Abstract
As we face an alarming increase in bacterial resistance to current antibacterial chemotherapeutics, expanding the available therapeutic arsenal in the fight against resistant bacterial pathogens causing respiratory tract infections is of high importance. The antibacterial potency of macrolones, a novel class of macrolide antibiotics, against key respiratory pathogens was evaluated in vitro and in vivo. MIC values against Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Haemophilus influenzae strains sensitive to macrolide antibiotics and with defined macrolide resistance mechanisms were determined. The propensity of macrolones to induce the expression of inducible erm genes was tested by the triple-disk method and incubation in the presence of subinhibitory concentrations of compounds. In vivo efficacy was assessed in a murine model of S. pneumoniae-induced pneumonia, and pharmacokinetic (PK) profiles in mice were determined. The in vitro antibacterial profiles of macrolones were superior to those of marketed macrolide antibiotics, including the ketolide telithromycin, and the compounds did not induce the expression of inducible erm genes. They acted as typical protein synthesis inhibitors in an Escherichia coli transcription/translation assay. Macrolones were characterized by low to moderate systemic clearance, a large volume of distribution, a long half-life, and low oral bioavailability. They were highly efficacious in a murine model of pneumonia after intraperitoneal application even against an S. pneumoniae strain with constitutive resistance to macrolide-lincosamide-streptogramin B antibiotics. Macrolones are the class of macrolide antibiotics with an outstanding antibacterial profile and reasonable PK parameters resulting in good in vivo efficacy.
INTRODUCTION
Respiratory tract infections (RTI), like acute otitis media, acute sinusitis, and community-acquired pneumonia (CAP), are among the most common illnesses throughout the world, leading to high direct and indirect costs of care (estimated at over $40 billion for CAP alone [1]), absence from work, decreased productivity, and high levels of antibiotic consumption (2, 3). Lower RTI remain the fourth leading cause of death worldwide, and pneumonia led to more than 1.3 million deaths in children under 5 years of age in 2011 (4). In the majority of studies, in particular, in developed Western countries, Streptococcus pneumoniae is the most prevalent pathogen causing CAP, regardless of the severity of illness, demographic considerations, and underlying comorbidities (1, 5 – 7). In the Asia-Pacific region, however, other pathogens, like the Gram-negative organism Klebsiella pneumoniae, are more prevalent, but their prevalence has high intraregional variability (8).
The widespread antimicrobial resistance and dissemination of multidrug-resistant strains are of great concern and are among the major challenges in health care (9). Pneumococcal resistance patterns differ significantly among countries, particularly in terms of resistance to macrolide antibiotics. Erythromycin resistance is predominantly mediated by two mechanisms: ribosome modification by methylation, resulting in a phenotype of inducible resistance to macrolide-lincosamide-streptogramin B (iMLSb) or constitutive resistance to macrolide-lincosamide-streptogramin B (cMLSb), and the production of efflux pumps, resulting in the M phenotype. In addition, pneumococcal isolates with mutations in 23S rRNA domains II and V and ribosomal proteins L4 and L22 have been reported, but their clinical significance is still rather low (10 – 13). In Streptococcus pyogenes, inducible erm gene expression has three distinct patterns with different levels of resistance to macrolides with a 14-, 15-, or 16-membered macrolactone ring (14).
There is a major difference in erythromycin resistance patterns among isolates from the United States, Europe, and Asia. In the United States, macrolide efflux is the most common resistance mechanism, while the presence of the erm(B) gene prevails in Europe and some Asian countries. The highest rate of occurrence of macrolide resistance, exceeding 70%, was observed in Taiwan, South Korea, China, and Hong Kong (15 – 18). In a recent study analyzing pneumococcal isolates from children hospitalized for RTI in China, virtually all isolates were erythromycin resistant, further emphasizing the role of the dissemination of successful, highly resistant international clones (19). Resistance to macrolide antibiotics among S. pyogenes isolates from patients with invasive disease is also on the rise; for example, in Japan the rate of resistance almost doubled from 2006 to 2012, when the rate of resistance exceeded 50% (20).
As the incidence of infections caused by bacteria resistant to one or more antibiotic classes continually increases, there is a permanent need for the development of new, more effective, and safe antimicrobial drugs. However, drug discovery is a costly, time-consuming process with a very high attrition rate, and major players in the pharmaceutical industry have been reluctant to pursue research in the anti-infective area. This is reflected in the dwindling number of new antibacterials approved for use in humans, the number of which continually declined from 16 in the mid-1980s to just 2 in the period between 2002 and 2010 (21). The antibacterials currently in development pipelines consist almost entirely of compounds belonging to well-established classes of molecules with known mechanisms of action (22 – 24), with the exception of two GlaxoSmithKline (GSK) compounds, a peptide deformylase inhibitor for CAP and skin and soft tissue infections (SSTI), which is in phase II clinical studies, and a leucyl-tRNA synthase inhibitor for Gram-negative bacterial infections (25 – 27).
Owing to issues related to the global burden of antibiotic resistance, our research efforts were directed toward the discovery of novel macrolides to be developed as oral (p.o.) antibiotics for treating community-acquired bacterial infections, particularly those of the respiratory tract. The targeted profile encompassed activity against key RTI bacterial pathogens (multidrug-resistant S. pneumoniae [MDRSP], Haemophilus influenzae, Moraxella catarrhalis, Chlamydia pneumoniae, Legionella pneumophila, Mycoplasma pneumoniae, Staphylococcus aureus) and efficacy and safety comparable to those of azithromycin. Various chemical approaches have been employed, including the conjugation of conventional macrolides with different ligand groups in order to modify ribosome binding and overcome the resistance caused by methylation of the macrolide binding site. The targeted profile was achieved by the development of and iterative improvements to the novel class of macrolide compounds named macrolones (28 – 34). The improved antibacterial activity of a series of 4″-O-acyl-8a-lactame (8-a-lactame) analogues containing aromatic substituents (a 4-nitrophenyl, pyridyl, or quinolyl group) (35) prompted further exploration of the 4″ position on the cladinose sugar of azalides and other scaffolds. The major breakthrough was achieved by combining the macrolide scaffold with the quinolone moiety, and the first analogue, a conjugate of 8-a-lactame and ciprofloxacin, showed enhanced activity against resistant strains of S. pneumoniae and S. pyogenes (30). Additional modifications further advanced the antibacterial potency, and excellent activities superior to those of telithromycin against a variety of macrolide-susceptible and -resistant bacterial isolates were obtained. During the lead optimization process, a comprehensive study of the structure-activity relationship (SAR) was performed. Derivatization was broadened using other macrocyclic cores (including clarithromycin and azithromycin), linkers of various lengths, various types and numbers of heteroatoms (O or N), different bonds to the scaffold (i.e., ester, ether) position of the linker on the quinolone unit, and different quinolone moieties (28 – 34).
In the study described in this paper, the activities of representative macrolones were analyzed in more detail. The macrolones selected for further in vivo evaluation were characterized by a quinolone moiety (comprising a cyclopropyl at the N-1 position and a chlorine atom at the C-7 position) attached by an ester bond via different linkers to the 4″ position of the azithromycin scaffold. The in vivo efficacies of these compounds in a mouse model of pneumonia induced by either erythromycin-sensitive (Erys) strains or cMLSb strains of S. pneumoniae, as well as pharmacokinetics (PK) in mice, are discussed.
(The results described in this report were presented in part at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Denver, CO, USA, in 2013.)
MATERIALS AND METHODS
Compounds.
All compounds used were synthesized at the Pliva Research Institute (later, the GSK Research Centre Zagreb), and their synthesis and primary antibacterial profiles have been published in part elsewhere (28, 29, 36 – 38). The structures of all tested compounds and their respective numbers in patents and previous publications are given in Fig. S1 to S4 in the supplemental material for this article.
In vitro antibacterial activity.
The organisms tested represent relevant Gram-positive bacterial (S. pneumoniae, S. pyogenes, and S. aureus) and Gram-negative bacterial (H. influenzae) respiratory tract pathogens and were either sensitive or resistant to macrolide antibiotics. Macrolide resistance was due to two major mechanisms: the production of efflux pumps (M phenotype) or ribosome modification by methylation. Methylase expression was inducible (iMLSb phenotype) or constitutive (cMLSb phenotype). Strains were grouped according to species and resistance mechanism, with each group comprising at least 10 isolates to allow statistical relevance and for MIC90 determination. Additional epidemiologically relevant panels of 37 H. influenzae and 73 S. pneumoniae RTI-related isolates from Croatia were used for the broader profiling of compound 19.
MICs were determined by the broth microdilution method described by CLSI guidelines (39), except that in the medium used to grow Streptococcus strains, lysed blood was replaced by 5% horse serum. The compounds were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 5 mg/ml. Azithromycin and telithromycin were used as controls. Dilutions of the tested compounds in Mueller-Hinton broth were prepared using a Tecan Genesis RSP 150 liquid handling system. Bacteria were grown on appropriate agar plates (Becton Dickinson, USA): Columbia agar with 5% sheep blood for streptococci and M. catarrhalis, chocolate agar for H. influenzae, and Mueller-Hinton agar for staphylococci.
Induction studies.
The ability of compounds to induce the expression of erm genes in strains woth iMLSb was checked by the triple-disk method (40) on five isolates with different induction phenotypes and genotypes: three S. pyogenes isolates [iMLSb-A with erm(B) and iMLSb-B and iMLSb-C, both of which had the erm(A) gene] and two S. aureus isolates [which had the erm(C) and erm(B) genes, respectively]. The test was performed by placing a disk with 0.5 μg of the tested compound between clindamycin (2 μg) and rokitamycin (3 μg) disks, approximately 2 cm apart, on plates (Columbia agar with 5% sheep blood for S. pyogenes and Mueller-Hinton agar for S. aureus), and the plates were inoculated by evenly spreading the bacterial suspension. After incubation for 20 h at 37°C, a D-shaped zone toward the clindamycin or rokitamycin disk indicated the induction of gene expression by the tested compound, while round, regular zones signified a lack of induction.
To assess the influence of incubation in the presence of subinhibitory concentrations of the compounds on erm expression, bacteria were preincubated with low concentrations of the compounds for 2.5 h and washed, and their susceptibility was tested by a standard broth microdilution method according to CLSI guidelines (41), except that for S. pyogenes lysed blood was replaced by horse serum. Bacterial suspensions with an optical density of a 0.5 McFarland standard were prepared by direct suspension of colonies from fresh plates, and the suspended colonies were diluted in a 1:10 ratio in the appropriate medium (as described above) containing compounds at subinhibitory concentrations (1 to 0.001 μg/ml, depending on the compound and the bacterial strain). After 2.5 h at 37°C, inducers were removed by centrifugation at 2,500 × g, bacteria were resuspended in fresh medium, and MICs were determined.
