Abstract
Against extensively drug-resistant (XDR) Enterobacter cloacae, combination antibiotic therapy may be the only option. We investigated the activity of various antibiotics in combination with polymyxin B using time-kill studies (TKS). TKS were conducted with four nonclonal XDR E. cloacae isolates with 5 log10 CFU/ml bacteria against maximum, clinically achievable concentrations of polymyxin B alone and in two-drug combinations with 10 different antibiotics. A hollow-fiber infection model (HFIM) simulating clinically relevant polymyxin B and tigecycline dosing regimens was conducted for two isolates over 240 h. Emergence of resistance was quantified using antibiotic-containing (3× MIC) media. Biofitness and stability of resistant phenotypes were determined. All XDR E. cloacae isolates were resistant to all antibiotics except for polymyxin B (polymyxin B MIC, 1 to 4 mg/liter). All isolates harbored metallo-β-lactamases (two with NDM-1, two with IMP-1). In single TKS, all antibiotics alone demonstrated regrowth at 24 h, except amikacin against two strains and polymyxin B and meropenem against one strain each. In combination TKS, only polymyxin B plus tigecycline was bactericidal against all four XDR E. cloacae isolates at 24 h. In HFIM, tigecycline and polymyxin B alone did not exhibit any killing activity. Bactericidal kill was observed at 24 h for both isolates for polymyxin B plus tigecycline; killing was sustained for one isolate but regrowth was observed for the second. Phenotypically stable resistant mutants with reduced in vitro growth rates were observed. Polymyxin B plus tigecycline is a promising combination against XDR E. cloacae. However, prolonged and indiscriminate use can result in resistance emergence.
INTRODUCTION
Enterobacter cloacae isolates are nosocomial pathogens responsible for a diversity of infections. These organisms exhibit a remarkable adaptive capability and can acquire resistance to several antibiotics (1). All bacteria of the Enterobacter genus are intrinsically resistant to ampicillin and narrow-spectrum cephalosporins; in addition, resistance to carbapenems is often conferred through porin loss plus overexpression of beta-lactamases and/or by acquisition of plasmid-mediated carbapenemases (2). Over the past decade, there has been an exponentially increasing number of publications reporting the occurrence of Klebsiella pneumoniae carbapenemase (KPC) enzymes and New Delhi metallo-β-lactamase-1 (NDM-1) carbapenemases in E. cloacae (3). Strains carrying these enzymes often also harbor a diversity of resistance levels to other classes of antibiotics, resulting in the development of extensively drug-resistant (XDR) and pan-drug-resistant (PDR) E. cloacae.
The development of such extensive resistance, coupled with a lack of novel antibiotics in the drug development pipeline, has created a demand for a reevaluation of current antibiotic regimens (4). Polymyxin B is a polypeptide antibiotic commercially released in the 1950s (5). Despite being available for more than 50 years, it has only recently been revived as a last resort against extensively drug-resistant Gram-negative bacteria (XDR-GNB). The use of polymyxin B as a monotherapy against XDR-GNB is debatable. Increasing reports of polymyxin heteroresistance against several Gram-negative organisms have suggested that rapid resistance to polymyxins can develop as a result of polymyxin B monotherapy, and that polymyxin B is best administered as part of a combination therapy (6, 7). Unfortunately, to date the optimal antibiotic combinations against XDR E. cloacae remain unclear. While there is an increasing number of studies exploring effective combinations against XDR E. cloacae, most studies reported effective combinations in vitro against XDR E. cloacae using time-kill studies (TKS) or checkerboard assays, and none have validated the suggested combinations in a hollow-fiber infection model (HFIM), where the bacteria are subjected to clinically relevant fluctuating concentrations (8, 9).
