of 20 μl of the 2× glycerol dye mix, and the products were then analyzed by gel electrophoresis in 1% agarose for 3 h at 40 V. One unit of enzyme activity was defined as the amount of DNA gyrase that was used to completely supercoil 0.2 μg of relaxed pBR322 DNA within 1 h. One unit of either the WT or mutant reconstituted gyrase was sufficient to completely supercoil 0.2 μg of relaxed pBR322 DNA in the presence of 1 mM ATP (Fig. 2, lanes 5, 7, and 9), while neither the GyrA nor the GyrB subunit alone generated supercoiled DNA (Fig. 2, lanes 3, 4, 6, and 8). The lack of supercoiling activity in GyrA or GyrB alone indicated the absence of contamination of E. coli DNA gyrase.
FIG 2.
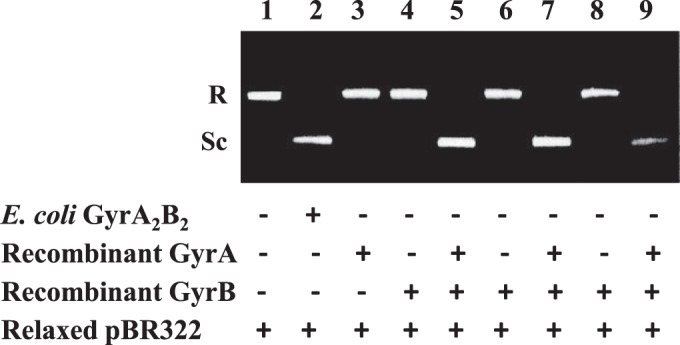
Supercoiling activities of recombinant M. tuberculosis GyrA and GyrB at 1 unit in the presence of relaxed pBR322 DNA and ATP. Lane 1, relaxed pBR322 DNA alone (negative control); lane 2, E. coli GyrA2B2 complex (Inspiralis) (positive control); lane 3, recombinant WT GyrA alone; lane 4, recombinant WT GyrB alone; lane 5, recombinant WT GyrA and GyrB complex; lane 6, recombinant GyrB-N538D alone; lane 7, recombinant WT GyrA and GyrB-N538D complex; lane 8, recombinant GyrB-E540D alone; lane 9, recombinant WT GyrA and GyrB-E540D complex. R and Sc, relaxed and supercoiled pBR322 DNA, respectively.
The inhibitory effects of quinolones on DNA gyrase were evaluated by the method described above in a reaction mixture containing 1 U of purified DNA gyrase in the absence or presence of 2-fold serial dilutions of each quinolone (0 to 400 μg/ml). The half-maximal inhibitory concentration was defined as the quinolone concentration that inhibits DNA gyrase supercoiling activity by 50% (IC50). The supercoiling activity was assessed by tracing the intensity of the bands corresponding to supercoiled pBR322 plasmids. All assays were performed in triplicate.
Data analysis.
Odds ratios were calculated to measure the association between the types of resistance and the patterns of resistance. Comparisons of MICs for isolates carrying different mutations were performed using the Mann-Whitney U test. Significant differences in IC50s were determined by independent t tests. All statistical analyses were performed with PASW Statistics 18 (SPSS Inc., Chicago, IL). P values of less than 0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The gyrA and gyrB sequence data have been deposited in GenBank under accession numbers KT339397 to KT339524 and KT339525 to KT339652, respectively.
RESULTS
Quinolone resistance patterns in pre-XDR-TB and XDR-TB isolates.
Among the 101 quinolone-resistant isolates, 96 (95.1%), 58 (57.4%), 49 (48.5%), and 19 (18.8%) were resistant to OFX, LVX, MXF, and GAT at 2 μg/ml, respectively. OFX-monoresistant isolates accounted for 41.2% of the pre-XDR-TB and 20.0% of the XDR-TB isolates. LVX- and MXF-monoresistant isolates were observed in 1.5% and 5.9% of the pre-XDR-TB isolates, respectively. Resistance to multiple quinolones was highly associated with XDR-TB (n = 24/30; 80.0%) compared to pre-XDR-TB (n = 36/71; 50.7%) (odds ratio, 3.89 [95% confidence interval {CI}, 1.42 to 10.66]; P = 0.008).
Gyrase mutations within the quinolone resistance-determining regions.