In vitro E. coli TnT assay.
The materials used in the in vitro E. coli transcription/translation (TnT) assay were part of an in vitro protein synthesis kit (E. coli S30 extract system for circular DNA; catalog number L 1020; Promega, USA). This kit contains all components needed for the efficient transcription and translation of a DNA template provided by the user. When the assay is employed for measurement of protein synthesis inhibition, plasmid pBESTluc, containing the firefly luciferase gene, is used as the DNA template. The intensity of the luminescence produced by the expression of luciferase correlates with the efficiency of protein synthesis and was quantified in the presence or absence of the tested compounds. The compounds were tested at five concentrations (10, 1, 0.5, 0.1, and 0.01 μM), and 50% inhibitory concentration (IC50) values were calculated. This method, however, does not discriminate transcription from translation inhibitors.
In vivo studies. (i) Animals.
Male C57BL/6J mice (Charles River, France) weighing 20 to 24 g were used for the pneumonia model. Pharmacokinetic studies were performed with male BALB/c mice weighing 20 to 24 g. Mice were kept on metal grids (6 mice per cage), maintained on a 12-h light/12-h dark cycle, and allowed free access to food (Mucedola, Italy) and tap water. All experimental animals were handled in accordance with the guidelines of the GlaxoSmithKline animal research ethics committee and were approved by the Croatian local government authorities.
(ii) Sample preparation and bioanalysis.
Hemolyzed blood samples (25 μl) in Eppendorf test tubes were treated by protein precipitation with the addition of 2 volumes of a mixture of acetonitrile-methanol (1:2) containing an internal standard (roxithromycin). A 1-mg/ml stock solution of each compound was prepared in DMSO, diluted in water, and spiked into blank rat blood to prepare duplicate standards with concentrations ranging from 5 to 25,000 ng/ml. One set of standards was analyzed at the beginning and one set of standards was analyzed at the end of testing of each sample batch. Quality control samples were prepared from separate stock solutions and analyzed at three concentrations (low, medium, high). The mixtures were centrifuged at 1,500 × g at 4°C for 10 min, and aliquots (0.01 ml) of the resulting supernatant fractions were transferred to a 96-well plate. Samples were analyzed with either a Sciex API 3000 or a Sciex API 2000 triple-quadrupole mass spectrometer (Sciex, Division of MDS Inc., Toronto, Ontario, Canada) coupled to a Hewlett-Packard high-pressure liquid chromatography (HPLC) system (HP1100; Hewlett-Packard) and a high-throughput screening HTS PAL CTC autosampler (CTC Analytics AG). Samples (5 μl) were injected onto an HPLC column [particle size, 3 μm; 2.0 by 50 mm; Phenomenex Luna C18(2) column] and eluted with a gradient at room temperature. The chromatographic conditions consisted of mobile phase A (1,000:1, vol/vol, acetonitrile and formic acid) and mobile phase B (1,000:1, vol/vol, water and formic acid), which were run over a 6-min gradient at a flow rate of 0.3 ml/min. A positive-ion mode with turbo spray, an ion source temperature of 400 to 450°C, and a dwell time of 300 to 400 ms were utilized for mass spectrometric detection. Quantitation was performed using multiple-reaction monitoring (MRM) at the specific transition for each compound. Linear regression plots of the compound-to-internal standard peak area ratio versus drug concentration were derived with 1/x or 1/x2 weighting. The dynamic range for the blood assay ranged from 0.005 μg/ml to 10 μg/ml. The accuracy for the quality control (QC) samples used during the analysis was below 24% (judged by the percent deviation from the nominal value) for at least two of three concentrations, and the precision was ≤20% (as judged by the relative standard deviation [SD]).
(iii) Pharmacokinetic analysis.
Noncompartmental analysis of all compounds was performed using WinNonlin Professional (version 4.0.1) software (Pharsight, Mountain View, CA). Individual blood concentrations and sample times for each animal were used in the analysis. Following administration of an intravenous (i.v.) bolus, the terminal elimination half-life (t1/2), the total area under the curve (AUC), the area under the concentration-time curve extrapolated to infinity, systemic blood clearance, and the steady-state volume of distribution were calculated by standard methods. After oral administration, the peak (maximum) concentration in plasma (Cmax) and the time to Cmax (Tmax) were taken directly from the individual profiles, and the areas under the curve and oral bioavailability were determined. Summarized pharmacokinetic parameters are reported as mean values ± SDs after both intravenous and oral administration.
(iv) In vivo pneumococcal pneumonia model in mice.
S. pneumoniae SP030 was used to induce pneumonia and was a clinical isolate provided by GSK, Verona, Italy. Erythromycin-resistant S. pneumoniae 1217 is a clinical isolate as well. The virulence was maintained by monthly intraperitoneal (i.p.) passage of the culture in C57BL/6J mice. Two mice were infected i.p. with 0.5 ml of an overnight culture grown in Todd-Hewitt broth (Difco). After 8 h, when the animals showed signs of illness, they were anesthetized using thiopental (Tiopental; PLIVA, Zagreb, Croatia) by the i.p. route, and the hearts were removed aseptically. Several drops of blood from the heart were added to a tube containing Todd-Hewitt broth medium and were also streaked on blood agar plates (Merck, Germany) for confirmation of the purity and identity of the inoculum. After overnight incubation under 5% CO2, the pure cultures of the bacteria (Todd-Hewitt broth; Difco) were stored in cryobanks (Mast Diagnostic, United Kingdom) at −80°C.
The bacteria used for infection were prepared by direct suspension of colonies from an overnight blood agar culture (Merck) in sterile tubes containing 5 ml of Todd-Hewitt broth (Difco). The suspension was adjusted to a turbidity equivalent to a 1 McFarland standard (bioMérieux, France), and after 24 h of incubation (under 5% CO2 at 37°C) the suspension was centrifuged at 1,500 rpm for 5 min and washed in sterile 0.9% (wt/vol) NaCl solution (Pliva), before being resuspended in sterile phosphate-buffered saline (PBS; pH 7.4; Sigma). In order to induce infection, approximately 106 CFU bacteria in logarithmic growth phase were used.
(v) Induction of pneumonia and bacterial counting.
After i.p. anesthesia with ketamine (Narketan, Vetoqinol)-xylazine (Rompun; Bayer), mice were infected by intranasal (i.n.) inoculation of 50 μl of PBS containing S. pneumoniae SP030, placed into cages, and allowed to awaken. The mice were observed for clinical signs of morbidity throughout the experiment. Compounds were given by the p.o. or i.p. route starting at 6 h postinfection (p.i.). Two dosing regimens (schedules) were applied, and doses of 10, 20, and 50 mg/kg of body weight each were administered twice (at 6 and 30 h postinfection, corresponding to once-daily dosing) or three times (at 6, 18, and 30 h postinfection, equaling a twice-daily regimen). Animals were sacrificed 44 h p.i. (14 h after the last dose), and the lungs were aseptically removed, washed in sterile PBS, pH 7.4 (to remove blood residues), and placed in a sterile petri dish (Falcon). The weight of the lungs of every sacrificed mouse was noted. Subsequently, the lungs were placed in Kartell test tubes containing sterile PBS (1:9, wt/vol) and homogenized with an Ultra Turrax homogenizer (Janke and Kunkel, IKA-Labortechnich, Germany). The homogenates were serially diluted, and 10-μl volumes were plated in triplicate onto blood agar (Merck). The plates were incubated at 37°C in 5% CO2 overnight to determine the number of S. pneumoniae bacteria by counting of the numbers of visible colonies. The limit of detection was 2 log10 CFU/ml of lung homogenates.
(vi) Determination of compound concentration in lung.
The concentrations of the compounds in lung homogenates were determined by a bioassay method, and Kocuria rhizophila Kovacs et al. ATCC 9341 (previously designated Sarcina lutea) was used as the test microorganism. An overnight culture of K. rhizophila bacteria was thoroughly mixed with warm (50°C) antibiotic agar no. 11 (Merck) in a 1:4 ratio and poured onto square bioassay plates. After the bacterium-agar mixture cooled, wells with an 8.5-mm diameter and a 4-mm depth were bored in the agar and filled in duplicate with 50 μl of lung homogenate or with 50 μl of the tested compounds diluted in phosphate buffer. For the standard curve, the compounds were loaded at concentrations of 0.1, 0.5, 1, 1.5, 2, 2.5, and 3 μg/ml. After the plates were allowed to cool for 30 min at 4°C, they were incubated at 37°C for 18 to 20 h. The concentration of compound in the lung homogenate was determined by comparison of the inhibition zones with the standard curve and is reported as micrograms of compound per gram of lung tissue.
(vii) Data analysis.
All the data for bacterial counts from the model of in vivo efficacy against pneumonia are presented as the median ± SD. The statistical significance of the number of CFU in the lungs of infected animals treated with the test compounds was assessed by the one-way analysis of variance (ANOVA) method (Kruskal-Wallis test) and Dunn's group comparison test using GraphPad Prism (version 4.03) software, and a P value of <0.05 was considered statistically significant.
RESULTS
Antibacterial activity.
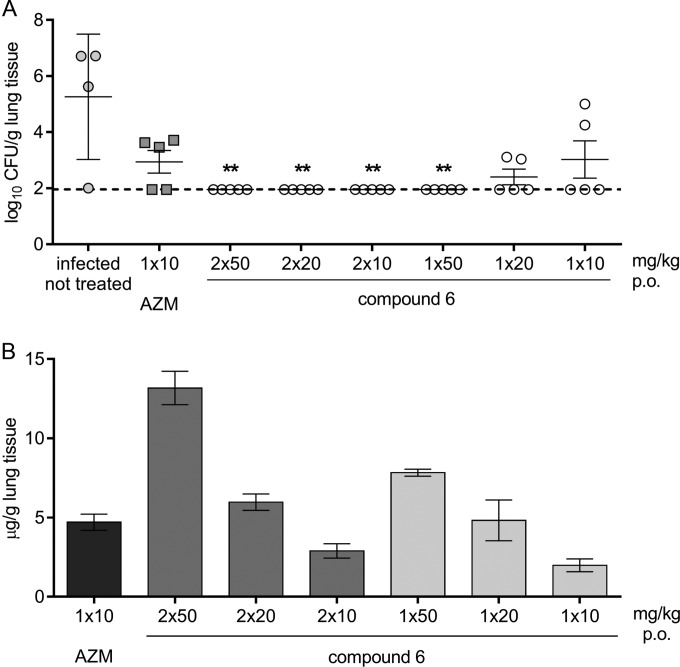
This paper describes 4″ ester derivatives of macrolides with a quinolone moiety that can be represented by the general formula given in Fig. 1.
