Abstract
A novel mcr variant, named mcr-1.2, encoding a Gln3-to-Leu functional variant of MCR-1, was detected in a KPC-3-producing ST512 Klebsiella pneumoniae isolate collected in Italy from a surveillance rectal swab from a leukemic child. The mcr-1.2 gene was carried on a transferable IncX4 plasmid whose structure was very similar to that of mcr-1-bearing plasmids previously found in Escherichia coli and K. pneumoniae strains from geographically distant sites (Estonia, China, and South Africa).
TEXT
Transferable polymyxin resistance mediated by the plasmid-borne mcr-1 gene has recently been described in Enterobacteriaceae, raising considerable concern (1). The mcr-1 gene product is a membrane-anchored enzyme able to modify the lipid A polymyxin target by addition of phosphoethanolamine, resulting in a reduction of affinity for polymyxins (1).
The mcr-1 gene has mostly been found in Escherichia coli isolates from animals, but also in those from human and food samples, and in isolates of other enterobacterial species, with a worldwide distribution (2). The gene has often been reported in strains susceptible to other antibiotics, but occasionally also in multidrug-resistant (MDR) strains (3 – 8), including members of high-risk epidemic lineages spreading in the clinical setting (9, 10). Thus far, however, it had never been found in Klebsiella pneumoniae strains of clonal group 258 (CG258), which is the lineage mainly responsible for the dissemination of KPC-type carbapenemases on the global scale (11, 12).
Here we describe the first detection of a novel mcr variant, named mcr-1.2, from an MDR KPC-producing K. pneumoniae strain belonging to sequence type 512 (ST512), a member of CG258.
Screening for KPC carbapenemase-producing Enterobacteriaceae in rectal swabs was carried out using the direct KPC screening test (DKST), based on direct plating of rectal swabs onto MacConkey agar in the presence of a meropenem disc and of a meropenem-plus-phenylboronic acid disc (13). Bacterial identification was carried out by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek-MS; bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibility was determined by reference broth microdilution (14) using custom plates (Trek Diagnostic Systems, Cleveland, OH, USA), and data were interpreted according to the EUCAST guidelines (EUCAST breakpoint tables v6.0). Whole-genome sequencing (WGS) and analysis were carried out as previously described (15). Plasmid finishing was achieved by a PCR-based strategy and Sanger sequencing. Sequence alignments were performed using Mauve (16), and physical maps were generated using Easyfig (17). Determination of the multilocus sequence type, plasmid replicons, and resistance gene content was performed in silico using online tools (http://www.genomicepidemiology.org/). Transfer of the colistin resistance determinant by conjugation was assayed on Mueller-Hinton (MH) agar plates (Oxoid, Basingstoke, United Kingdom) with an initial donor/recipient ratio of 0.1, using E. coli J53 (F− met pro Azir [azide resistance]) as the recipient (18). After incubation at 35°C for 16 h, transconjugants were selected on MH agar supplemented with colistin (2 μg/ml) and sodium azide (150 μg/ml). The transfer frequency was expressed as the number of transconjugants per recipients (t/r). Transfer of mcr to transconjugants was confirmed by PCR targeting the mcr gene (10).
K. pneumoniae KP-6884 was isolated in 2014 from a rectal surveillance swab obtained from an Italian child admitted to the pediatric onco-hematology ward of Pisa University Hospital. The strain was preliminarily identified as a putative KPC producer by the DKST method. The child was receiving a cycle of anticancer chemotherapy for acute lymphoblastic leukemia and had previously been admitted twice to the same ward (1 and 2 months earlier). Of note, the child had not received colistin before the first isolation of KP-6884.
KP-6884 showed an MDR phenotype, including resistance to β-lactams, fluoroquinolones, trimethoprim-sulfamethoxazole, gentamicin, and colistin, remaining susceptible only to amikacin and tigecycline (Table 1). As such, KP-6884 exhibited an unusual phenotype (resistance to gentamicin and trimethoprim-sulfamethoxazole and susceptibility to amikacin) compared to that of the most prevalent carbapenem-resistant (CRE) KPC-producing K. pneumoniae strain circulating in our setting (19, 20).
TABLE 1.