All pre-XDR-TB and XDR-TB isolates had at least one mutation within the QRDRs (Table 2). The detected mutations resulted in nine different amino acid alterations in GyrA (i.e., G88A, A90V, S91P, and D94A/G/H/N/V/Y) and three different amino acid alterations in GyrB (i.e., D533A, N538D, and E540D). Single mutations in GyrA were identified in 92.3% of quinolone-resistant isolates. Single mutations in GyrB accounted for 3.3% of quinolone-resistant isolates. Double mutations in GyrA alone or in both GyrA and GyrB were observed in 3.3% and 1.1% of quinolone-resistant isolates, respectively. The most prevalent alteration occurred at position 94 in GyrA (72.7% of all isolates). D94G was the most frequent variation (42.4%) and was significantly associated with XDR-TB (odds ratio, 2.88 [95% CI 1.15 to 7.22]; P = 0.024). A90V and D94A substitutions in GyrA were the second (17.4%) and third (11.9%) most common mutations, respectively. None of the quinolone-susceptible isolates contained mutations within the QRDRs.
TABLE 2.
Mutations within QRDR-A and QRDR-B in pre-XDR-TB and XDR-TB isolates
| Mutation(s)a | No. of isolatesb |
% of all isolates | |
|---|---|---|---|
| Pre-XDR-TB | XDR-TB | ||
| GyrA | |||
| G88A | 1 | 1.1 | |
| A90V | 12 | 4 | 17.4 |
| S91P | 1 | 1.1 | |
| D94A | 11 | 11.9 | |
| D94G | 22 | 17 | 42.4 |
| D94H | 2 | 2 | 4.3 |
| D94N | 5 | 3 | 8.7 |
| D94V | 1 | 1.1 | |
| D94Y | 3 | 1 | 4.3 |
| A90V + D94A | 2 | 2.2 | |
| A90V + D94N | 1 | 1.1 | |
| GyrB | |||
| N538D | 1 | 1.1 | |
| E540D | 2 | 2.2 | |
| GyrA + GyrB | |||
| D94A + D533A | 1 | 1.1 | |
GyrA S95T was found in all isolates.
Sequential isolates with an identical gyrase mutation that were taken from a single patient were counted as only one isolate.
Gyrase polymorphisms outside the quinolone resistance-determining regions.
In total, 20 mutations were found outside the QRDRs of GyrA and GyrB (Table 3). Among these mutations, six were identified solely in quinolone-susceptible isolates, seven were found solely in quinolone-resistant isolates, five were detected in both quinolone-susceptible and quinolone-resistant isolates, and two (i.e., GyrA E21Q and G668D) were present in all isolates studied. We identified two synonymous mutations (i.e., GyrA I614I and L653L) and three nonsynonymous mutations (i.e., GyrA A384V, GyrB M330I, and V340L) that were previously reported to be lineage-specific mutations (31, 32).
TABLE 3.
Mutations outside QRDR-A and QRDR-B in quinolone-susceptible and quinolone-resistant M. tuberculosis isolates
| Gene and nucleotide position(s) | Allele change(s) | Amino acid substitution(s)a | No. of isolatesb |
Spoligotype(s)c | Previously reported lineage-specific mutations (reference[s]) | |
|---|---|---|---|---|---|---|
| QS | QR | |||||
| gyrA | ||||||
| 165 | G/A | V55V | 1 | EAI1_SOM | NDd | |
| 996 | C/T | Y332Y | 1 | EAI6_BGD1 | ND | |
| 1151 and 1842 | C/T and T/C | A384V and I614I | 8 | 2 | EAI1_SOM, EAI2_NTB, EAI5, EAI6_BGD1, Manu1 | Lineage 1 (31, 32) |
| 1468 | C/A | H490N | 1 | Beijing | ND | |
| 1542 | G/A | E514E | 1 | 1 | Beijing | ND |
| 1820 | G/A | R607H | 1 | Beijing | ND | |
| 1959 | G/C | L653L | 1 | EAI2_NTB | Lineage 1.2.1 (31) | |
| 2106 | C/A | G720G | 1 | EAI5 | ND | |
| 2238 | C/T | G746G | 2 | T2 | ND | |
| gyrB | ||||||
| 261 | G/A | E87E | 11 | Manu_ancestor | ND | |
| 990 | G/C | M330I | 8 | 2 | EAI1_SOM, EAI2_NTB, EAI5, EAI6_BGD1, Manu1 | Lineage 1 (31, 32) |
| 1018 | G/T | V340L | 1 | LAM9 | Lineage 4.3.4.2 (31) | |
| 1457 | C/A | S486Ye | 1 | Beijing | ND | |
| 1651 | G/A | G551Re | 1 | 2 | Beijing | ND |
| 1933 | G/C | G645S | 1 | EAI1_SOM | ND | |
| 1966 | G/T | D656Y | 1 | Beijing | ND | |
| 2082 | C/T | D694D | 1 | EAI1_SOM | ND | |
MICs.