FIG 1.
General formula for the macrolone compounds. Structures vary with respect to the macrolide scaffold (erythromycin-derived 8-a-lactame, erythromycin oxime, clarithromycin, clarithromycin carbamate, azithromycin, and roxithromycin), linker length, the type and number of heteroatoms (O or N) in the linker, and the position of the linker on the quinolone unit. X, O, N or OCH2CH2; Y, O or N; Z, halogen atom (Cl or F); R1, methyl ester of quinolone carboxylic acid.
Macrolones were divided into subclasses according to the macrolide scaffold (azithromycin, clarithromycin, clarithromycin carbamate, 8-a-lactame, erythromycin oxime, and roxithromycin), linker length, heteroatom composition, and the influence of these structural features on the antibacterial properties of the compounds was examined. Compared to the “gold standard” of macrolide therapy, azithromycin, all tested compounds showed significant potency against relevant bacterial respiratory tract pathogens. In Table 1, strains are grouped according to species and the macrolide resistance mechanism, and the compounds' MIC90 values are presented.
TABLE 1.
Antibacterial activities of novel macrolone compounds against key respiratory pathogens, grouped by species and resistance mechanisma
| Compound (compound no.) | MIC90 (μg/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. pneumoniae |
S. pyogenes |
S. aureus |
H. influenzae (n = 12) | ||||||||
| Erys (n = 10) | M (n = 13) | MLSb (n = 13) | Erys (n = 10) | M (n = 12) | iMLSb (n = 14) | cMLbS (n = 13) | Erys (n = 11) | M (n = 10) | iMLSb (n = 12) | ||
| Azithromycin | 0.03 | 8 | >64 | 0.06 | 4 | >64 | >64 | 1 | >64 | >64 | 2 |
| Clarithromycin | 0.03 | 8 | >64 | ≤0.015 | 4 | >64 | >64 | 0.125 | 32 | >64 | 16 |
| Telithromycin | ≤0.015 | 0.5 | 0.5 | ≤0.015 | 0.5 | 4 | 32 | 0.06 | 0.25 | 0.06 | 2 |
| Clindamycin | ≤0.015 | 0.03 | >64 | 0.03 | 0.03 | 0.3 | >64 | 0.06 | 0.125 | 0.125 | 32 |
| Ciprofloxacin | 0.5 | 1 | 1 | 0.25 | 0.5 | 1 | 0.5 | >64 | 0.5 | >64 | ≤0.015 |
| azi-NN-Cl (1) | 0.125 | 0.5 | 0.125 | 0.06 | 1 | 0.25 | 0.5 | 2 | 1 | 2 | 8 |
| azi-NMeN-Cl (2) | ≤0.015 | ≤0.015 | 0.25 | ≤0.015 | 0.25 | 0.125 | 1 | 1 | 2 | 2 | 8 |
| azi-NmeN (3) | ≤0.015 | 0.03 | 0.03 | ≤0.015 | 0.25 | 0.25 | 2 | 2 | 2 | 2 | 8 |
| azi-NEtN-Cl (4) | 0.03 | 1 | 1 | 0.06 | 2 | 0.25 | 2 | 1 | 4 | 4 | 32 |
| azi-NN-Cl-Qme (5) | ≤0.015 | 0.06 | 0.06 | 0.03 | 0.5 | 0.25 | 0.25 | 1 | >8 | 64 | 16 |
| azi-NMeN-Cl-Qme (6) | ≤0.015 | 0.06 | 0.06 | 0.06 | 0.03 | 0.25 | 1 | 4 | 16 | 32 | 16 |
| azi-ON-Cl (7) | ≤0.015 | ≤0.015 | 0.125 | ≤0.015 | 0.03 | ≤0.015 | 0.25 | 0.5 | 0.5 | 0.5 | 2 |
| azi-ON (8) | ≤0.015 | 0.06 | 1 | ≤0.015 | 0.06 | 0.03 | 1 | 0.5 | 1 | 1 | 2 |
| clari-ON-Cl (9) | ≤0.015 | ≤0.015 | 0.25 | ≤0.015 | ≤0.015 | 0.03 | 0.5 | 0.5 | 0.5 | 1 | 4 |
| clari-ON (10) | ≤0.015 | ≤0.015 | 4 | ≤0.015 | 0.25 | 0.06 | 2 | 4 | 0.5 | 2 | 8 |
| claric-ON-Cl (11) | ≤0.015 | ≤0.015 | 0.25 | ≤0.015 | ≤0.015 | 0.06 | 0.5 | 0.5 | 1 | 2 | 4 |
| claric-ON (12) | ≤0.015 | 0.03 | 0.125 | ≤0.015 | ≤0.015 | 0.06 | 0.25 | 0.5 | 1 | 2 | 2 |
| azi-OO-Cl (13) | ≤0.015 | ≤0.015 | 0.5 | ≤0.015 | 0.03 | ≤0.015 | 1 | 0.5 | 1 | 0.5 | 2 |
| azi-OO (14) | ≤0.015 | 0.03 | 0.25 | ≤0.015 | 0.06 | ≤0.015 | 1 | 0.25 | 0.5 | 1 | 2 |
| clari-OO-Cl (15) | ≤0.015 | 0.06 | 1 | ≤0.015 | 0.06 | 0.06 | 2 | 0.5 | 0.5 | 1 | 8 |
| clari-OO (16) | ≤0.015 | ≤0.015 | 0.5 | ≤0.015 | ≤0.015 | ≤0.015 | 1 | 1 | 0.25 | 4 | 8 |
| claric-OO-Cl (17) | ≤0.015 | ≤0.015 | 0.25 | ≤0.015 | ≤0.015 | 0.03 | 0.5 | 1 | 1 | 1 | 8 |
| claric-OO (18) | ≤0.015 | 0.03 | 0.25 | ≤0.015 | 0.125 | 0.125 | 1 | 0.5 | 1 | 4 | 4 |
| azi-OON-Cl (19) | ≤0.015 | ≤0.015 | 0.25 | ≤0.015 | ≤0.015 | ≤0.015 | 0.5 | 0.5 | 0.5 | 0.5 | 2 |
| azi-OON (20) | ≤0.015 | 0.03 | 0.5 | ≤0.015 | 0.06 | ≤0.015 | 0.25 | 0.25 | 0.25 | 0.25 | 8 |
| azi-OON-F (21) | ≤0.015 | 0.03 | 1 | 0.03 | 0.25 | 0.06 | 4 | 0.5 | 0.5 | 1 | 32 |
| clari-OON-Cl (22) | ≤0.015 | 0.06 | 4 | 0.03 | 0.125 | 2 | 2 | 1 | 2 | 8 | 8 |
| claric-OON-Cl (23) | ≤0.015 | ≤0.015 | 0.125 | ≤0.015 | ≤0.03 | ≤0.015 | 0.25 | 0.25 | 0.5 | 1 | 4 |
| 8a-OON-Cl (24) | ≤0.015 | ≤0.015 | 0.125 | ≤0.015 | 0.03 | 0.03 | 0.5 | 0.25 | 0.25 | 0.25 | 4 |
| eox-OON-Cl (25) | ≤0.015 | ≤0.015 | 0.5 | ≤0.015 | ≤0.015 | ≤0.015 | 0.5 | 0.25 | 0.25 | 0.5 | 2 |
| rox-OON-Cl (26) | 0.06 | 0.125 | 2 | 0.06 | 0.06 | 0.25 | 2 | 0.5 | 1 | 4 | 4 |
The structures of all compounds are provided in the supplemental material. n, number of isolates tested; Erys, erythromycin susceptible; iMLSb, inducible resistance to macrolide, lincosamide, and streptogramin B antibiotics; cMLSb, constitutive resistance to macrolide, lincosamide, and streptogramin B antibiotics; M, efflux-mediated macrolide. Boldface data represent compounds that were evaluated in vivo. The abbreviations of the compounds represent the scaffold heteroatom composition of the linker and the presence of the halogen atom at the quinolone moiety. Macrolide scaffolds are as follows: azi, azithromycin; clari, clarithromycin; claric, clarithromycin carbamate; 8-a, 8-a-lactame; eox, erythromycin oxime; rox, roxithromycin.
In addition to erythromycin-sensitive isolates of S. pneumoniae, S. pyogenes, and S. aureus, the tested macrolones showed potency against isolates with different erythromycin resistance mechanisms. Despite their exceptional activity against streptococci, compounds with two nitrogen atoms in the linker (compounds 1 to 6) still did not show sufficient potency against H. influenzae, as compound 1 (azi-NN-Cl) exhibited an MIC90 of 8 μg/ml. Potency in the range of that of azithromycin and telithromycin, i.e., 2 μg/ml, is targeted. Methylation of the nitrogen atom in the linker did not impair the potency of compounds 2 and 3, but ethylation (compound 4) resulted in decreased activity against H. influenzae and a slightly lower efficiency against S. aureus strains with the iMLSb and M phenotypes. Esterification of 3-quinolone carboxylic acid (compound 6 [azi-NMeN-Cl-QMe, where Me represents methyl]) further diminished the potency against H. influenzae and S. aureus, regardless of the resistance phenotype, while the compound retained high potency against streptococci.
Introduction of one oxygen atom (derivatives 7 to 12) or two oxygen atoms (compounds 13 to 18) into the linker enhanced the potency against both H. influenzae and S. aureus. Compounds on the azithromycin scaffold (compounds 7 and 13) had MIC90 values of 2 μg/ml against H. influenzae; at the same time, they maintained high potency against all resistant S. pneumoniae and S. pyogenes strains with all resistance phenotypes tested, including S. pyogenes strains with cMLSb. Removal of the halogen atom at position 7 of the quinolone moiety (compounds 8 and 14) did not influence the potency against H. influenzae; however, activity against S. pneumoniae erm(B) strains was somewhat decreased (MIC90s, 0.125 μg/ml and 1 μg/ml for compound 7 and its dehalogenated analogue 8, respectively). Derivatives on a 14-membered clarithromycin scaffold were less active against H. influenzae (MIC90, 8 μg/ml), and removal of the halogen reduced the potency against S. aureus with iMLSb. Compound 12, a clarithromycin 11,12-carbamate derivative (claric-ON), had good overall potency. Nevertheless, in both compound groups with NO and OO linkers and halogenated derivatives on an azithromycin scaffold, compounds 7 and 13, were selected as the most potent.