In this study, we (i) investigated the in vitro activity of various antibiotics in combination with polymyxin B using TKS against drug-resistant E. cloacae using clinically relevant antibiotic concentrations, (ii) validated the findings in an HFIM, where the organisms will be subjected to clinically relevant fluctuating drug concentrations, and (iii) detected further emergence of resistance of these XDR E. cloacae strains when subjected to the clinically relevant fluctuating drug concentrations and characterized the phenotypic changes and overall biofitness associated with this further emergence of resistance development.
(This study was presented in part at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 9 to 12 September 2012, and the 23rd European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 27 to 30 April 2013.)
MATERIALS AND METHODS
Bacterial isolates.
Four nonclonal clinical carbapenem-resistant E. cloacae strains (ECL9800, ECL15118, ECL2197, and ECL6217) employed in this study were identified from a 1,700-bed tertiary public hospital in Singapore from 2009 to 2011, collected as part of a national surveillance study. Genus identity was previously determined using Vitek 2 ID-GN cards (bioMérieux, Inc., Hazelwood, MO). The isolates were stored at −70°C, and fresh isolates were subcultured twice on 5% blood agar plates (Thermo Scientific, Malaysia) for 24 h at 35°C before each experiment.
Susceptibility testing and molecular mechanisms of resistance.
MICs of all tested antibiotics were determined for all isolates using commercial custom-made broth microdilution panels (Trek Diagnostics, East Grinstead, United Kingdom), performed in accordance with the manufacturer's recommendations. The studies were conducted in duplicate and were repeated at least once on a separate day. Fresh isolates were subcultured twice on 5% blood agar plates for 24 h at 35°C before MIC testing. A multiplex PCR assay with five different primer pairs was employed to detect genes encoding commonly acquired metallo-β-lactamases (MBLs) (blaVIM, blaIMP, blaSIM, blaGIM, and blaSPM) (10). The presence of genes encoding extended-spectrum beta-lactamases (ESBLs), plasmid-mediated AmpCs, NDM, and KPCs were determined using PCR (10). Changes in porin gene expression (OmpC and OmpF) were determined using reverse transcriptase PCR, and the presence of efflux pumps was determined using the efflux pump inhibitor phenyl-arginine-β-naphthylamide (PAβN) (50 μg/ml) (11, 12).
Antimicrobial agents.
A total of 11 antibiotics were employed (Table 1). Stock solutions of all antimicrobial agents except tigecycline and rifampin were prepared in sterile water, aliquoted, and stored at −70°C. Tigecycline in solution was freshly prepared before each experiment. Rifampin was dissolved in dimethyl sulfoxide (DMSO) and was then serially diluted in sterile water to the desired final drug concentration. The final DMSO concentration (<1%, vol/vol) had no effect on E. cloacae growth. Prior to each experiment, the drug was thawed and diluted to the desired concentrations with cation-adjusted Mueller-Hinton II broth (Ca-MHB) (BBL, Sparks, MD).
TABLE 1.
Simulated antibiotic dosing regimens and corresponding drug concentrations
| Drug(s) | Simulated dosing regimen | Concn (mg/liter) | Reference |
|---|---|---|---|
| Amikacin | 15-20 mg/kg of body wt every 24 h | 80 | 13 |
| Cefepime | 2 g every 8 h | 200 | 14 |
| Levofloxacin | 750 mg every 24 h | 8 | 15 |
| Rifampin | 600 mg every 12 h | 2 | 16 |
| Tigecycline | 100 mg every 12 h | 2 | 17 |
| Meropenem | 2 g every 8 h (infused over 3 h) | 64 | 18 |
| Polymyxin B | 30,000 IU/kg/day or at least 1 MU every 12 h | 2 | 19 |
| Imipenem | 1 g every 6 h (infused over 0.5 h) | 32 | 20 |
| Doripenem | 1 g every 8 h (infused over 4 h) | 13 | 21 |
| Aztreonam | 6 g every 24 h (infused over 24 h) | 18 | 22 |
| Piperacillin-tazobactam | 4.5 g every 6 h (infused over 4 h) | 75/15 | 23 |
TKS.