MICs against randomly selected clinical isolates harboring one of three most frequent GyrA mutations, i.e., D94G (n = 6), A90V (n = 5), or D94A (n = 5), or bearing a GyrB mutation, i.e., N538D (n = 1) or E540D (n = 4), are shown in Table 4. The MICs of all quinolones tested against the clinical isolates carrying gyrase mutations within the QRDRs were higher than those against M. tuberculosis H37Rv, and 5 clinical strains without mutations in the QRDRs of gyrA or gyrB. MICs were clearly distinct between susceptible and resistant strains. DC-159a demonstrated the lowest MIC among the isolates tested. The fourth-generation (i.e., MXF and GAT) and third-generation (i.e., LVX and SPX) quinolones exhibited better antimycobacterial activities against either wild-type or mutant isolates than the second-generation quinolones (i.e., OFX and CIP). Interestingly, these findings were different in isolates harboring GyrB-E540D, which conferred high-level resistance particularly to later-generation quinolones, i.e., SPX, MXF, GAT, and DC-159a (8- to 16-fold increases in MICs relative to those against M. tuberculosis H37Rv) but caused low-level resistance to early-generation quinolones, i.e., OFX, LVX, and CIP (2- to 4-fold increase) (P < 0.01). The isolates bearing GyrA-D94G or GyrB-N538D mutations had a tendency to show higher MICs for all quinolones tested (8- to 32-fold increase) than those bearing GyrA-A90V or GyrA-D94A mutations (2- to 8-fold increase) (P < 0.01).
TABLE 4.
MICs of quinolones tested against clinical M. tuberculosis isolates harboring different gyrase mutations
| Mutation (no. of isolates) | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|
| OFX | LVX | CIP | SPX | MXF | GAT | DC-159a | |
| None, wild-type M. tuberculosis H37Rv | 1 | 0.5 | 0.5 | 0.25 | 0.25 | 0.125 | 0.06 |
| None, quinolone susceptible (5) | 1–2 | 0.25–0.5 | 0.5–1 | 0.25 | 0.125–0.25 | 0.06–0.125 | 0.03–0.06 |
| GyrA D94G (6) | 8–16 | 4–8 | 8–16 | 2 | 2–8 | 2–4 | 0.25–0.5 |
| GyrB N538D (1) | 8 | 8 | 8 | 2–4 | 2 | 2 | 0.25 |
| GyrA A90V (5) | 8 | 2–4 | 2–8 | 0.5–2 | 0.5–1 | 0.5–1 | 0.125 |
| GyrA D94A (5) | 2–8 | 2–8 | 2–8 | 1 –2 | 0.5–2 | 0.25–1 | 0.06–0.25 |
| GyrB E540D (4) | 2 | 1–2 | 1–2 | 2 | 4 | 2 | 0.5 |
Boldface indicates an increase in MIC of >8-fold.
Determination of inhibitory effects on DNA supercoiling activities and IC50s of quinolones.
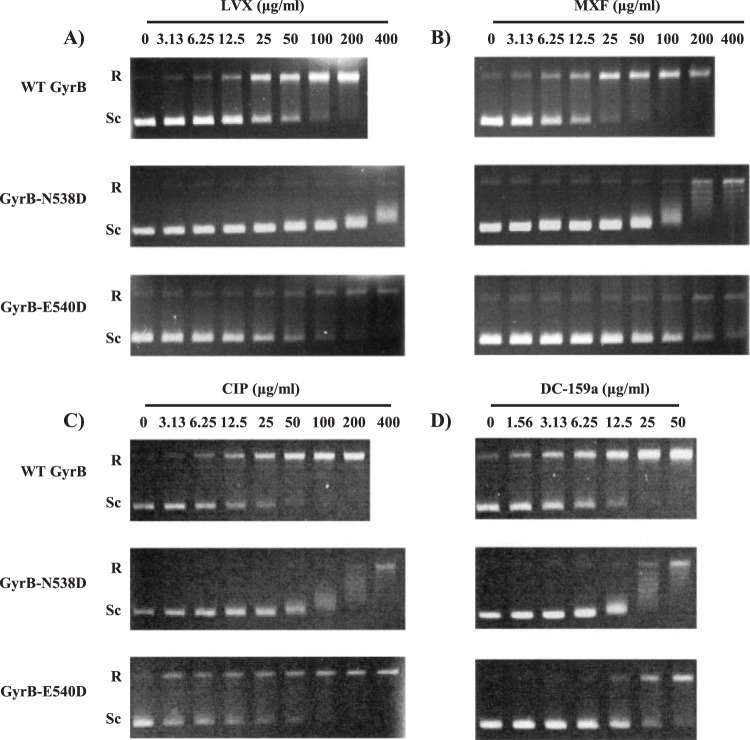
The specific activities of WT GyrA, WT GyrB, GyrB-N538D, and GyrB-E540D were 1.7 × 104, 1.0 × 104, 7.5 × 103, and 4.8 × 103 U/mg, respectively. As summarized in Table 5, we performed a DNA supercoiling inhibition assay to elucidate an uncharacterized role of GyrB-E540D in quinolone resistance and calculated the IC50 of each quinolone, which is listed from high to low values. Representative results on the inhibitory effects of LVX, MXF, CIP, and DC-159a on DNA supercoiling are shown in Fig. 3. Each quinolone demonstrated concentration-dependent inhibitory activities. Overall, linear regression analysis revealed a good correlation between IC50s and MICs (r2 = 0.91). DC-159a exhibited the lowest IC50 of all quinolones tested against DNA gyrase bearing either WT GyrB or mutant GyrB (N538D and E540D). Intriguingly, introduction of the E540D mutation in the GyrB subunit remarkably increased IC50s, particularly for later-generation quinolones, i.e., SPX, MXF, GAT, and DC-159a (2.9- to 11.2-fold increases), to the same levels as those observed in GyrB-N538D mutant enzymes (2.3- to 11.7-fold increases) (P > 0.05), but it had a less significant impact on IC50s for early-generation quinolones, i.e., OFX, LVX, and CIP (1.5- to 2-fold increases) than GyrB-N538D (7.1- to 16.1-fold increases) (P < 0.01).