Activity against S. aureus improved for compounds with an elongated OON linker (compounds 19 to 26). The MIC90 of compound 19 (azi-OON-Cl) was 0.5 μg/ml against strains both with the M phenotype and those with the iMLSb phenotype, as well as against Erys isolates. Simultaneously, this compound maintained excellent potency against streptococci and showed an MIC90 of 2 μg/ml against H. influenzae. In contrast to azithromycin derivatives with shorter linkers, dehalogenation of compounds 19 and 20 reduced their effectiveness against H. influenzae. When the attachment position of the linker to the quinolone subunit was changed from position 6 to position 7, in which case there was a fluorine atom at position 6 (compound 21, azi-OON-F), the potency against H. influenzae decreased dramatically (MIC90, 32 μg/ml). As compound 19 showed the most favorable antibacterial profile, the influence of the macrolide core within the molecule on antibacterial activity was investigated by the use of various scaffolds and retention of the same linker quinolone moiety. Among the macrolide scaffolds profiled, there were clarithromycin, clarithromycin 11,12-carbamate, 8-a-aza-8a-homoerithromycin (8-a-lactame), erythromycin oxime, and roxithromycin. The least potent molecule was a clarithromycin derivative, compound 22, while an erythromycin oxime derivative, compound 25, had potency similar to that of the azithromycin analogue compound 19.
Overall, it was shown that the azithromycin core, the presence of oxygen in the linker, and the presence of a free carboxylic acid on the quinolone contribute positively to antibacterial activity.
The antibacterial activity of compound 19 against recent clinical isolates of RTI-related S. pneumoniae (n = 73) (Table 2) and H. influenzae (n = 37) (Table 3) was profiled further. Among the pneumococci, the erm(B) gene was confirmed to be present in 21 isolates, the mef gene was confirmed to be present in 11 isolates, and 1 isolate had both the erm(B) gene and the mef gene, while 40 isolates were erythromycin susceptible. As this is an epidemiologically relevant set of isolates, cumulative results are presented independently of the macrolide resistance phenotypes or incidence. Compound 19 was highly potent against all tested S. pneumoniae isolates, with the MIC90 being ≤0.015 μg/ml, and the telithromycin MIC90 was 0.125 μg/ml. Regarding H. influenzae, azithromycin and telithromycin were more active against a given set of isolates, with the MIC90 being 2 μg/ml, whereas the MIC90 of compound 19 was 4 μg/ml.
TABLE 2.
Antibacterial activity of compound 19 (azi-OON-Cl) and standard antibiotics against recent RTI-related clinical isolates of S. pneumoniae collected in Croatiaa
| Compound | MIC (μg/ml) |
||
|---|---|---|---|
| 50% | 90% | Range | |
| 19 (azi-OON-Cl) | ≤0.015 | ≤0.015 | ≤0.015–0.125 |
| Telithromycin | ≤0.015 | 0.125 | ≤0.015–0.5 |
| Azithromycin | ≤0.125 | >64 | ≤0.125–>64 |
| Clindamycin | ≤0.125 | >64 | ≤0.125–>64 |
| Tetracycline | ≤0.125 | 32 | ≤0.125–>64 |
| Penicillin G | ≤0.015 | 2 | ≤0.015–8 |
| Cefotaxime | ≤0.015 | 1 | ≤0.015–8 |
| Moxifloxacin | 0.125 | 0.125 | 0.06–0.25 |
Data are for 73 S. pneumoniae isolates.
TABLE 3.
Antibacterial activity of compound 19 (azi-OON-Cl) and standard antibiotics against recent RTI-related clinical isolates of H. influenzae collected in Croatiaa
| Compound | MIC (μg/ml) |
|||
|---|---|---|---|---|
| 50% | 90% | Range | Geometric mean | |
| 19 (azi-OON-Cl) | 2 | 4 | 2–4 | 2.38 |
| Telithromycin | 2 | 2 | 1–4 | 1.51 |
| Azithromycin | 1 | 2 | 0.5–2 | 1.16 |
| Clarithromycin | 8 | 8 | 2–16 | 5.76 |
| Clindamycin | 8 | 16 | 2–>16 | 6.89 |
| Ampicillin | 0.125 | 1 | 0.125–>16 | |
| Cefotaxime | ≤0.03 | ≤0.03 | ≤0.03–0.125 | |
| Moxifloxacin | ≤0.03 | ≤0.03 | ≤0.03 | |
Data are for 37 H. influenzae isolates.
Among the S. aureus strains tested, two Erys and seven iMLSb isolates were resistant to quinolone antibiotics, but representative macrolones demonstrated very high potencies against these isolates (Table 4).
TABLE 4.
Activity of macrolones against quinolone-resistant S. aureus isolatesa
| Phenotype | Isolate | Genotype | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound 1 | Compound 7 | Compound 13 | Compound 19 | CIP | AZM | TEL | |||
| Erys | B 0935 | 2 | 0.25 | 0.25 | 0.125 | >64 | 0.5 | 0.06 | |
| Erys | B 0936 | 2 | 0.25 | 0.125 | 0.25 | >64 | 0.5 | 0.06 | |
| iMLSb | B 0940 | erm(C) | 1 | 0.25 | 0.25 | 0.5 | >64 | >64 | ≤0.015 |
| iMLSb | B 0941 | erm(A) | 2 | 0.25 | 0.25 | 0.5 | >64 | >64 | 0.06 |
| iMLSb | B 0942 | erm(C) | 2 | 0.125 | 0.25 | 0.5 | 32 | >64 | 0.06 |
| iMLSb | B 0943 | erm(A) | 2 | 0.25 | 0.25 | 0.5 | 8 | >64 | 0.03 |
| iMLSb | B 0944 | erm(A) | 2 | 0.5 | 0.5 | 0.5 | 32 | >64 | 0.03 |
| iMLSb | B 0950 | erm(A) | 2 | 0.25 | 0.5 | 0.5 | 16 | >64 | 0.03 |
| iMLSb | B 0538 | erm(C) | 2 | 0.25 | 0.25 | 0.5 | 32 | >64 | 0.03 |
The majority of isolates showed inducible resistance to macrolides as well. Erys, erythromycin susceptible; iMLSb, inducible resistance to macrolide, lincosamide and streptogramin B antibiotics; CIP, ciprofloxacin; AZM, azithromycin; TEL, telithromycin.
As macrolone compounds possess a quinolone moiety attached to a macrolide core, it was important to assess whether they are inhibitors of protein synthesis. Therefore, their efficiency in an E. coli in vitro transcription/translation assay was determined. Macrolones acted as typical protein synthesis inhibitors, with IC50s being in the higher nanomolar range (Table 5). Conversely, ciprofloxacin was inactive in this assay, with an IC50 of >50 μM.
TABLE 5.
Inhibition of prokaryotic protein synthesis, determined by E. coli transcription/translation assay
| Compound (compound no.) | IC50 (μM) |
|---|---|
| azi-NN-Cl (1) | 0.28 |
| azi-ON-Cl (7) | 0.31 |
| azi-ON (8) | 0.49 |
| clari-ON-Cl (9) | 0.86 |
| claric-NO-Cl (11) | 0.73 |
| claric-NO (12) | 0.68 |
| azi-OO-Cl (13) | 0.58 |
| azi-OO (14) | 0.47 |
| claric-OO-Cl (17) | 0.87 |
| azi-OON-Cl (19) | 0.19 |
| Telithromycin | 0.31 |
| Azithromycin | 0.32 |
| Erythtomycin | 0.45 |
| Ciprofloxacin | >50 |
Induction studies.
The propensity of the compounds to induce methyltransferase expression in strains with inducible resistance was checked by the disk diffusion method against five isolates (two S. aureus and three S. pyogenes isolates) with different resistance genes and induction profiles (Table 6). The macrolones did not induce erm expression in any of the tested strains, whereas erythromycin, azithromycin, and clarithromycin were the strong inducers. Telithromycin induced erm expression only in an S. pyogenes erm(B) isolate.
TABLE 6.
Induction potential of azithromycin, telithromycin, and compounds 13 and 19 by triple-disk diffusion method
| Strain | Phenotype | Genotype | Induction potentiala |
|||
|---|---|---|---|---|---|---|
| AZM | TEL | Compound 13 | Compound 19 | |||
| S. pyogenes | iMLSb-C | erm(A) | ** | ° | ° | ° |
| S. pyogenes | iMLSb-B | erm(A) | ** | ° | ° | ° |
| S. pyogenes | iMLSb-A | erm(B) | ** | ** | ° | ° |
| S. aureus | iMLSb | erm(C) | ** | ° | ° | ° |
| S. aureus | iMLSb | erm(A) | ** | ° | ° | ° |
All compounds were tested by use of a 0.5-μg/disk. AZM, azithromycin; TEL, telithromycin; **, induction toward erm expression for clindamycin and rokitamycin resistance; °, no induction.
Two strains [S. pyogenes iMLSb-A with the erm(B) gene and S. aureus iMLSb with the erm(C) gene] were selected to determine the impact of incubation in the presence of subinhibitory concentrations of the compounds on erm induction (Table 7).
TABLE 7.
Induction of inducibly expressed erm genes after incubation with subinhibitory concentrations of erythromycin, telithromycin, and compound 19a
| Strain | Inducer | Inducer concn (μg/ml) | Inoculum size (no. of CFU/ml) | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| AZM | ERY | CLI | CIP | TEL | Compound 19 | ||||
| S. aureus B 0949 erm(C) iMLSb | 1.3E+06 | >256 | >256 | 0.06 | 0.25 | 0.25 | 0.5 | ||
| ERY | 1 | 1.4E+05 | >256 | >256 | >8 | 0.25 | >8 | 8 | |
| ERY | 0.1 | 1.0E+06 | >256 | >256 | >8 | 0.25 | >8 | 4 | |
| ERY | 0.01 | 1.4E+06 | >256 | >256 | >8 | 0.25 | >8 | 4 | |
| TEL | 0.01 | 9.0E+05 | >256 | >256 | 0.125 | 0.25 | 0.125 | 0.25 | |
| 19 | 0.1 | 1.4E+05 | >256 | >256 | 0.06 | 0.25 | 0.25 | 0.25 | |
| 5.0E+05 | >256 | >256 | 0.25 | 1 | 8 | 0.03 | |||
| S. pyogenes B 0895 erm(B) iMLSb-A | ERY | 0.1 | 1.0E+05 | >256 | >256 | >8 | 1 | >32 | 1 |
| ERY | 0.01 | 8.0E+05 | >256 | >256 | >8 | 1 | >32 | 1 | |
| ERY | 0.001 | 8.0E+05 | >256 | >256 | >8 | 1 | >32 | 1 | |
| TEL | 0.1 | 8.0E+04 | >256 | >256 | >8 | 1 | >32 | 0.5 | |
| TEL | 0.01 | 2.0E+05 | >256 | >256 | >8 | 1 | 4 | 0.03 | |
| 19 | 0.001 | 1.5E+06 | >256 | >256 | 0.125 | 1 | 8 | ≤0.015 | |
AZM, azithromycin; ERY, erythromycin; CLI, clindamycin; CIP, ciprofloxacin; TEL, telithromycin.