TKS was performed for each antibiotic individually and in 2-drug combination with polymyxin B, using clinically achievable unbound concentrations (13 – 23). The simulated steady-state free peak concentrations of the following antibiotics and their corresponding representative doses are shown in Table 1. To perform TKS, an overnight bacterial culture was prepared using Ca-MHB and incubated at 35°C until log-phase growth. The bacteria were further diluted, and 15 ml was transferred to sterile flasks, each containing 1 ml of drug(s) at 16 times the target concentration. This gave a final inoculum concentration of approximately 5 log10 CFU/ml (1 × 105 CFU/ml to 5 × 105 CFU/ml) in each flask. The flasks were incubated in a shaker water bath at 35°C, and serial samples were obtained in duplicate at 0, 2, 4, 8, and 24 h. Each sample was centrifuged at 10,000 × g for 15 min (Eppendorf 5417R centrifuge; Hamburg, Germany) and reconstituted with sterile normal saline to its original volume to minimize drug carryover effect. Total bacterial count was quantified by depositing serial 10-fold dilutions of the broth sample onto Mueller-Hinton agar (MHA) plates using a spiral plater (Interscience, St. Nom La Breteche, France). Inoculated plates were incubated at 35°C for 18 to 24 h and enumerated visually. The lower limit of detection for the colony counts was 2.6 log10 CFU/ml.
HFIM.
To validate the activity of bactericidal combinations against the XDR E. cloacae strains in our TKS, an HFIM in which the bacteria were exposed to clinically relevant fluctuating drug concentrations over time (due to repeated dosing and constant elimination) was employed. A schematic diagram of the HFIM has been described previously (24). The antibiotic(s) was directly injected into the central reservoir to clinically achievable peak concentrations. Fresh, drug-free Ca-MHB was continuously infused from the diluent reservoir into the central reservoir to simulate drug elimination in humans, with equal volumes of drug-containing medium removed concurrently to maintain an isovolumetric system. Bacteria were inoculated into the extracapillary compartment of the hollow-fiber cartridge (Fibercell Systems, Inc., Frederick, MD). The bacteria were confined to the extracapillary compartment but exposed to the fluctuating drug concentration(s) in the central reservoir by means of an internal circulatory pump in the bioreactor loop. The experimental setup was slightly modified for the combination setup to account for the different half-lives (t1/2) of the two antibiotics (25).
Experimental setup.
Two XDR E. cloacae strains (ECL15118 and ECL9800) were employed in the HFIM testing. HFIM testing was conducted for up to 240 h in a humidified incubator set at 35°C. To prepare the inoculum, overnight cultures of the isolates were prepared as described above, and 20 ml of each E. cloacae suspension was inoculated into the extracapillary compartment of the hollow-fiber cartridge. The final inoculum concentration in the extracapillary compartment was approximately 5 log10 CFU/ml (1 × 105 CFU/ml to 5 × 105 CFU/ml). The most promising combination regimen based on the activity in TKS was selected for the HFIM experiment. For objective comparison, the respective single-drug regimens of the selected combination and a placebo control were tested. Each infection model was subjected to different drug exposures, simulating steady-state pharmacokinetic profiles of unbound drug corresponding to clinical doses simulated in the TKS, with maintenance doses to reattain the targeted concentrations. The effectiveness of the different regimens was compared based on the observed viable bacterial burden over time, as described below.
Microbiological response.
To determine the effect of the drug exposures on the total and resistant bacterial burden over time in the HFIM, serial samples were obtained from each infection model at 0 (baseline), 4, 8, 12, 24 (predose), 28, 32, 36, 48, 60, 72, 84, 96, 120, 144, 168, 192, 216, and 240 h in duplicate and quantified as described above. For total bacterial population, the samples were cultured on drug-free MHA plates, while for resistant bacterial subpopulations, the samples were cultured on MHA supplemented with the exposed agent at 3 times the MIC of the organism.
Pharmacokinetic validation and drug assay.