MICs for K. pneumoniae KP-6884, E. coli J53 (pMCR1.2-IT), and E. coli J53
| Antibiotic | MIC (μg/ml) (category) |
||
|---|---|---|---|
| KP-6884a | J53 (pMCR1.2-IT) | J53 | |
| Amoxicillin-clavulanic acid | >8/2 (R) | 8/2 | 8/2 |
| Cefotaxime | >4 (R) | 0.12 | 0.12 |
| Ceftazidime | >128 (R) | 0.5 | 0.5 |
| Cefepime | >32 (R) | ≤1 | ≤1 |
| Ertapenem | >1 (R) | ≤1 | ≤1 |
| Imipenem | >16 (R) | ≤1 | ≤1 |
| Meropenem | 64 (R) | ≤0.12 | ≤0.12 |
| Piperacillin-tazobactam | >128/4 (R) | ≤2/4 | ≤2/4 |
| Ciprofloxacin | >2 (R) | ≤0.06 | ≤0.06 |
| Amikacin | 8 (S) | ≤4 | ≤4 |
| Gentamicin | >4 (R) | ≤1 | ≤1 |
| Trimethoprim-sulfamethoxazole | >4/76 (R) | ≤0.5/9.5 | ≤0.5/9.5 |
| Tigecycline | 0.5 (S) | 0.5 | 0.25 |
| Colistin | 8 (R) | 8 | ≤0.5 |
R, resistant; S, susceptible.
To investigate the mechanisms of resistance of KP-6884, a WGS approach was adopted. The draft genome was de novo assembled in 91 scaffolds (largest scaffold, 544,125 bp; N50, 270,452 bp; L50, 8; average GC, 57.09%), with an estimated genome size of 5,626,271 bp and average coverage of 80×. A total of 5,631 coding DNA sequences were identified using the PGAP annotation pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok).
In silico analysis of the draft genome confirmed the identification of KP-6884 as K. pneumoniae sensu stricto (21) and revealed that it belonged to ST512.
Screening for acquired resistance determinants revealed the presence of genes encoding β-lactamases (blaTEM-1, blaSHV-11, and blaKPC-3), aminoglycoside-modifying enzymes (aadA2, aadA5, aacA4, and aacC2d), sulfonamide resistance (sul1), trimethoprim resistance (dfrA17), and colistin resistance (a new mcr gene variant). Compared to mcr-1 (1), the sole allelic variant thus far described, the mcr gene from KP-6884 carried a missense mutation at position 8 (A→T) resulting in a Gln-to-Leu change in the N-terminal protein region. A BLAST search, using this novel allelic variant, here referred to as mcr-1.2, showed no perfect match with any of the mcr genes present in the nr/wgs databases (last accessed 11 May 2016). Overall, the acquired resistome of KP-6884 was consistent with the resistance phenotype, including the unusual gentamicin resistance justified by the presence of the aacC2d gene (Table 1). Other possible causes of colistin resistance, mediated by mutations in chromosomal genes (22, 23), were excluded by the analysis of sequence data. In detail, mgrB, pmrAB, phoPQ, and crrAB sequences did not show any genetic alteration previously or potentially associated with colistin resistance (the sequences were identical to those of colistin-susceptible strains).
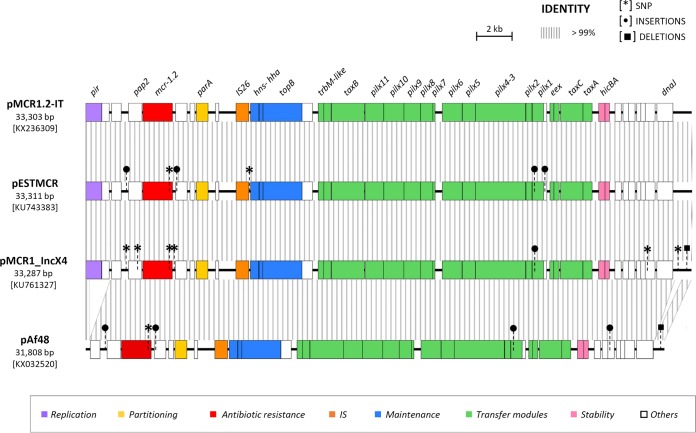
Bioinformatic analysis revealed the presence of several plasmid replicons, including ColE, IncFIA-FIB, IncFII-FIB, IncX3, and IncX4. The latter was located on the same contig as mcr-1.2. Plasmid finishing resulted in a 33.3-kb-long circular molecule, representing the complete sequence of the IncX4 plasmid carrying mcr-1.2, named pMCR1.2-IT (Fig. 1). Overall, pMCR1.2-IT was very similar to other previously sequenced IncX4 plasmids carrying mcr-1, namely, pMCR1-IncX4 (GenBank accession no. KU761327) (24), pESTMCR (GenBank accession no. KU743383), and pAf48, from a clinical K. pneumoniae isolate from China, an E. coli isolate from pig sludge in Estonia, and an E. coli clinical isolate from South Africa (GenBank accession no. KX032520), respectively (25) (Fig. 1). In all these plasmids, the mcr gene was embedded in the same genetic environment, with a downstream gene encoding the PAP2 transmembrane protein and without an upstream ISApl1, unlike what has been observed in other non-IncX4 plasmids (24) (Fig. 1). These findings underscored the broad intercontinental distribution of this type of resistance plasmid.