Pharmacokinetics in mice.
Relevant PK data for mice after administration of the tested compounds are given in Table 8. Values are given as the total drug concentration in blood samples after a single intravenous bolus dose (target dose, 2 mg/kg) or oral dose (target dose, 10 mg/kg).
TABLE 8.
Major pharmacokinetic parameters of selected macrolones in mice after intravenous and oral administrationa
| Compound | Intravenous bolusb |
Oral administrationc |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cmax (ng/ml) | AUC0–t (ng · h/ml) | AUC0–∞ (ng · h/ml) | CL (ml/min/kg) | Vss (liters/kg) | MRT (h) | t1/2 (h) | Cmax (ng/ml) | Tmaxd (h) | AUC0–t (ng · h/ml) | AUC0–∞ (ng · h/ml) | DNAUC0-∞ (ng · h/ml/mg/kg) | t1/2 (h) | F (%) | |
| 1e | 2,453 ± 788 | 3,307 ± 821 | ND | 5.3 ± 1.5 | 2.5 ± 0.4 | 8.0 ± 0.8 | 15.9 ± 4.3 | 49.0 ± 16.0 | 0.5 ± 0.1 | 124.7 ± 12.5 | ND | 12.5 ± 1.3 | ND | <1 |
| 7 | 781 ± 630 | 1,109 ± 242 | 1,349 ± 306 | 26.7 ± 4.5 | 15.0 ± 8.3 | 9.0 ± 3.6 | 8.2 ± 2.0 | 10.0 ± 8.0 | 2.0 ± 0.6 | 58.0 ± 73.9 | 98.4 ± 141.2 | 10.5 ± 15.0 | 8.5 ± 12 | 2 ± 2 |
| 19 | 1,887 ± 1,797 | 1,650 ± 227 | 1,794 ± 248 | 17.9 ± 2.7 | 8.2 ± 1.7 | 7.6 ± 0.9 | 7.9 ± 0.7 | 8.0 ± 3.0 | 2.0 ± 0.0 | 42.3 ± 7.1 | 47.3 ± 5.4 | 4.9 ± 0.6 | 2.5 ± 0.2 | <1 |
| 6 | 487 ± 173 | 2,998 ± 1,288 | ND | 12.9 ± 6.3 | 6.8 ± 2.9 | 9.0 ± 0.5 | 12.8 ± 2.3 | 140 ± 44 | 4.0 ± 0.0 | 1,767.8 ± 597.7 | ND | 176.8 ± 59.8 | 15.0 ± 3.0 | 12 ± 4 |
Three animals per group were tested. Abbreviations: Cmax, maximum concentration in plasma; AUC0–t, area under the plasma concentration-time curve from time zero to time t; AUC0–∞, area under the plasma concentration-time curve from time zero to infinity; CL, clearance; Vss, volume of distribution at steady state; MRT, mean residence time; t1/2, half-life; Tmax, time to Cmax; DNAUC0–∞, dose-normalized area under the plasma concentration-time curve from time zero to infinity; F, bioavailability; ND, not determined.
Target dose, 2 mg/kg.
Target dose, 10 mg/kg.
Tmax is expressed as the median.
Compound 1 administered at a dose of 1 mg/kg.
Selected macrolones on an azithromycin core with different heteroatom compositions in the linker (compound 1 [azi-NN-Cl], compound 7 [azi ON-Cl], compound 19 [azi-OON-Cl], and compound 6 [azi-NMeN-Cl-QMe]) were characterized by low to moderate systemic clearance (27% of liver blood flow for compound 7), a large volume of distribution at steady state (from 2.5 liters/kg for compound 1 to 15 liters/kg for compound 7), and a long half-life. Compounds 1, 7, and 19, however, had poor oral bioavailability. On the other hand, compound 6 had a much better profile after oral administration, resulting in an oral bioavailability of 12% and the highest oral exposure (area under the plasma concentration-time curve from time zero to time t, 1,767.8 ± 597.7 ng · h/ml) that was more than 10-fold higher than that of the other compounds tested.
The in vivo efficacy of the compounds was assessed in a mouse model of pneumococcal pneumonia induced by S. pneumoniae SP030 Erys by evaluation of bacterial eradication from the lungs of treated animals. Due to their poor oral bioavailability, compounds 1, 7, and 19 were administered i.p. in two (at a 24-h interval) or three (at a 12-h interval) doses starting at 6 h postinfection. Azithromycin was used as a comparator. Animals were sacrificed 14 h after the last dose, and bacterial counts in the lungs were determined. All compounds fully eradicated the bacteria to the limit of detection (<2 log10 CFU), and no viable pneumococci were identified in lung homogenates (Fig. 2; see also Fig. S5 and S6 in the supplemental material). In addition, measurable quantities of compounds were detected by bioassay in lung homogenates.
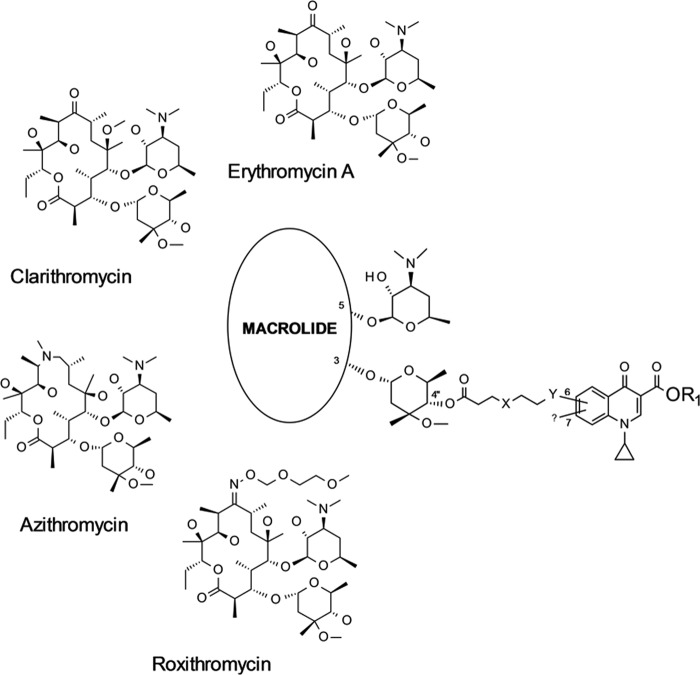
FIG 2.
Efficacy of compound 19 (azi-OON-Cl) in a mouse model of pneumonia induced by S. pneumoniae SP030 Erys. Compound and azithromycin were administered i.p. The MIC values of compound 19 and azithromycin were ≤0.015 and 0.03 μg/ml, respectively. (A) Number of CFU per gram of lung tissue. A statistically significant reduction in the number of CFU is designated by asterisks (***, P < 0.0001). (B) Concentration of compound in lung tissue 14 h after the last dose, given as mean values ± SEMs. AZM, azithromycin; cpd 19, compound 19.
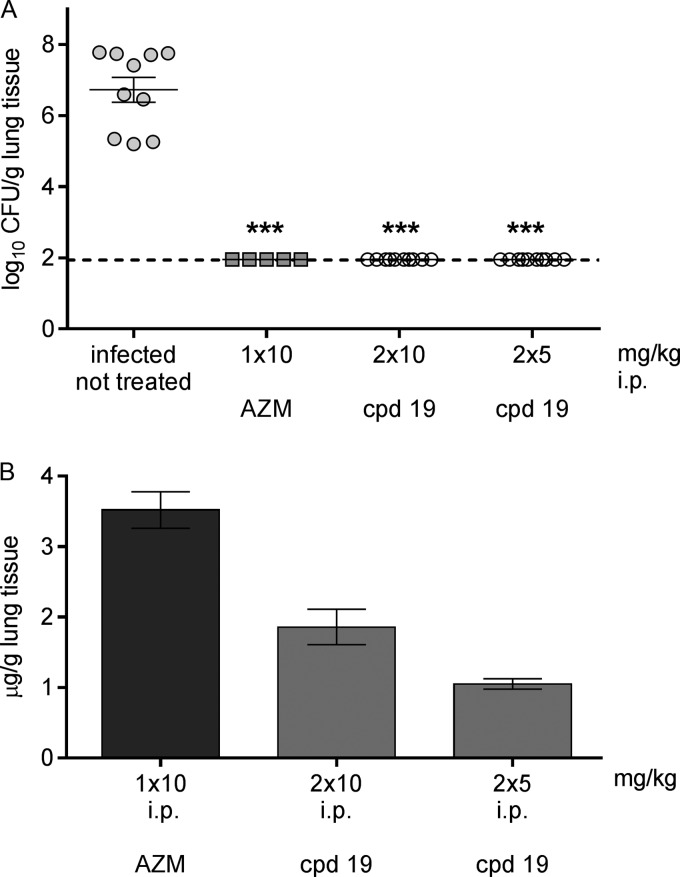
As the overall antibacterial profiles of compounds 7 and 19 (having one and two oxygen atoms in the linker, respectively) were superior to those of compound 1 (azi-NN-Cl), they were tested further in a model of pneumonia induced by resistant strain S. pneumoniae B 1217 with a constitutively expressed erm(B) gene (Fig. 3). In this case, the comparators were azithromycin and telithromycin. The MICs of both compounds for this strain were ≤0.015 μg/ml, that of azithromycin was >64 μg/ml, and that of telithromycin was 0.03 μg/ml. Azithromycin did not significantly reduce the number of CFU in the lungs, while telithromycin gave good eradication, yet the decrease in the number of CFU did not reach statistical significance. On the contrary, the macrolones were fully effective and compound 19 cleared the bacteria from the lungs to the limit of detection in all animals, and compound 7 did so in 4/5 animals.
FIG 3.
Efficacy of compounds 19 (azi-OON-Cl) and 7 (azi-ON-Cl) in a model of mouse pneumonia induced by S. pneumoniae B 1217 with MLSb. All compounds were administered i.p. The MICs of the tested compounds were as follows: ≤0.015 μg/ml for compounds 7 and 19, 0.06 μg/ml for telithromycin, and >64 μg/ml for azithromycin. (A) Number of CFU per gram of lung tissue. A statistically significant reduction in the number of CFU is designated by asterisks (**, P < 0.001). (B) Concentration of compound in lung tissue 14 h after the last dose, given as mean values ± SEMs. AZM, azithromycin; TEL, telithromycin; cpd 19, compound 19; cpd 7, compound 7.