Serial drug samples were obtained from the central reservoir of the infection models and kept frozen at −70°C until analysis (within 1 month from the completion of the hollow-fiber infection model studies). Drug concentrations in these samples were assayed using a validated liquid chromatography-mass spectrophotometry (LC-MS) method. Assay of tigecycline was performed on an Agilent 1290 Infinity ultraperformance liquid chromatography (UPLC) system (Santa Clara, CA) interfaced with an Agilent 6430 triple-quadrupole mass spectrometer (QQQ MS) equipped with an electrospray ionization (ESI) source (Santa Clara, CA), as described in a previous study (26). For polymyxin B, the LC-MS consisted of an Agilent 1290 Infinity UPLC (Santa Clara, CA, USA) interfaced with the AB SCIEX QTRAP 5500 tandem mass spectrometer (MS/MS) (Framingham, MA, USA) equipped with an ESI source. The column and autosampler temperatures were maintained at 60°C and 4°C, respectively. The chromatographic column (Acquity UPLC BEH HILIC column; 1.7 μm volume, 2.1 mm by 100 mm; Waters, Milford, MA, USA) was operated with mobile phases consisting of 0.1% formic acid and 10 mM ammonium formate in acetonitrile (solvent A) and water (solvent B) delivered at 0.45 ml/min. The optimized elution conditions were a linear gradient of 5 to 35% B (0 to 2.5 min), isocratic conditions of 95% B (2.51 to 3.5 min), and reequilibration to 5% (3.51 to 4 min). Chromatographic peak integration was performed using the Analyst software. The concentration-time profiles of both antibiotics were modeled by fitting a one-compartment linear model to the observations using ADAPT II software (27).
Emergence-of-resistance studies.
To characterize the resistant strains isolated during the course of HFIM testing, resistant isolates recovered from antibiotic-supplemented plates (at baseline and throughout the experiment) were stored, and MIC testing with the agent used was repeated on at least two bacterial colonies to determine the presence of resistance. Susceptibility to the other antimicrobial agents was performed using commercial custom-made broth microdilution panels, as described above, to look for potential changes in resistance. To determine the stability of the resistant phenotypes, resistant isolates (if present) were subjected to 20 days of passaging on drug-free MHA and MHA supplemented with the exposed antibiotic at 3 times the MIC, and susceptibility testing for all of the antimicrobial agents was performed at the 10-day and 20-day time points. Finally, the growth kinetics of the resistant isolates was compared to that of the original isolate using in vitro time-growth studies. To carry out the time-growth studies, overnight culture of the isolates was prepared as described above, and 24 ml of the suspension was transferred to 50-ml sterile conical flasks until the final concentration of the bacterial suspension in each flask was approximately 5 log10 CFU/ml (1 × 105 CFU/ml to 5 × 105 CFU/ml). The flasks were then incubated in a shaker water bath at 35°C, serial samples were obtained from each flask at 0 (baseline), 1, 2, 3, 4, 5, 6, and 24 h after incubation, and bacterial load was determined by quantitative cultures as described above. The exponential growth of the bacterial population over 24 h was analyzed using an adapted mathematical model (28).
Definitions and pharmacodynamic endpoints.
XDR was defined as nonsusceptibility to at least one agent in all but two or fewer antimicrobial categories (29). Bactericidal activity (primary endpoint) was defined as a 3 log10 CFU/ml decrease (99.9% kill) in the colony count from the initial inoculum at 24 h (30). Synergy (secondary endpoint) was defined as a 2 log10 CFU/ml decrease in the colony count by the drug combination compared with its most active constituent and a 2 log10 CFU/ml decrease from the initial inoculum at 24 h, while indifference was defined as a 2 log10 CFU/ml change at 24 h by the combination compared with that by the most active single agent (30).
RESULTS
Susceptibility testing and molecular mechanisms of resistance.