TABLE 5.
IC50s of seven quinolones for wild-type and mutant GyrB of M. tuberculosis
| Quinolone | Substituents |
IC50 (μg/ml, mean ± SD) |
IC50 ratio |
P valuea | |||||
|---|---|---|---|---|---|---|---|---|---|
| R-1 | R-7 | R-8 | WT | GyrB-N538D | GyrB-E540D | GyrB-N538D/WT | GyrB-E540D/WT | ||
| OFX | Bridge N-1–C-8 | Methyl piperazine | Bridge N-1–C-8 | 54.4 ± 6.2 | >400 | 93.2 ± 8.5 | NDb | 1.7 | <0.001 |
| LVX | Bridge N-1–C-8 | Methyl piperazine | Bridge N-1–C-8 | 22.5 ± 2.2 | 363.3 ± 20.3 | 46.4 ± 2.5 | 16.1 | 2.0 | 0.001 |
| CIP | Cyclopropyl | Piperazine | H | 21.8 ± 5.9 | 154.5 ± 1.2 | 32.1 ± 9.8 | 7.1 | 1.5 | 0.002 |
| MXF | Cyclopropyl | Azabicyclo | OCH3 | 11.5 ± 0.6 | 95.3 ± 14.6 | 99.3 ± 6.0 | 8.3 | 8.7 | 0.688 |
| SPX | Cyclopropyl | Dimethyl piperazine | F | 9.4 ± 1.5 | 109.8 ± 10.5 | 104.5 ± 13.7 | 11.7 | 11.2 | 0.651 |
| GAT | Cyclopropyl | Methyl piperazine | OCH3 | 9.2 ± 1.2 | 98.2 ± 2.8 | 76.5 ± 18.8 | 10.7 | 8.3 | 0.096 |
| DC-159a | Fluorinated cyclopropyl | Methyl pyrrolidine | OCH3 | 5.7 ± 0.7 | 13.5 ± 4.2 | 16.1 ± 4.3 | 2.3 | 2.9 | 0.497 |
Significant differences between the IC50 of GyrB N538D and the IC50 of GyrB E540D were determined by an independent t test. A P value of less than 0.05 was considered statistically significant.
ND, not determined.
FIG 3.
Representative data on the inhibitory effects of levofloxacin (LVX) (A), moxifloxacin (MXF) (B), ciprofloxacin (CIP) (C), and DC-159a (D) on DNA supercoiling activities. The supercoiling activities against DNA gyrase complexes bearing WT GyrB, GyrB-N538D, and GyrB E540D in the presence of the indicated concentration of each quinolone are shown. R and Sc, relaxed and supercoiled pBR322 DNA, respectively.
DISCUSSION
As can be seen from the phenotypic profiles of quinolone susceptibility and MIC results, later-generation quinolones, i.e., SPX, MXF, GAT, and DC-159a, exhibited better in vitro activity against M. tuberculosis than early-generation quinolones, i.e., OFX, LVX, and CIP. Our findings underscore the possibility of evaluating later-generation quinolones for the treatment of ofloxacin-resistant TB (33, 34) and as a salvage therapy in patients with MDR-TB (35). In addition, our results imply that susceptibility to OFX may not necessarily indicate susceptibility to all quinolones when OFX is used as a representative quinolone in susceptibility testing (17). We thus suggest that the susceptibility to each quinolone should be tested as recommended by WHO (36) because the quinolones are not completely cross-resistant. However, it should be noted that the percentage of later-generation quinolone-resistant isolates and the level of cross-resistance may be underestimated in this study, because the critical concentration of 2 μg/ml for LVX, MXF, and GAT was used instead of 1.0, 0.5, and 1.0 μg/ml, respectively (24, 29).