FIG 1.
Linear map of plasmid pMCR1.2-IT and comparison with previously sequenced mcr-1-carrying IncX4 plasmids. Homologous segments are indicated by striped shading, representing ≥99% sequence identity. Genes encoding proteins of known functions are in different colors, as detailed in the legend. Differences at the sequence level, including single nucleotide polymorphisms (SNPs), insertions (IS), and deletions, are shown by different symbols. Compared to pMCR1.2-IT, pESTMCR, pMCR1-incX4, and pAf48 differed in 2, 6, and 1 SNPs, respectively. In pESTMCR, SNPs were located within the mcr-1 gene and in an intergenic region adjacent to hns (n = 1). In pMCR1-IncX4, SNPs were located within the mcr-1 gene, within pap2 (n = 1), in intergenic regions (n = 2), in a gene coding for a hypothetical protein (n = 1), and in the iteron region (n = 1); in genes coding for the PAP2 transmembrane protein and for MCR, nucleotide substitutions led to missense mutations. In pAf48, the single SNP was located within the mcr gene; deletions of 1,160 bp and of 388 bp were present in the replication region, affecting the pir gene and a region upstream of dnaJ, respectively. Other differences consisted of the following: (i) a 22-bp deletion located in the iteron region in pMCR1-incX4; (ii) an insertion of a single nucleotide downstream of the gene coding for the PAP2 protein and of 5 nucleotides within a gene encoding a hypothetical protein upstream of mcr-1 in pESTMCR and pAf48; (iii) a 48-bp insertion within a gene encoding a hypothetical protein located between hicA and dnaJ and a 2-bp deletion within the iteron region in pAf48; and (iv) a single nucleotide insertion within the pilX1 gene in pMCR1-incX4, pESTMCR, and pAf48.
Gene transfer experiments demonstrated that the mcr-1.2 gene could be transferred by conjugation from KP-6884 to E. coli J53 with a frequency of 5 × 10−5 (t/r). The colistin MIC of transconjugants showed a 16-fold increase (Table 1), confirming that MCR-1.2 was functional. The MICs of transconjugants for all other agents were unchanged.
Concluding remarks.
To the best of our knowledge, this is the first description of a novel MCR-1 functional variant and also the first time that an mcr-type gene has been found to be associated with an ST512 KPC-3-producing high-risk clone of K. pneumoniae. Until now, only a few cases of human infections caused by carbapenemase-producing (NDM-type, VIM-1, KPC-2, OXA-48), mcr-positive strains of E. coli and K. pneumoniae have been reported (3 – 6, 24, 26).
Considering that the clinical use of polymyxins is essentially restricted to the treatment of invasive infections caused by carbapenem-resistant and extensively drug-resistant Gram-negative nonfermenters, the emergence of transferable colistin resistance among CRE is a cause of serious concern, especially in settings of high CRE endemicity. In Pisa University Hospital, where CRE strains have been endemic since 2010 (13), the rate of colistin resistance among carbapenem-resistant K. pneumoniae isolates of clinical origin was 15% in the year 2014.
Strain KP-6884, however, was responsible for colonization of an immunocompromised patient in whom the strain did not cause an invasive infection. Therefore, the pathogenic potential of KP-6884 and of strains with similar features should be further investigated in appropriate infection models.
