Abstract
Here, we present the full sequences of three mcr-1-carrying plasmids isolated from extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli. The plasmids belong to three different replicon types and are 34,640 bp, 209,401 bp, and 247,885 bp in size. We describe for the first time a composite transposon containing mcr-1 localized on a multidrug-resistant (MDR) IncHI2 plasmid harboring additional determinants of resistance to six different classes of antibiotics, including the ESBL gene blaCTX-M-1, and heavy metal resistance.
TEXT
The recent description of the plasmid-mediated colistin resistance gene, mcr-1, in strains isolated from food animals, food, and humans in China was a trigger for an avalanche of retrospective studies investigating the presence of this specific gene (1). Since then, mcr-1 has been found almost all over the world, and the earliest evidence for its presence dates back to the 1980s (2). The wide spread of mcr-1 and the finding that this resistance marker is often associated with multidrug-resistant Enterobacteriaceae, e.g., extended-spectrum-β-lactamase (ESBL) producers or carbapenemase producers, is of great concern to public health (3).
The mcr-1 gene has so far been associated with different plasmid replicon types, such as IncI2, IncHI2, IncP, IncFIB, and IncX4 (1, 4, 5, 6, 7). However, only very limited data about the complete sequences of such plasmids and the genetic structure surrounding the mcr-1 gene are available. In this study, we present the complete sequences of three distinct mcr-1-harboring plasmids isolated from ESBL-producing Escherichia coli isolates originating from river water (E. coli OW3E1, B1:ST359, and SHV-12), imported vegetables from Thailand (E. coli H226B, A:ST167, and CTX-M-55), and imported poultry meat from Italy (E. coli S38, B1:ST602, and CTX-M-1) (8, 9).
The mcr-1-harboring plasmids were transferred by transformation experiments into E. coli DH5α, and colistin-resistant transformants were selected on LB agar supplemented with 2 mg/liter colistin (Sigma, St. Louis, MO, USA). The mcr-1-harboring plasmids pOW3E1, pH226B, and pS38 were extracted using the Large-Construct kit (Qiagen, Hombrechtikon, Switzerland) according the manufacturer's protocol. The plasmids were sequenced on a PacBio RS2 device (Pacific Biosciences, Menlo Park, CA, USA) with a 10-kb size-selected insert library and P6/C4 chemistry. pOW3E1 and pH226B were multiplexed with four other plasmids (data not shown) in a barcoded library (symmetric 384-barcode set) and run on two single-molecule real-time sequencing (SMRT) cells; pS38 was prepared as a single library and sequenced on another SMRT cell.
Debarcoding, whitelisting, and de novo assembly (using the HGAP3 algorithm) were performed using SMRTanalysis version 2.3 (Pacific Biosciences). The HGAP3 settings were kept at the defaults, except for the expected genome size, which was set between 60 bp and 200 bp. The HGAP3 analysis produced complete plasmid sequences of the multiplexed samples with at least 50-fold coverage over the entire molecules. Plasmid pS38 was sequenced individually with 2,250-fold coverage. The plasmid sequence was automatically annotated using the online Rapid Annotation Subsequencing Technology (RAST) service and CLC Main Workbench version 7.7 (CLC bio, Aarhus, Denmark) (10). Automated annotation was manually refined using the BLASTn and BLASTp programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Plasmid pOW3E1 is 34,640 bp in size, with a G+C content of 42.3% (Table 1). The plasmid backbone is similar to that of other IncX4 plasmids, including a type IV secretion system (pilX operon), which is associated with this plasmid group (data not shown). Interestingly, no resistance determinants other than the mcr-1 gene were located on pOW3E1.
TABLE 1.
Features of mcr-1-harboring plasmids described in this study
| Plasmid | Size (bp) | Inc type | Additional antimicrobial resistance gene(s) (agent) | Further resistance determinant(s) |
|---|---|---|---|---|
| pOW3E1 | 34,640 | IncX4 | None | None |
| pH226B | 209,401 | IncHI1 | None | Heavy metals (Zn2+, Cu2+, or Ag2+) |
| pS38 | 247,885 | IncHI2 | estX3, aadA2, aadA1a, aadA1b (aminoglycosides) | Heavy metals (Hg2+), tellurite, quaternary ammonium compounds |
| cmlA1 (chloramphenicol) | ||||
| sul3, dfrA1b (sulfonamides) | ||||
| mefB (macrolide) | ||||
| blaCTX-M-1 (β-lactam) | ||||
| tetA (tetracycline) |
Plasmid pH226B is 209,401 bp in size, with a G+C content of 46.5% (Table 1). It consists of a typical IncHI1 backbone and shares many open reading frames (ORFs) with the reference plasmid of the IncHI1 group R27 (GenBank accession number AF250878). Besides the mcr-1 gene, a Tn3 family transposon carrying heavy metal resistance determinants (e.g., for Zn2+, Cu2+, or Ag2+) is present on pH226B as well as a remotely located tcuABC operon that enables the use of tricarballylate as a carbon/energy source.