All isolates were resistant to penicillins, cephalosporins, carbapenems, fluoroquinolones, and aminoglycosides (data not shown). No interpretative standards are provided by the Clinical and Laboratory Standards Institute (CLSI) for Enterobacteriaceae against polymyxin B or tigecycline. The MICs of the four isolates for polymyxin B and tigecycline, as well as molecular mechanisms of resistance, are shown in Table 2. Of note, all four XDR E. cloacae isolates harbored genes encoding metallo-beta-lactamases (two with blaIMP-1, two with blaNDM-1). Reductions in OmpC and OmpF porin expression were observed in all four isolates. Addition of PAβN resulted in a decrease in levofloxacin MICs in three strains, suggesting the presence of efflux pumps in these strains.
TABLE 2.
Polymyxin B and tigecycline MICs (mg/liter) and mechanisms of resistance in the 4 XDR E. cloacae strains
| Enterobacter cloacae strain | MIC (mg/liter) |
Resistance mechanism(s) | |
|---|---|---|---|
| Polymyxin B MIC | Tigecycline MIC | ||
| ECL15118 | 1 | 2 | SHV, TEM, NDM; porin loss |
| ECL9800 | 4 | 4 | SHV, TEM, IMP-1; presence of efflux; porin loss |
| ECL6217 | 1 | 0.5 | NDM; presence of efflux; porin loss |
| ECL2197 | 1 | 1 | SHV, TEM, IMP-1; presence of efflux; porin loss |
TKS.
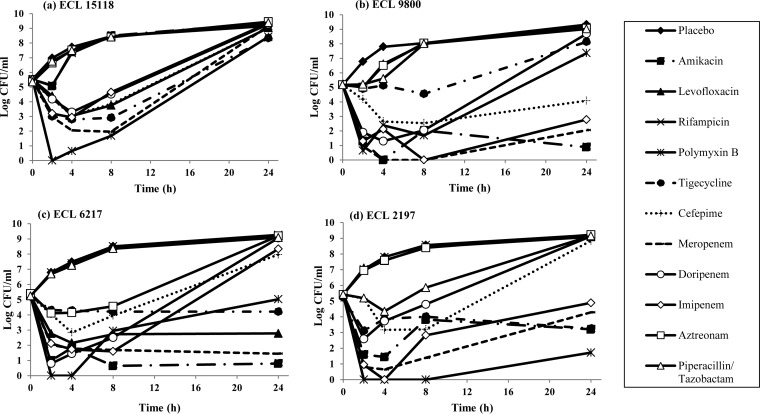
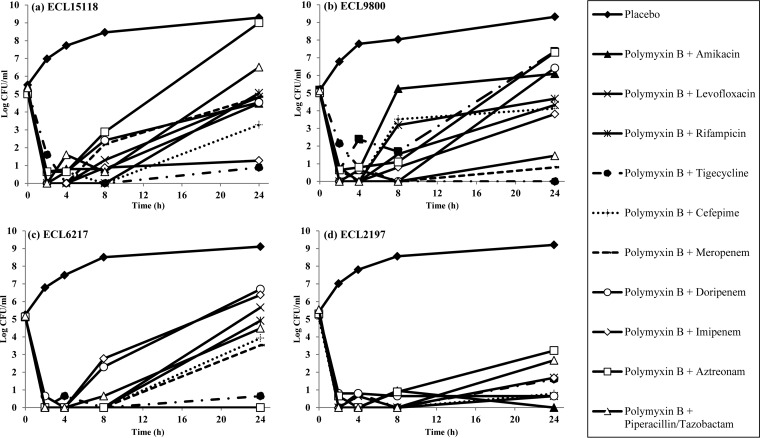
In single-drug TKS, all antibiotics alone demonstrated regrowth at 24 h, except amikacin against ECL9800 and ECL6217 and meropenem and polymyxin B against ECL6217 and ECL2197, respectively (Fig. 1). In combination time-kill studies, few two-drug combinations (2 to 3 combinations for each strain) were found to be bactericidal against ECL15118, ECL9800, and ECL6217. In contrast, a large number of two-drug combinations showed bactericidal activity against ECL2197. Polymyxin B plus tigecycline was the only combination that was consistently bactericidal against all four E. cloacae isolates, and it demonstrated synergism against 3/4 strains. Polymyxin plus amikacin, polymyxin plus meropenem, and polymyxin plus imipenem were bactericidal against 2/4 isolates and demonstrated synergism in 2/4, 1/4, and 1/4 isolates, respectively (Fig. 2).