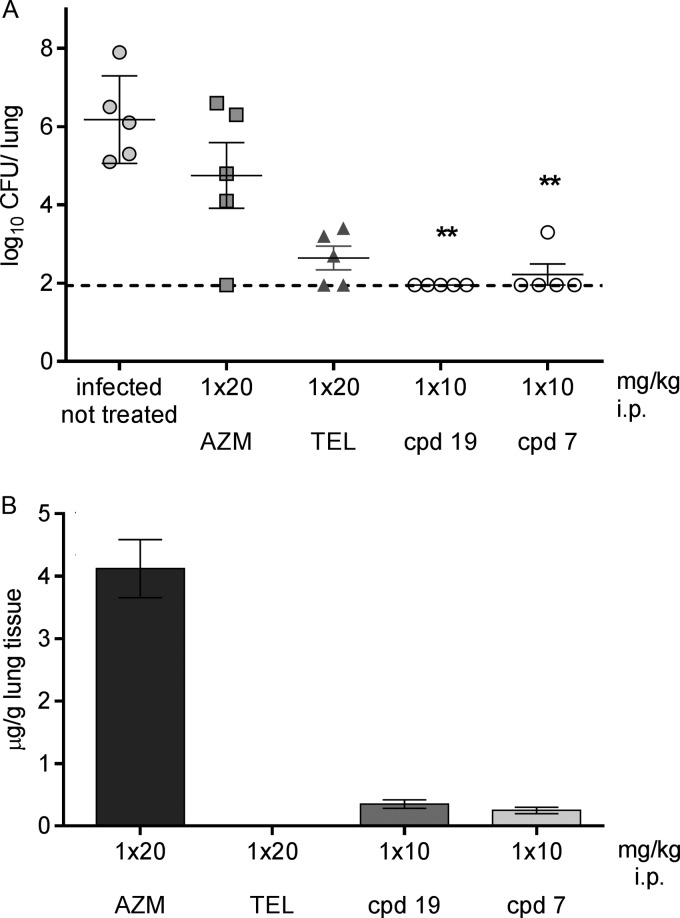
As compound 6 (azi-NMeN-Cl-QMe) had oral bioavailability (F) of 12%, it was tested in the model of pneumonia induced by S. pneumoniae Erys and was administered by the p.o. route (Fig. 4). Two dosing regimens were applied. Namely, doses of 10, 20, and 50 mg/kg each were administered twice (at 6 and 30 h postinfection, corresponding to once-daily dosing) or three times (at 6, 18, and 30 h postinfection, equaling a twice-daily regimen). When applied three times, bacteria could not be recovered from the lungs of treated animals independently of the dose given, while with twice-daily administration, full efficiency was observed after the highest dose (50 mg/kg) and the reduction in CFU counts was comparable to that achieved with azithromycin at a 20-mg/kg dose.
FIG 4.
Efficacy of compound 6 (azi-NMeN-Cl-QMe) in a mouse model of pneumonia induced by S. pneumoniae SP030 Erys. Compound 6 and azithromycin were administered per os. The MIC values of compound 6 and azithromycin were ≤0.015 and 0.03 μg/ml, respectively. (A) Number of CFU per gram of lung tissue. A statistically significant reduction in the number of CFU is designated by asterisks (**, P < 0.001). (B) Concentration of compound in lung tissue 14 h after the last dose, given as mean values ± SEMs.
DISCUSSION
The results of in vitro and in vivo evaluations of the investigated macrolones clearly point to the potential of this class of compounds as novel antibacterial agents. The structure-activity relationships of compounds within a specific class of macrolones and among a group of compounds including a broader set of chemically more diverse analogues were discussed elsewhere (28 – 34). The study described here focused on fine-tuning the biological properties of the compounds by linker modification.
The antibacterial potencies of macrolones against key bacterial pathogens that are involved in RTI and that have different macrolide resistance phenotypes were superior to those of currently marketed macrolide antibiotics (Table 1). The introduction of oxygen into the linker improved the activity against H. influenzae compared to the activities of analogues with two nitrogen atoms, while their excellent activity against streptococci was retained. Elongation of the linker by addition of an oxygen atom (OON in compounds 19 to 26) was also favorable for overall potency. It is important to note the excellent activity of macrolones against erythromycin-resistant S. pyogenes strains with inducible or constitutive erm gene expression. This provides a clear advantage over the ketolide telithromycin, as it is not sufficiently active against S. pyogenes erm(B) strains (42, 43). More recently, solithromycin, a fluoroketolide antibiotic with an improved antimicrobial profile covering macrolide-resistant respiratory pathogens, including S. pyogenes erm(B) strains, has been developed and is in phase III clinical development for the treatment of moderate to moderately severe community-acquired bacterial pneumonia (CABP) (44 – 46). However, due to safety issues, the use of telithromycin has been restricted, while a new drug application (NDA) for another ketolide derivative, cethromycin, was denied by FDA in 2009. The most critical issue is rare but potentially fatal hepatotoxicity that has been extensively documented for telithromycin (this led to the addition of a “black box warning,” the strictest warning by the U.S. Food and Drug Administration [FDA] that appears on a prescription drug's label and is designed to call attention to serious or life-threatening risks) (47 – 49). It has not been reported for cethromycin or solithromycin, but safety data for these drugs are limited, so it is not yet clear if hepatotoxicity is a ketolide class effect. This issue heavily impedes prospects for ketolide use for the treatment of mild to moderate pneumonia (47). Selection of the azithromycin core as the major scaffold for macrolone compounds can potentially circumvent safety issues that might arise from the selection of the cores of other macrolide compounds. In addition, azithromycin is well-known for its accumulation in neutrophils and in tissues as well as sustained activity in targeted organs (i.e., lungs) (50, 51). The overall favorable PK and safety properties of azithromycin made it a major scaffold of choice for the development of macrolones.
The MICs obtained for streptococci with cMLSb, although low, were higher than those obtained for sensitive isolates. As macrolones exhibit a macrolide mode of action (by selective inhibition of bacterial protein synthesis through interactions with the ribosome), it is most likely that both parts of molecule (the scaffold and the linker quinolone) bind to the bacterial ribosome and that additional interactions not seen with the standard macrolide antibiotics are formed by the quinolone moiety. It is possible that additional interactions lead to the improved activity against strains with cMLSb compared to the activities of related antibiotics. Nonetheless, it is also reasonable to assume that the changed interaction of the macrolide core with the methylated ribosome influences to a certain degree the overall activity of compounds, thus resulting in MIC values severalfold higher than those for other resistant strains.
Inducible macrolide resistance is of strong clinical significance. Depending on the study and the geographical region concerned, macrolide resistance has been detected in 30 to 90% of S. pyogenes isolates (40, 52). The geographic variability of macrolide resistance in S. aureus isolates was even greater and depended on whether the strains studied were methicillin sensitive or resistant, as well as the region, and the rates of macrolide resistance in S. aureus isolates has been found to vary from 10 to 90% (53, 54). The propensity of macrolones to induce erm gene expression was measured by two methods: the disk diffusion method and preincubation of resistant strains with a potential inducer and subsequent MIC determination. The lowest concentration of inducer needed to promote erm gene expression by disk diffusion was found for each strain tested and was as low as 0.001 μg of erythromycin per disk against the most sensitive, erm(A)-harboring S. aureus strain. This indicates the importance of maintaining high purity requirements for the tested compounds, as even traces of some impurities (starting macrolides, for example) can impact the result and lead to the induction of resistance genes. Our lead compounds at a concentration of 0.5 μg/disk did not induce resistance genes in any of the strains by disk diffusion, while telithromycin induced gene expression in an S. pyogenes erm(B) strain with an iMLS-A phenotype. When the above-mentioned strains were preincubated in the presence of subinhibitory concentrations of inducers, compound 19 did not induce erm gene expression, additionally demonstrating the lack of its macrolide resistance induction potential.
The ribosomal exit tunnel plays an important role in the modulation of the translation process through specific interactions with the leading peptides of nascent proteins that regulate ribosome stalling, initial secondary structure formation of synthesized proteins, and, as a consequence, gene expression at the translation level (55 – 60). It has been shown that macrolide antibiotics do not inhibit the synthesis of all peptides equally but, rather, show specific inhibitory effects depending on the peptide (61). As it is known that the induction profiles of genes belonging to the erm family depend primarily on the attenuator sequence, i.e., the leading peptide (62), it is plausible to assume that the lack of induction in the presence of macrolones arises from their ability to establish unique interactions with the ribosomal exit tunnel and/or different leading peptides of proteins. Nevertheless, to ascertain the definite absence of induction, it is necessary to test the interaction of macrolones with a wider panel of bacterial stains. Ketolides were considered noninducing antibiotics, but focused in vitro studies have shown their potential to induce the erm(C) gene (63). Although the S. aureus strain with inducible expression of the erm(C) gene used in this study did not exhibit reduced susceptibility after incubation in the presence of subinhibitory concentrations of telithromycin, induction was detected for S. pyogenes isolates with the erm(B) gene. It is worth noting that induction of erm(B) gene expression in this strain by erythromycin and higher concentration of telithromycin (0.1 μg/ml) resulted in a significant increase in the MIC values for clindamycin (>8 μg/ml) and telithromycin (>32 μg/ml), while compound 19, despite substantial increase, retained activity with an MIC value of 1 μg/ml. Preincubation with compound 19 did not induce erm gene expression.
As macrolones possess a quinolone moiety attached to the macrolide core, it was important to determine whether they are protein synthesis inhibitors. Since these compounds achieved IC50s in the higher nanomolar range in in vitro transcription/translation studies, a result comparable to that for macrolide antibiotics, protein synthesis inhibition can be claimed to be their basic mechanism of action. However, the possibility of additional activity that would be specific for quinolones cannot be definitely excluded by application of only this method. Assays for assessment of the inhibition of DNA gyrase and topoisomerase IV have been performed for a number of macrolone class representatives, and none of them have been shown to have inhibitory properties (data not shown), which clearly indicates that macrolones do not exhibit their antibacterial activity through the quinolone mode of action. The lack of a quinolone mode of action is further corroborated by the full potency of macrolones against quinolone-resistant S. aureus strains.