Plasmid pS38, extracted from an E. coli isolate originating from imported poultry meat, represents a large multidrug-resistant (MDR) plasmid (247,885 bp; G+C content, 46.7%) with high similarity in its backbone to the IncHI2 reference plasmid R478 (accession no. NC_005211) (Table 1). The MDR region of this plasmid includes two class 1 integrons carrying transposons, In641 (estX3, psp, aadA2, cmlA1, aadA1a, and qacH2) and In369 (dfrA1b and aadA1b), conferring resistance to aminoglycosides, chloramphenicol, quaternary ammonium compounds, and trimethoprim. Between these integrons, the macrolide efflux pump gene mefB, the ESBL gene blaCTX-M-1, and the sulfonamide resistance gene sul3 are present. Downstream of integron In369, two transposons follow, the first carrying mercury resistance determinants and the second carrying tetracycline resistance determinants. Furthermore, a tellurite resistance operon is present on pS38.
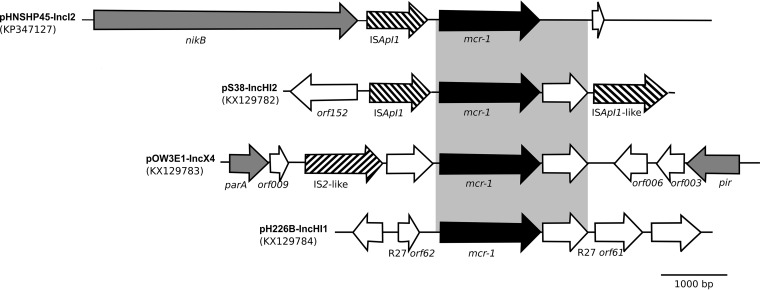
The mcr-1 genes were found to be located within various genetic contexts in the sequenced plasmids (Fig. 1). In all three plasmids, a 735-bp ORF encoding a hypothetical protein with similarities to a PAP2 superfamily protein was detected immediately downstream of the mcr-1 gene (both together are here referred to as the mcr-1 element).
FIG 1.
Genetic structures surrounding the mcr-1 gene in plasmids of this study compared to those in plasmid pHNSHP45. The arrows indicate open reading frames (orf), with black, stippled, gray, and white arrows representing mcr-1, insertion (IS) elements, ORFs with known function, and ORFs with unknown function, respectively.
In pOW3E1, the mcr-1 gene is inserted approximately 1.5 kb upstream of the gene encoding the replicon initiation protein and is found close to an IS2-like insertion element. An ISApl1 element is absent. In pH226B, the mcr-1 element is embedded in the IncHI1 backbone between two genes that were highly similar to those annotated as orf61 and orf62 on the IncHI1 reference plasmid R27. No mobile element was present upstream or downstream nearby. In pS38, the mcr-1 element is flanked by two ISApl1 elements (composite transposon) and is located directly upstream of the MDR region.
To our knowledge, this is the first description of an MDR IncHI2 plasmid harboring a composite transposon containing the mcr-1 gene and determinants for resistance to six different classes of antibiotics, including the ESBL gene blaCTX-M-1, and additional heavy metal resistance determinants. The fact that the mcr-1 gene is found to be part of a composite transposon is highly worrisome, since accelerated dissemination by illegitimate recombination of the whole transposon to either plasmids or chromosomes is likely.
Furthermore, we present for the first time an IncHI1 plasmid carrying the mobile colistin resistance determinant. The presence of heavy metal resistance determinants on pS38 and pH226B might lead to coselection, for example by zinc, which is widely used as a feed additive for fattening pigs.
Since the mcr-1 element was found on different plasmid types and the ISApl1 element was not always present, we hypothesize that the mcr-1 gene may have been mobilized independently several times. In the case of pH226B, however, where the mcr-1 gene is embedded in the middle of the backbone without any evidence for the presence of mobile elements, no conclusions concerning the mobilization of this gene can be made.
Accession number(s).
The GenBank accession numbers for pS38, pOW3E1, and pH226B are KX129782, KX129783, and KX129784, respectively.
ACKNOWLEDGMENTS
We are grateful to the staff of the Functional Genomics Center Zurich, particularly Anna Bratus, Andrea Patrignani, and Weihong Qi, for excellent technical support and bioinformatics support.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Shen Z, Wang Y, Shen Y, Shen J, Wu C. 2016. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 3.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbock L, Chakraborty T, for the RESET consortium. 2016. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhi C, Lv L, Yu LF, Doi Y, Liu JH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Lienhard R, Kieffer N, Clerc O, Poirel L. 2016. Plasmid-mediated colistin-resistant Escherichia coli in bacteremia in Switzerland. Clin Infect Dis 15:1322–1323. doi: 10.1093/cid/ciw124. [DOI] [PubMed] [Google Scholar]
- 6.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 18 April 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 7.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1 -harboring plasmids from extended spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4371–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zurfluh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. 2016. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother 60:2594–2595. doi: 10.1128/AAC.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zogg AL, Zurfluh K, Nüesch-Inderbinen M, Stephan R. 2016. Characteristics of ESBL-producing Enterobacteriaceae and meticillin-resistant Staphylococcus aureus (MRSA) isolated from Swiss and imported raw poultry meat collected at retail level. Schweiz Arch Tierheilk; http://www.gstsvs.ch/fileadmin/media/pdf/SAT_open/SAT_06_2016_Zogg.pdf. [DOI] [PubMed] [Google Scholar]
- 10.Aziz RK, Bartles D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]