FIG 1.
Microbiological response of ECL15118 (a), ECL9800 (b), ECL6217 (c), and ECL2197 (d) over 24 h when exposed to various single antibiotics in TKS. In single-drug TKS, amikacin (filled square) was bactericidal against ECL9800 and ECL6217 at 24 h, while meropenem (dash) and polymyxin B (asterisk) were bactericidal at 24 h against ECL6217 and ECL2197, respectively.
FIG 2.
Microbiological response of ECL15118 (a), ECL9800 (b), ECL6217 (c), and ECL2197 (d) over 24 h when exposed to polymyxin B-containing combinations in TKS. Polymyxin B in combination with tigecycline (filled circle) was the only combination that was consistently bactericidal against all four E. cloacae isolates at 24 h.
Hollow-fiber infection model.
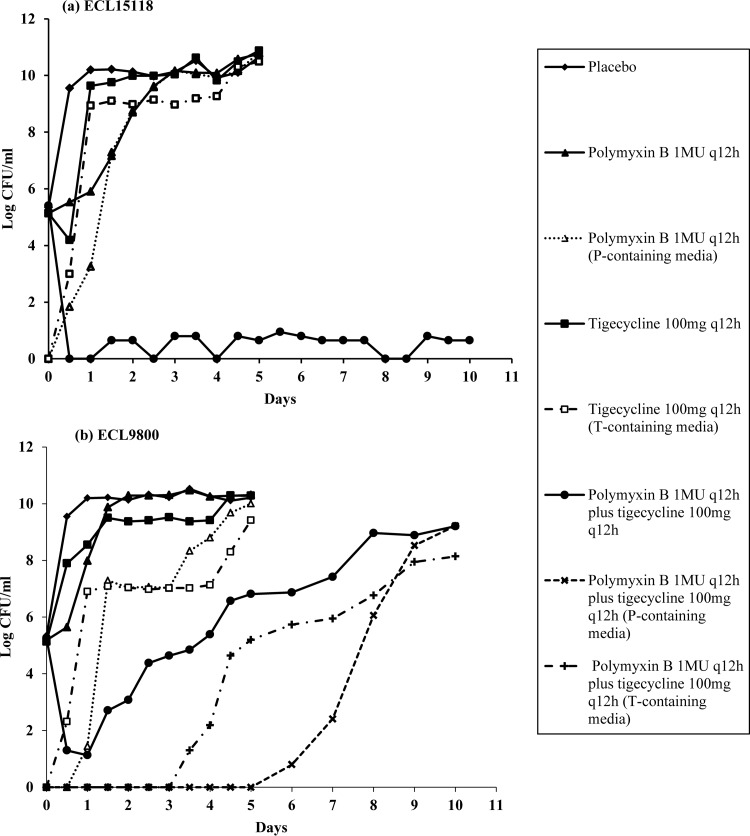
Polymyxin B plus tigecycline was chosen for HFIM studies, as it was the only two-drug combination that was consistently bactericidal against all four E. cloacae isolates. The time courses of bacterial burden for ECL15118 and ECL9800 against polymyxin B and tigecycline are shown in Fig. 3. Overall, observations in HFIM were in agreement with findings in the TKS: against both isolates, both tigecycline and polymyxin B alone did not exhibit any killing activity despite repeated dosing. In comparison, when polymyxin B was used in combination with tigecycline, bactericidal killing was seen at 24 h for both isolates; this suppression was sustained for ECL15118 for up to 240 h. For ECL9800, a slow but apparent regrowth was evident despite repeated dosing, reaching 9 log10 CFU/ml at 240 h. This increase in total bacterial burden was accompanied by an increase in polymyxin B- and tigecycline-resistant subpopulations in the respective drug-supplemented media.