Based on the fact that quinolone resistance in M. tuberculosis occurs by introduction of mutations in gyrA and gyrB (15), we attempted to determine how gyrase mutations are related to the quinolone-resistance phenotypes of pre-XDR-TB and XDR-TB Thai clinical isolates. The prevalence of gyrA mutations in clinical isolates from quinolone-resistant tuberculosis varies across countries, ranging from 64.5% to 94.1% (37 – 40). In this study, we found a relatively high prevalence of mutations in QRDR-A, at 96.7% among isolates examined, and the most frequent mutations occurred in GyrA at D94 (72.7%). Because the D94 residue, which anchors the water-magnesium ion bridge with a conserved C3/C4 keto acid moiety of quinolones, plays an important role in stabilizing the quinolone molecule in the QBP, an amino acid substitution at this position will exaggerate the deleterious effect of the binding between most quinolones and DNA gyrase (41, 42). In addition, the high frequency of D94 mutations in clinical isolates from pre-XDR and XDR-TB may be due to the positive epistasis between D94 mutations and mutations conferring rifampin resistance leading to the progression of MDR-TB to XDR-TB (43).
Furthermore, our results underscore the differences in quinolone resistance levels caused by different mutations in GyrA, i.e., D94G, A90V, and D94A. Similarly, other studies have reported that clinical isolates carrying D94G exhibit higher levels of resistance to OFX, MXF, and GAT than isolates carrying A90V or D94A (16, 17, 38). Recently, Rigouts et al. showed strong evidence for a higher rate of failure/relapse in the presence of the D94G mutation (70.0%) than in that of the D94A mutation (11.1%) when patients were treated with GAT (38). One possible explanation for this difference is that the GyrA-D94G mutation may increase bacterial fitness while simultaneously lowering quinolone susceptibility (43, 44).
In addition to the mutations in QRDR-A, we observed two quinolone resistance-associated mutations in QRDR-B, i.e., N538D and E540D. Whereas the N538D mutation is a well-characterized mutation causing high-level resistance across quinolones (20, 21), there is little information on the role of the E540D mutation in quinolone-resistant tuberculosis. Previously, Malik et al. demonstrated that E540D transductants uniquely exhibit resistance to MXF and not to CIP, LVX, or OFX (21). Here, we first described a novel characteristic of clinical isolates carrying the E540D mutation, which confers high-level resistance (8- to 16-fold increases in MICs) not only to MXF but also to SPX and GAT, while the E540D mutation maintained low-level resistance (2- to 4-fold increases in MICs) to OFX, LVX, and CIP. These fold differences in MICs were approximately the same as described by Malik et al., even though the MICs against M. tuberculosis H37Rv were relatively 2-fold higher in this study (21).
These findings were confirmed by the DNA gyrase supercoiling inhibition assay and may be explained by a structure-activity relationship. In principle, the addition of the cyclopropyl moiety at N-1 and the substituents at C-8 in later-generation quinolones led to the improvement in antimycobacterial activities (45 – 47). Interestingly, this enhanced the activity of the later-generation quinolones against WT GyrB and the GyrB-N538D mutant but not the GyrB-E540D mutant. The likely reason for this occurrence is that replacement of glutamic acid, a larger amino acid residue, at position 540 with aspartic acid, a smaller amino acid residue, may disturb hydrogen bonds between the amino acid residue at position 540 and the C-7 substituents of quinolones and may reduce the QBP size, leading to a steric effect of the C-8 substituents, e.g., 8-fluoro and 8-methoxy groups (21, 48).
Consideration of structure-activity relationships revealed three structural features that are related to the alteration of supercoiling-inhibitory activities of the quinolones that were tested. First, the addition of N-1-cyclopropyl, C-8-fluoro, and C-8-methoxy substituents led to lower IC50s against DNA gyrase bearing either WT GyrB or GyrB-N538D. Second, in contrast, the presence of C-8-fluoro or C-8-methoxy moieties did not necessarily decrease the IC50s against the GyrB-E540D mutant enzyme. Lastly, the addition of fluorine to N-1-cyclopropyl and the presence of the C-8-methyl pyrrolidine group in DC-159a were able to decrease the IC50s against either WT GyrB or mutant GyrB (N538D and E540D).
Protein structure analysis of DNA gyrase interacting with quinolones is required to further explain this paradoxical effect. Nonetheless, the characteristic resistance pattern of clinical isolates carrying the GyrB-E540D mutation raises awareness of using later-generation quinolones for the treatment of MDR-TB with low-level resistance to OFX (36). In addition, these findings reveal that gyrB QRDR mutations may have significant roles in quinolone resistance even if they are rare mutations. Likewise, Von Groll et al. noted that gyrB mutations should be considered in determining resistance to quinolones, especially to 8-methoxyfluoroquinolones (48). Due to insufficient information regarding the patients' medical history, we are not able to identify whether the GyrB-E540D mutation was acquired from previous exposure to 8-methoxyfluoroquinolones.