Since the unique pharmacological properties of azithromycin (long half-life, good tissue penetration, cellular accumulation, side effects fewer and milder than those of other antibiotics) and its favorable safety profile are well recognized and macrolones with an azithromycin core have excellent in vitro profiles superior to those of other macrolones, these analogues, which have varying linker lengths and heteroatom compositions different from those of azithromycin, were selected for in vivo evaluation. Macrolones are molecules with low to moderate systemic clearance, a long half-life, a large volume of distribution (indicating good tissue penetration), and very poor oral bioavailability. As these rather high-molecular-weight compounds show good stability in liver microsomes (data not shown) and have good solubility, the limited oral bioavailability is likely due to poor permeation. In an attempt to improve the gastrointestinal absorption of compound 1, compound 6 was designed to be more lipophilic by methylation of the central nitrogen atom in the linker and esterification of 3-quinolone carboxylic acid, as it is known that decreased hydrophilicity improves the transfer of molecules through biological membranes (64). These modifications resulted in an enhanced oral bioavailability of 12% (compound 1 had an F of <1%) and a substantial increase in oral exposure (compound 6 had an AUC more than 10 times higher than that of compound 1 [AUC, 1,767.8 ± 597.7 ng · h/ml and 124.7 ± 12.5 ng · h/ml, respectively]) and better tissue penetration (volume of distribution at steady state, 6.8 ± 2.9 and 2.5 ± 0.4 liters/kg for compounds 6 and 1, respectively). In addition, this modification led to a more than 7-fold increase in the accumulation of compound 6 in human polymorphonuclear leukocytes (65) (compounds 1 and 6 are designated compounds 27 and 31, respectively, in reference 65). Compound 6 has a low systemic clearance and a long half-life of 12.8 ± 2.3 h, comparable to that of other macrolones. The changes introduced were, however, detrimental for potency against S. aureus and H. influenzae.
Due to the low oral bioavailability of the macrolone molecules in mice, the in vivo efficacy of compounds 1, 7, and 19 was assessed after i.p. application in the mouse pneumococcal pneumonia model. In a model of pneumonia induced by S. pneumoniae SP030 Erys, administration of two or three doses resulted in full bacterial eradication, and no viable colonies could be recovered from the lungs of treated animals, regardless of the compound or dosing regimen. Although a dose-response is not clearly visible at the level of CFU counts (due to the high efficacy of the tested analogues), compounds accumulated in lung tissue in a dose-dependent manner in measurable microgram quantities. When tested in a model of pneumonia induced by S. pneumoniae B 1217 with cMLSb, compounds 7 and 19 proved to be fully efficacious as well. In the model of pneumonia induced by S. pneumoniae SP030 Erys, compound 6, which had an F of 12%, achieved a level of exposure by oral administration sufficient for successful bacterial clearance. Pharmacokinetic studies in rat (29) revealed that compound 19 exhibited improved exposure and oral bioavailability in comparison to the data for mice reported here, while compound 7 did not.
In a preliminary in vitro toxicity assessment (e.g., cytotoxicity for HepG2 and THP-1 cell lines), macrolones displayed acceptable safety profiles (33).
In conclusion, the results of an in vitro and in vivo evaluation of macrolones clearly differentiate them from currently available macrolide antibiotics. The in vitro antibacterial profiles of the macrolones against key respiratory tract pathogens are superior to those of marketed macrolide drugs, including the ketolide telithromycin.
Supplementary Material
ACKNOWLEDGMENTS
We thank all team members of the New Antibacterial Macrolides Program/Project from the Pliva Research Institute, Zagreb, Croatia; GSK R&D, Verona, Italy; GSK R&D, Harlow, United Kingdom; and GSK R&D Collegeville, PA, USA, for intensive collaborative research efforts, of which the current report represents just a part. In particular, we thank Andrea Fajdetić, Sulejman Alihodžić, Adrijana Vinter, and Ivana Palej Jakopovoć for synthesis of the compounds and Višnja Majzel for exceptional technical assistance and support.
The presented research was funded and supported by the resources available at Pliva and GlaxoSmithKline R&D facilities.
Željko Kelnerić, Nerrisa Simon, and John Broskey are no longer employees of GlaxoSmithKline. The present addresses of the other authors are given on the first page of the article.
We have no conflicts of interest to declare.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00524-16.
REFERENCES
- 1.Niederman MS. 2009. Community-acquired pneumonia: the U.S. perspective. Semin Respir Crit Care Med 30:179–188. doi: 10.1055/s-0029-1202937. [DOI] [PubMed] [Google Scholar]
- 2.Broulette J, Yu H, Pyenson B, Iwasaki K, Sato R. 2013. The incidence rate and economic burden of community-acquired pneumonia in a working-age population. Am Health Drug Benefits 6:494–503. [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolau D. 2004. Treatment with appropriate antibiotic therapy in community-acquired respiratory tract infections. Am J Manag Care 10(12 Suppl):S381–S388. [PubMed] [Google Scholar]
- 4.Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiguro T, Takayanagi N, Yamaguchi S, Yamakawa H, Nakamoto K, Takaku Y, Miyahara Y, Kagiyama N, Kurashima K, Yanagisawa T, Sugita Y. 2013. Etiology and factors contributing to the severity and mortality of community-acquired pneumonia. Intern Med 52:317–324. doi: 10.2169/internalmedicine.52.8830. [DOI] [PubMed] [Google Scholar]
- 6.Torres A, Blasi F, Peetermans WE, Viegi G, Welte T. 2014. The aetiology and antibiotic management of community-acquired pneumonia in adults in Europe: a literature review. Eur J Clin Microbiol Infect Dis 33:1065–1079. doi: 10.1007/s10096-014-2067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodhead M. 2009. The European vision of community-acquired pneumonia. Semin Respir Crit Care Med 30:136–145. doi: 10.1055/s-0029-1202932. [DOI] [PubMed] [Google Scholar]
- 8.Song JH, Thamlikitkul V, Hsueh PR. 2011. Clinical and economic burden of community-acquired pneumonia amongst adults in the Asia-Pacific region. Int J Antimicrob Agents 38:108–117. [DOI] [PubMed] [Google Scholar]
- 9.Rossolini GM, Mantengoli E, Montagnani F, Pollini S. 2010. Epidemiology and clinical relevance of microbial resistance determinants versus anti-Gram-positive agents. Curr Opin Microbiol 13:582–588. doi: 10.1016/j.mib.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Canu A, Malbruny B, Coquemont M, Davies TA, Appelbaum PC, Leclercq R. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother 46:125–131. doi: 10.1128/AAC.46.1.125-131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wierzbowski AK, Nichol K, Laing N, Hisanaga T, Nikulin A, Karlowsky JA, Hoban DJ, Zhanel GG. 2007. Macrolide resistance mechanisms among Streptococcus pneumoniae isolated over 6 years of Canadian Respiratory Organism Susceptibility Study (CROSS) (1998-2004). J Antimicrob Chemother 60:733–740. doi: 10.1093/jac/dkm273. [DOI] [PubMed] [Google Scholar]
- 12.Setchanova L, Kostyanev T, Alexandrova A, Mitov I, Nashev D, Kantardjiev T. 2013. Microbiological characterization of Streptococcus pneumoniae and nontypeable Haemophilus influenzae isolates as primary causes of acute otitis media in Bulgarian children before the introduction of conjugate vaccines. Ann Clin Microbiol Antimicrob 12:6. doi: 10.1186/1476-0711-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins PA, Chochua S, Jackson D, Beall B, McGee L. 2015. Mobile elements and chromosomal changes associated with MLS resistance phenotypes of invasive pneumococci recovered in the United States. Microb Drug Resist 21:121–129. doi: 10.1089/mdr.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seppala H, Nissinen A, Yu Q, Huovinen P. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J Antimicrob Chemother 32:885–891. doi: 10.1093/jac/32.6.885. [DOI] [PubMed] [Google Scholar]
- 15.Bozdogan B, Bogdanovich T, Kosowska K, Jacobs MR, Appelbaum PC. 2004. Macrolide resistance in Streptococcus pneumoniae: clonality and mechanisms of resistance in 24 countries. Curr Drug Targets Infect Disord 4:169–176. doi: 10.2174/1568005043340821. [DOI] [PubMed] [Google Scholar]
- 16.Farrell DJ, Couturier C, Hryniewicz W. 2008. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT year 5 (2003-2004). Int J Antimicrob Agents 31:245–249. doi: 10.1016/j.ijantimicag.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Felmingham D, Canton R, Jenkins SG. 2007. Regional trends in [beta]-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J Infect 55:111–118. doi: 10.1016/j.jinf.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins S, Brown S, Farrell D. 2008. Trends in antibacterial resistance among Streptococcus pneumoniae isolated in the USA: update from PROTEKT US years 1-4. Ann Clin Microbiol Antimicrob 7:1. doi: 10.1186/1476-0711-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng Q, Zhang T, Ding Y, Tao Y, Lin Y, Wang Y, Black S, Zhao G. 2014. Molecular characterization and antimicrobial susceptibility of Streptococcus pneumoniae isolated from children hospitalized with respiratory infections in Suzhou, China. PLoS One 9:e93752. doi: 10.1371/journal.pone.0093752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wajima T, Morozumi M, Chiba N, Shouji M, Iwata S, Sakata H, Ubukata K. 2013. Associations of macrolide and fluoroquinolone resistance with molecular typing in Streptococcus pyogenes from invasive infections, 2010-2012. Int J Antimicrob Agents 42:447–449. doi: 10.1016/j.ijantimicag.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 21.Cole ST. 2014. Who will develop new antibacterial agents? Philos Trans R Soc Lond B Biol Sci 369:20130430. doi: 10.1098/rstb.2013.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbanat D, Morrow B, Bush K. 2008. New agents in development for the treatment of bacterial infections. Curr Opin Pharmacol 8:582–592. doi: 10.1016/j.coph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 24.Butler MS, Cooper MA. 2011. Antibiotics in the clinical pipeline in 2011. J Antibiot 64:413–425. doi: 10.1038/ja.2011.44. [DOI] [PubMed] [Google Scholar]