FIG 3.
Microbiological responses of ECL15118 (a) and ECL9800 (b) against polymyxin B monotherapy, tigecycline monotherapy, and polymyxin B in combination with tigecycline in a hollow-fiber infection model.
Pharmacokinetic validation.
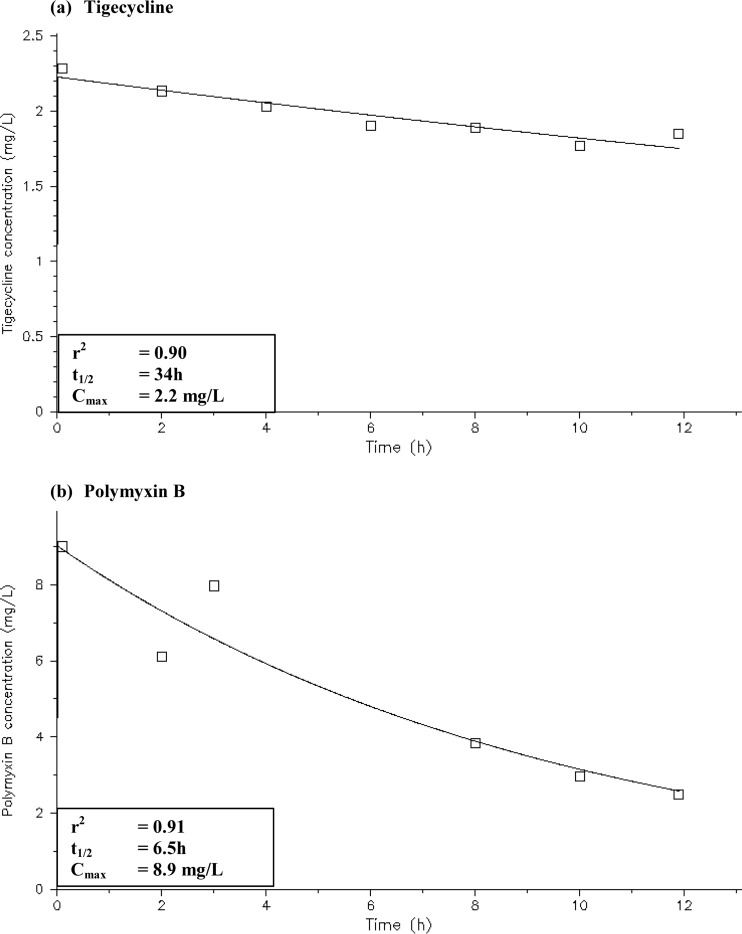
Simulated drug exposures in the infection models were satisfactory (Fig. 4). Simulated drug exposures had an r2 value of 0.90 to 0.91; the simulated polymyxin B and tigecycline half-lives were 6.5 h and 34 h, respectively. Intraday and interday accuracy for the lower-quality control and higher-quality control samples had a coefficient of variation (CV) of <5% (data not shown).
FIG 4.
Pharmacokinetic profiles of the infection models tigecycline at 100 mg every 12 h (a) and polymyxin B at 30,000 IU/kg/day or at least 1 MU every 12 h (b).
Emergence-of-resistance studies.