Forty-five percent of mutations outside the QRDRs found in this study either were previously proved to be neutral variants, e.g., GyrB S486Y and G551R (20), or were well-known lineage-specific markers, e.g., GyrA A384V and GyrB M330I (31, 32), while whether the remaining 55% of mutations outside the QRDRs, e.g., GyrB G645S and D656Y, are related to quinolone resistance is still unclear since they coexisted with mutations in the QRDRs. Therefore, additional analysis may be needed to determine the unclarified roles of these mutations in quinolone resistance (18 – 21). In addition, one synonymous mutation, GyrB-E87E, may be a potential genetic marker for the Manu-ancestor clade (Spoligo-International-Type 523), which is a rare genotype causing the clonal spread of XDR-TB in western Thailand (27, 49).
Currently, the clinical applications of whole-genome sequencing are being expanded to produce genotypic drug susceptibility profiles faster than phenotypic susceptibility results can be obtained (50, 51). The construction of a database of resistance-related mutations is required for the reliable genotypic prediction of drug resistance. We demonstrate that mutations within not only QRDR-A but also QRDR-B should be considered potential determinants of quinolone resistance (14, 18, 19), while mutations outside the QRDRs are likely linked to the genotypic background of M. tuberculosis (31, 32). Furthermore, knowing the gyrase mutations not only is relevant for the prediction of quinolone resistance but also may be useful for estimating levels of resistance to various quinolones (16, 17, 40), which can aid in the selection of the appropriate quinolone and its dosing.
ACKNOWLEDGMENTS
This work was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (grant no. PHD/0328/2552 to A.D. [student] and A.C. [advisor]). Financial support for the laboratories was also obtained from the Drug Resistant Tuberculosis Research Fund of the Siriraj Foundation, the Research and Development Fund of the Faculty of Medicine Siriraj Hospital, Mahidol University, and the Japan Science and Technology/National Science and Technology Development Agency (JST/NSTDA, grant no. P-12-01777). A.C. is supported by the Chalermprakiat grant from the Faculty of Medicine Siriraj Hospital, Mahidol University.
This work is dedicated to the late HRH Princess Galyanivadhana Krom Laung Narathiwas Rajnakarindth, the patron of the Drug Resistant Tuberculosis Research Fund, Siriraj Foundation, on the occasion of her 92nd birthday.
REFERENCES
- 1.WHO. 2015. Global tuberculosis report 2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.WHO. 2009. Treatment of tuberculosis: guidelines, 4th ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Anderson LF, Tamne S, Watson JP, Cohen T, Mitnick C, Brown T, Drobniewski F, Abubakar I. 2013. Treatment outcome of multi-drug resistant tuberculosis in the United Kingdom: retrospective-prospective cohort study from 2004 to 2007. Euro Surveill 18(40):pii=20601 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20601. [DOI] [PubMed] [Google Scholar]
- 4.Andriole VT. 2005. The quinolones: past, present, and future. Clin Infect Dis 41:S113–S119. doi: 10.1086/428051. [DOI] [PubMed] [Google Scholar]
- 5.Van Deun A, Maug AK, Salim MA, Das PK, Sarker MR, Daru P, Rieder HL. 2010. Short, highly effective, and inexpensive standardized treatment of multidrug-resistant tuberculosis. Am J Respir Crit Care Med 182:684–692. doi: 10.1164/rccm.201001-0077OC. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad Z, Minkowski A, Peloquin CA, Williams KN, Mdluli KE, Grosset JH, Nuermberger EL. 2011. Activity of the fluoroquinolone DC-159a in the initial and continuation phases of treatment of murine tuberculosis. Antimicrob Agents Chemother 55:1781–1783. doi: 10.1128/AAC.01514-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Disratthakit A, Doi N. 2010. In vitro activities of DC-159a, a novel fluoroquinolone, against Mycobacterium species. Antimicrob Agents Chemother 54:2684–2686. doi: 10.1128/AAC.01545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed I, Jabeen K, Inayat R, Hasan R. 2013. Susceptibility testing of extensively drug-resistant and pre-extensively drug-resistant Mycobacterium tuberculosis against levofloxacin, linezolid, and amoxicillin-clavulanate. Antimicrob Agents Chemother 57:2522–2525. doi: 10.1128/AAC.02020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kam KM, Yip CW, Cheung TL, Tang HS, Leung OC, Chan MY. 2006. Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb Drug Resist 12:7–11. doi: 10.1089/mdr.2006.12.7. [DOI] [PubMed] [Google Scholar]
- 10.Chaiprasert A, Srimuang S, Tingtoy N, Makhao N, Sirirudeeporn P, Tomnongdee N, Theankeaw O, Charoensook S, Leechawengwongs M, Prammananan T. 2014. Second-line drug susceptibilities of multidrug-resistant tuberculosis strains isolated in Thailand: an update. Int J Tuberc Lung Dis 18:961–963. doi: 10.5588/ijtld.13.0197. [DOI] [PubMed] [Google Scholar]
- 11.Prammananan T, Arjratanakool W, Chaiprasert A, Tingtoy N, Leechawengwong M, Asawapokee N, Leelarasamee A, Dhiraputra C. 2005. Second-line drug susceptibilities of Thai multidrug-resistant Mycobacterium tuberculosis isolates. Int J Tuberc Lung Dis 9:216–219. [PubMed] [Google Scholar]
- 12.Tagliani E, Cabibbe AM, Miotto P, Borroni E, Toro JC, Mansjo M, Hoffner S, Hillemann D, Zalutskaya A, Skrahina A, Cirillo DM. 2015. Diagnostic performance of the new version (v2.0) of GenoType MTBDRsl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J Clin Microbiol 53:2961–2969. doi: 10.1128/JCM.01257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambur OH, Davidsen T, Frye SA, Balasingham SV, Lagesen K, Rognes T, Tonjum T. 2009. Genome dynamics in major bacterial pathogens. FEMS Microbiol Rev 33:453–470. doi: 10.1111/j.1574-6976.2009.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantel A, Petrella S, Veziris N, Brossier F, Bastian S, Jarlier V, Mayer C, Aubry A. 2012. Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother 56:1990–1996. doi: 10.1128/AAC.06272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piton J, Petrella S, Delarue M, André-Leroux G, Jarlier V, Aubry A, Mayer C. 2010. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One 5:e12245. doi: 10.1371/journal.pone.0012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willby M, Sikes RD, Malik S, Metchock B, Posey JE. 2015. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5427–5434. doi: 10.1128/AAC.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devasia R, Blackman A, Eden S, Li H, Maruri F, Shintani A, Alexander C, Kaiga A, Stratton CW, Warkentin J, Tang YW, Sterling TR. 2012. High proportion of fluoroquinolone-resistant Mycobacterium tuberculosis isolates with novel gyrase polymorphisms and a gyrA region associated with fluoroquinolone susceptibility. J Clin Microbiol 50:1390–1396. doi: 10.1128/JCM.05286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantel A, Petrella S, Matrat S, Brossier F, Bastian S, Reitter D, Jarlier V, Mayer C, Aubry A. 2011. DNA gyrase inhibition assays are necessary to demonstrate fluoroquinolone resistance secondary to gyrB mutations in Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:4524–4529. doi: 10.1128/AAC.00707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S, Willby M, Sikes D, Tsodikov OV, Posey JE. 2012. New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS One 7:e39754. doi: 10.1371/journal.pone.0039754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard. CLSI document M24-A. Clinical and Laboratory Standards Institue, Wayne, PA. [PubMed] [Google Scholar]
- 23.WHO. 2008. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 24.WHO. 2012. Updated interim critical concentrations for first-line and second-line DST. http://www.stoptb.org/wg/gli/assets/documents/Updated%20critical%20concentration%20table_1st%20and%202nd%20line%20drugs.pdf. Accessed 6 May 2016.