Accession numbers.
The draft genome of K. pneumoniae KP-6884 and the complete sequence of pMCR1.2-IT have been deposited at DDBJ/EMBL/GenBank under accession no. LXQO00000000 and KX236309, respectively.
REFERENCES
- 1.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Poirel L. 2016. Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin Microbiol Infect 22:398–400. doi: 10.1016/j.cmi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Falgenhauer L, Waezsada S-E, Yao Y, Imirzalioglu C, Sbohrer AK, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T7; RESET consortium. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. 2016. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 16:281. doi: 10.1016/S1473-3099(16)00006-2. [DOI] [PubMed] [Google Scholar]
- 5.Du H, Chen L, Tang Y-W, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 6.Mulvey MR, Mataseje LF, Robertson J, Nash JHE, Boerlin P, Toye B, Irwin R, Melano RG. 2016. Dissemination of the mcr-1 colistin resistance. Lancet Infect Dis 16:289–290. doi: 10.1016/S1473-3099(16)00067-0. [DOI] [PubMed] [Google Scholar]
- 7.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 20 June 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the USA. Antimicrob Agents Chemother doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giufrè M, Monaco M, Accogli M, Pantosti A, Cerquetti M, PAMURSA Study Group. 3 June 2016. Emergence of the colistin resistance mcr-1 determinant in commensal Escherichia coli from residents of long-term-care facilities in Italy. J Antimicrob Chemother doi: 10.1093/jac/dkw195. [DOI] [PubMed] [Google Scholar]
- 9.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:30085–30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 10.Cannatelli A, Giani T, Antonelli A, Principe L, Luzzaro F, Rossolini GM. 2016. First detection of the mcr-1 colistin resistance gene in Escherichia coli, Italy. Antimicrob Agents Chemother 60:3257–3258. doi: 10.1128/AAC.00246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 12.Rossolini GM. 2015. Extensively drug-resistant carbapenemase-producing Enterobacteriaceae producing carbapenemases: an emerging challenge for clinicians and healthcare systems. J Intern Med 277:528–531. doi: 10.1111/joim.12350. [DOI] [PubMed] [Google Scholar]
- 13.Giani T, Tascini C, Arena F, Ciullo I, Conte V, Leonildi A, Menichetti F, Rossolini GM. 2012. Rapid detection of intestinal carriage of Klebsiella pneumoniae producing KPC carbapenemase during an outbreak. J Hosp Infect 81:119–122. doi: 10.1016/j.jhin.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute (CLSI). 2015. M07-A10. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically; approved standard, 10th ed. CLSI, Wayne, PA. [Google Scholar]
- 15.Arena F, Henrici De Angelis L, Pieralli F, Di Pilato V, Giani T, Torricelli F, D'Andrea MM, Rossolini GM. 2015. Draft genome sequence of the first hypermucoviscous Klebsiella quasipneumoniae subsp. quasipneumoniae isolate from a bloodstream infection. Genome Announc 3:e00952-15. doi: 10.1128/genomeA.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sennati S, Riccobono E, Di Pilato V, Villagran AL, Pallecchi L, Bartoloni A, Rossolini GM. 2016. pHN7A8-related multiresistance plasmids (blaCTX-M-65, fosA3 and rmtB) detected in clinical isolates of Klebsiella pneumoniae from Bolivia: intercontinental plasmid dissemination? J Antimicrob Chemother 71:1732–1734. doi: 10.1093/jac/dkv506. [DOI] [PubMed] [Google Scholar]
- 19.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R; AMCLI-CRE Survey Participants, Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill 18:20489 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489. [PubMed] [Google Scholar]
- 20.Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, Network EuSCAPE-Italy, Grundmann H, Pantosti A, Rossolini GM. 2014. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill 19:20939. doi: 10.2807/1560-7917.ES2014.19.42.20939. [DOI] [PubMed]
- 21.Brisse S, Passet V, Grimont PAD. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 22.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang Y-W, Kreiswirth BN, Chen L, Du H. 20 June 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates, South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang YW, Kreiswirth BN, Du H, Chen L. 23 May 2016. Detection of mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae (CRE) from different hospitals in China. Antimicrob Agents Chemother doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]