- 25.Coates A, Halls G. 2011. Antibiotics in phase II and III clinical trials, p 167–183. In Coates ARM. (ed), Antibiotic resistance, 211 ed Springer, Berlin, Germany. [Google Scholar]
- 26.Corey R, Naderer OJ, O'Riordan WD, Dumont E, Jones LS, Kurtinecz M, Zhu JZ. 2014. Safety, tolerability, and efficacy of GSK1322322 in the treatment of acute bacterial skin and skin structure infections. Antimicrob Agents Chemother 58:6518–6527. doi: 10.1128/AAC.03360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pucci MJ, Bush K. 2013. Investigational antimicrobial agents of 2013. Clin Microbiol Rev 26:792–821. doi: 10.1128/CMR.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fajdetic A, Cipcic Paljetak H, Lazarevski G, Hutinec A, Alihodzic S, Djerek M, Stimac V, Andreotti D, Sunjic V, Berge JM, Mtak S, Dumic M, Lociuro S, Holmes DJ, Marsic N, Erakovic Haber V, Spaventi R. 2010. 4″-O-([omega]-Quinolylamino-alkylamino)propionyl derivatives of selected macrolides with the activity against the key erythromycin resistant respiratory pathogens. Bioorg Med Chem 18:6559–6568. doi: 10.1016/j.bmc.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 29.Fajdetic A, Vinter A, Cipcic Paljetak H, Padovan J, Jakopovic IP, Kapic S, Alihodzic S, Filic D, Modric M, Kosutic-Hulita N, Antolovic R, Schoenfeld ZI, Mutak S, Erakovic Haber V, Spaventi R. 2011. Synthesis, activity and pharmacokinetics of novel antibacterial 15-membered ring macrolones. Eur J Med Chem 46:3388–3397. doi: 10.1016/j.ejmech.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Hutinec A, Djerek M, Lazarevski G, Sunjic V, Cipcic Paljetak H, Alihodzic S, Erakovic Haber V, Dumic M, Marsic N, Mutak S. 2010. Novel 8a-aza-8a-homoerythromycin–4″-(3-substituted-amino)propionates with broad spectrum antibacterial activity. Bioorg Med Chem Lett 20:3244–3249. doi: 10.1016/j.bmcl.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 31.Kapic S, Cipcic Paljetak H, Palej Jakopovic I, Fajdetic A, Ilijas M, Stimac V, Brajsa K, Holmes DJ, Berge J, Alihodzic S. 2011. Synthesis of macrolones with central piperazine ring in the linker and its influence on antibacterial activity. Bioorg Med Chem 19:7281–7298. doi: 10.1016/j.bmc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Matanovic Skugor M, Stimac V, Palej I, Lugaric D, Cipcic Paljetak H, Filic D, Modric M, Djilovic I, Gembarovski D, Mutak S, Erakovic Haber V, Holmes DJ, Ivezic-Schoenfeld Z, Alihodzic S. 2010. Synthesis and biological activity of 4″-O-acyl derivatives of 14- and 15-membered macrolides linked to [omega]-quinolone-carboxylic unit. Bioorg Med Chem 18:6547–6558. doi: 10.1016/j.bmc.2010.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Palej Jakopovic I, Kragol G, Forrest AK, Frydrych CSV, Stimac V, Kapic S, Matanovic Skugor M, Ilijas M, Cipcic Paljetak H, Jelic D, Holmes DJ, Hickey DMB, Verbanac D, Erakovic Haber V, Alihodzic S. 2010. Synthesis and properties of macrolones characterized by two ether bonds in the linker. Bioorg Med Chem 18:6578–6588. doi: 10.1016/j.bmc.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Kapic S, Cipcic Paljetak H, Alihodzic S, Antolovic R, Erakovic Haber V, Jarvest RL, Holmes DJ, Broskey JP, Hunt E. 2010. 6-Alkylquinolone-3-carboxylic acid tethered to macrolides synthesis and antimicrobial profile. Bioorg Med Chem 18:6569–6577. doi: 10.1016/j.bmc.2010.06.048. [DOI] [PubMed] [Google Scholar]
- 35.Stimac V, Alihodzic S, Lazarevski G, Mutak S, Marusic Istuk Z, Fajdetic A, Palej I, Cipcic Paljetak H, Padovan J, Tavcar B, Erakovic Haber V. 2009. Synthesis and biological properties of 4″-O-acyl derivatives of 8a-aza-8a-homoerythromycin. J Antibiot (Tokyo) 62:133–144. doi: 10.1038/ja.2009.1. [DOI] [PubMed] [Google Scholar]
- 36.Alihodzic S, Andreotti D, Berdik A, Bientinesi I, Biondi S, Ciraco M, Damiani F, Djerek M, Dumic M, Erakovic V. May 2003. Macrolides. Patent WO03042228.
- 37.Alihodzic S, Berdik A, Berge JM, Jarvest RL, Mutak S. November 2004. Macrolides substituded at the 4″-position. Patent WO2004101585.
- 38.Alihodzic S, Mutak S, Pavlovic D, Palej I, Stimac V, Kapic S, Vinter A, Matanovic Skugor M. November 2005. Ester linked macrolides useful for the treatment of microbial infections. Patent WO2005108412.
- 39.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Giovanetti E, Montanari MP, Mingoia M, Varaldo PE. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother 43:1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07-A6. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Tamayo J, Perez-Trallero E, Gomez-Garces JL, Alos JI, on behalf of the Spanish Group for the Study of Infection in the Primary Health Care Setting. 2005. Resistance to macrolides, clindamycin and telithromycin in Streptococcus pyogenes isolated in Spain during 2004. J Antimicrob Chemother 56:780–782. doi: 10.1093/jac/dki286. [DOI] [PubMed] [Google Scholar]
- 43.Banche G, Roana J, Allizond V, Andreotti S, Malabaila A, Li Vigni N, Mandras N, Scalas D, Tullio V, Carlone NA, Savoia D, Gaido E, Barbui A, Cuffini AM. 2008. In vitro compared activity of telithromycin and azithromycin against northwest Italian isolates of Streptococcus pyogenes and Streptococcus pneumoniae with different erythromycin susceptibility. Lett Appl Microbiol 47:309–314. doi: 10.1111/j.1472-765X.2008.02423.x. [DOI] [PubMed] [Google Scholar]
- 44.Farrell DJ, Castanheira M, Sader HS, Jones RN. 2010. The in vitro evaluation of solithromycin (CEM-101) against pathogens isolated in the United States and Europe (2009). J Infect 61:476–483. doi: 10.1016/j.jinf.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Farrell DJ, Sader HS, Castanheira M, Biedenbach DJ, Rhomberg PR, Jones RN. 2010. Antimicrobial characterisation of CEM-101 activity against respiratory tract pathogens, including multidrug-resistant pneumococcal serogroup 19A isolates. Int J Antimicrob Agents 35:537–543. doi: 10.1016/j.ijantimicag.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 46.Oldach D, Clark K, Schranz J, Das A, Craft JC, Scott D, Jamieson BD, Fernandes P. 2013. Randomized, double-blind, multicenter phase 2 study comparing the efficacy and safety of oral solithromycin (CEM-101) to those of oral levofloxacin in the treatment of patients with community-acquired bacterial pneumonia. Antimicrob Agents Chemother 57:2526–2534. doi: 10.1128/AAC.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgopapadakou NH. 2014. The wobbly status of ketolides: where do we stand? Expert Opin Investig Drugs 23:1313–1319. doi: 10.1517/13543784.2014.954036. [DOI] [PubMed] [Google Scholar]
- 48.Brinker AD, Wassel RT, Lyndly J, Serrano J, Avigan M, Lee WM, Seeff LB. 2009. Telithromycin-associated hepatotoxicity: clinical spectrum and causality assessment of 42 cases. Hepatology 49:250–257. doi: 10.1002/hep.22620. [DOI] [PubMed] [Google Scholar]
- 49.Ross DB. 2007. The FDA and the case of Ketek. N Engl J Med 356:1601–1604. doi: 10.1056/NEJMp078032. [DOI] [PubMed] [Google Scholar]
- 50.Bosnar M, Kelneric Z, Munic V, Erakovic V, Parnham MJ. 2005. Cellular uptake and efflux of azithromycin, erythromycin, clarithromycin, telithromycin, and cethromycin. Antimicrob Agents Chemother 49:2372–2377. doi: 10.1128/AAC.49.6.2372-2377.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foulds G, Shepard RM, Johnson RB. 1990. The pharmacokinetics of azithromycin in human serum and tissues. J Antimicrob Chemother 25(Suppl A):73–82. [DOI] [PubMed] [Google Scholar]
- 52.Kozlov RS, Bogdanovitch TM, Appelbaum PC, Ednie L, Stratchounski LS, Jacobs MR, Bozdogan B. 2002. Antistreptococcal activity of telithromycin compared with seven other drugs in relation to macrolide resistance mechanisms in Russia. Antimicrob Agents Chemother 46:2963–2968. doi: 10.1128/AAC.46.9.2963-2968.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcinak JF, Frank AL. 2006. Epidemiology and treatment of community-associated methicillin-resistant Staphylococcus aureus in children. Expert Rev Anti Infect Ther 4:91–100. doi: 10.1586/14787210.4.1.91. [DOI] [PubMed] [Google Scholar]
- 54.Cetin ES, Gunes H, Kaya S, Aridogan BC, Demirci M. 2008. Macrolide-lincosamide-streptogramin B resistance phenotypes in clinical staphylococcal isolates. Int J Antimicrob Agents 31:364–368. doi: 10.1016/j.ijantimicag.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 55.Lu J, Hua Z, Kobertz WR, Deutsch C. 2011. Nascent peptide side chains induce rearrangements in distinct locations of the ribosomal tunnel. J Mol Biol 411:499–510. doi: 10.1016/j.jmb.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Deutsch C. 2014. Regional discrimination and propagation of local rearrangements along the ribosomal exit tunnel. J Mol Biol 426:4061–4073. doi: 10.1016/j.jmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J, Deutsch C. 2005. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol 12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 58.Lu J, Deutsch C. 2008. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. 2009. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol 330:1005–1014. doi: 10.1016/S0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 61.Starosta AL, Karpenko VV, Shishkina AV, Mikolajka A, Sumbatyan NV, Schluenzen F, Korshunova GA, Bogdanov AA, Wilson DN. 2010. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol 17:504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 62.Weisblum B. 2000. Resistance to macrolide-lincosamide-streptogramin antibiotics, p 694–710. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 1st ed ASM Press, Washington, DC. [Google Scholar]
- 63.Bailey M, Chettiath T, Mankin AS. 2008. Induction of erm(C) expression by noninducing antibiotics. Antimicrob Agents Chemother 52:866–874. doi: 10.1128/AAC.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patrick G. 2001. Quantitative structure-activity relationships, p 125–139. In Stanbury H. (ed), Medicinal chemistry, 1st ed Bios Scientific Publishers Ltd., Oxford, United Kingdom. [Google Scholar]
- 65.Munic K, Kostrun VS, Fajdetic A, Bosnar M, Kelneric Z, Stepanic V, Erakovic HV. 2013. Structure-property relationship for cellular accumulation of macrolones in human polymorphonuclear leukocytes (PMNs). Eur J Pharm Sci 49:206–219. doi: 10.1016/j.ejps.2013.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.