- 25.van Helden PD, Victor TC, Warren RM, van Helden EG. 2001. Isolation of DNA from Mycobactetirum tuberculosis. In Parish T, Stoker NG (ed), Mycobcateirum tuberculosis protocols. Humana Press, Totowa, NJ. [Google Scholar]
- 26.Hall TA. 1999. BioEdit: a user-friendly biological seqeunce alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 27.Disratthakit A, Meada S, Prammananan T, Thaipisuttikul I, Doi N, Chaiprasert A. 2015. Genotypic diversity of multidrug-, quinolone- and extensively drug-resistant Mycobacterium tuberculosis isolates in Thailand. Infect Genet Evol 32:432–439. doi: 10.1016/j.meegid.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 28.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CLSI. 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed CLSI document M24-A2. Clinical and Laboratory Standards Institue, Wayne, PA. [PubMed] [Google Scholar]
- 30.Onodera Y, Tanaka M, Sato K. 2001. Inhibitory activity of quinolones against DNA gyrase of Mycobacterium tuberculosis. J Antimicrob Chemother 47:447–450. doi: 10.1093/jac/47.4.447. [DOI] [PubMed] [Google Scholar]
- 31.Coll F, McNerney R, Guerra-Assuncao JA, Glynn JR, Perdigao J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson KR, Tierney DB, Jeon CY, Mitnick CD, Murray MB. 2010. Treatment outcomes among patients with extensively drug-resistant tuberculosis: systematic review and meta-analysis. Clin Infect Dis 51:6–14. doi: 10.1086/653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo KW, Lee SD, Kim WS, Kim DS, Shim TS. 2014. Treatment outcomes and moxifloxacin susceptibility in ofloxacin-resistant multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 18:39–43. doi: 10.5588/ijtld.13.0307. [DOI] [PubMed] [Google Scholar]
- 35.Seung KJ, Becerra MC, Atwood SS, Alcantara F, Bonilla CA, Mitnick CD. 2014. Salvage therapy for multidrug-resistant tuberculosis. Clin Microbiol Infect 20:441–446. doi: 10.1111/1469-0691.12335. [DOI] [PubMed] [Google Scholar]
- 36.WHO. 2014. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 37.Mokrousov I, Otten T, Manicheva O, Potapova Y, Vishnevsky B, Narvskaya O, Rastogi N. 2008. Molecular characterization of ofloxacin-resistant Mycobacterium tuberculosis strains from Russia. Antimicrob Agents Chemother 52:2937–2939. doi: 10.1128/AAC.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigouts L, Coeck N, Gumusboga M, de Rijk WB, Aung KJ, Hossain MA, Fissette K, Rieder HL, Meehan CJ, de Jong BC, Van Deun A. 2016. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 71:314–323. doi: 10.1093/jac/dkv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh P, Jain A, Dixit P, Prakash S, Jaiswal I, Venkatesh V, Singh M. 2015. Prevalence of gyrA and B gene mutations in fluoroquinolone-resistant and -sensitive clinical isolates of Mycobacterium tuberculosis and their relationship with MIC of ofloxacin. J Antibiot (Tokyo) 68:63–66. doi: 10.1038/ja.2014.95. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Lu J, Wang Y, Pang Y, Zhao Y. 2014. Prevalence and molecular characterization of fluoroquinolone-resistant Mycobacterium tuberculosis isolates in China. Antimicrob Agents Chemother 58:364–369. doi: 10.1128/AAC.01228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aldred KJ, Blower TR, Kerns RJ, Berger JM, Osheroff N. 2016. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1525055113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blower TR, Williamson BH, Kerns RJ, Berger JM. 2016. Crystal structure and stability of gyrase–fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1525047113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borrell S, Teo Y, Giardina F, Streicher EM, Klopper M, Feldmann J, Müller B, Victor TC, Gagneux S. 2013. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health 2013:65–74. doi: 10.1093/emph/eot003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcusson LL, Frimodt-Møller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog 5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aubry A, Pan XS, Fisher LM, Jarlier V, Cambau E. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob Agents Chemother 48:1281–1288. doi: 10.1128/AAC.48.4.1281-1288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renau TE, Gage JW, Dever JA, Roland GE, Joannides ET, Shapiro MA, Sanchez JP, Gracheck SJ, Domagala JM, Jacobs MR, Reynolds RC. 1996. Structure-activity relationships of quinolone agents against mycobacteria: effect of structural modifications at the 8 position. Antimicrob Agents Chemother 40:2363–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renau TE, Sanchez JP, Gage JW, Dever JA, Shapiro MA, Gracheck SJ, Domagala JM. 1996. Structure-activity relationships of the quinolone antibacterials against mycobacteria: effect of structural changes at N-1 and C-7. J Med Chem 39:729–735. doi: 10.1021/jm9507082. [DOI] [PubMed] [Google Scholar]
- 48.Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, Palomino JC, da Silva PA. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 53:4498–4500. doi: 10.1128/AAC.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Regmi SM, Chaiprasert A, Coker OO, Disratthakit A, Prammananan T, Suriyaphol P, Yik-Ying T, Twee Hee O. 2015. Draft genome sequence of an extensively drug-resistant Mycobacterium tuberculosis Manu-Ancestor Spoligo-International Type 523 isolate from Thailand. Genome Announcements 3:e01589–14. doi: 10.1128/genomeA.01589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Outhred AC, Jelfs P, Suliman B, Hill-Cawthorne GA, Crawford ABH, Marais BJ, Sintchenko V. 2015. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother 70:1198–1202. doi: 10.1093/jac/dku508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witney AA, Gould KA, Arnold A, Coleman D, Delgado R, Dhillon J, Pond MJ, Pope CF, Planche TD, Stoker NG, Cosgrove CA, Butcher PD, Harrison TS, Hinds J. 2015. Clinical application of whole-genome sequencing to inform treatment for multidrug-resistant tuberculosis cases. J Clin Microbiol 53:1473–1483. doi: 10.1128/JCM.02993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]