The MICs of isolates recovered from antibiotic-supplemented plates are shown in Table 3. As expected, an increase in MICs to the respective antibiotics was observed in isolates postexposure to polymyxin B alone [ECL15118(post-P), ≥32 mg/liter; ECL9800(post-P), 8 mg/liter] and tigecycline alone [ECL15118(post-T), 8 mg/liter, ECL9800(post-T), 8 mg/liter]. Cross-resistance to other antimicrobial agents in the screening panel was not observed. For ECL9800 isolates recovered postexposure to polymyxin B in combination with tigecycline [ECL9800(post-P+T)], an increase in MIC to both polymyxin B (16 mg/liter) and tigecycline was observed (16 mg/liter). Susceptibilities to all other antibiotics remained the same. MICs of the postexposure isolates remained similar at day 10 and day 20 of serial passage on drug-free media, which confirmed the stability of the resistant phenotypes in the postexposure mutant strains.
TABLE 3.
Emergence of resistance studiesa
| Strain | Kg (h−1) | MIC |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin | Levofloxacin | Rifampin | Polymyxin B | Tigecycline | Cefepime | Meropenem | Doripenem | Imipenem | Aztreonam | Ampicillin-sulbactam | Piperacillin-tazobactam | ||
| ECL15118 | 1.57 | ≥128 | ≥64 | ≥64 | 1 | 2 | ≥64 | ≥32 | ≥16 | ≥32 | ≥128 | ≥128/4 | ≥256/4 |
| ECL15118 (post-polymyxin B exposure) | 1.44 | ≥128 | ≥64 | ≥64 | ≥32 | 1 | ≥64 | ≥32 | ≥16 | ≥32 | ≥128 | ≥128/4 | ≥256/4 |
| ECL15118 (post-tigecycline exposure) | 1.57 | ≥128 | ≥64 | ≥64 | 1 | 8 | ≥64 | ≥32 | ≥16 | ≥32 | ≥128 | ≥128/4 | ≥256/4 |
| ECL9800 | 1.66 | 16 | ≥64 | ≥64 | 4 | 4 | 256 | ≥32 | ≥16 | 16 | ≥128 | ≥128/4 | ≥256/4 |
| ECL9800 (post-polymyxin B exposure) | 1.5 | 16 | ≥64 | ≥64 | 8 | 4 | 256 | ≥32 | ≥16 | 16 | ≥128 | ≥128/4 | ≥256/4 |
| ECL9800 (post-tigecycline exposure) | 1.56 | 16 | ≥64 | ≥64 | 4 | 8 | 256 | ≥32 | ≥16 | 16 | ≥128 | ≥128/4 | ≥256/4 |
| ECL9800 (post-polymyxin B plus tigecycline exposure) | 1.4 | 16 | ≥64 | ≥64 | 16 | 16 | 256 | ≥32 | ≥16 | 16 | ≥128 | ≥128/4 | ≥256/4 |
Shown are the susceptibility and growth rate of ECL15118 and ECL9800, pre- and post-exposure to polymyxin B, tigecycline, and polymyxin B in combination with tigecycline.
The mean best-fit growth rate constant (Kg) for ECL15118 and ECL9800 was 1.57 h−1 and 1.66 h−1, respectively (Table 3). A similar growth rate was observed in the ECL15118(post-T) strains; in the ECL15118(post-P) strains, however, a lower Kg of 1.44 h−1 was observed, suggesting the development of a substantial biofitness deficit as resistance to polymyxin B developed. For ECL9800, lower Kg values were observed for all postexposure mutant strains [ECL9800(post-P), 1.50 h−1; ECL9800(post-T), 1.56 h−1; ECL9800(post-P+T), 1.40 h−1], suggesting loss of biofitness in all strains after drug exposure as resistance to either polymyxin B and/or tigecycline developed.
DISCUSSION
The emergence of novel beta-lactamases with direct carbapenem-hydrolyzing capabilities in Enterobacteriaceae constitutes an important public health threat (1). Genes encoding these beta-lactamases are usually associated with various mobile genetic structures, permitting their ease of dissemination into different Enterobacteriaceae species (2). As infections caused by carbapenem-resistant Enterobacteriaceae (CRE) are associated with limited treatment options, combination antimicrobial therapy has been increasingly employed as the first-line therapy against CRE (2). However, the selection of the appropriate combination is a challenging process; in addition to achieving rapid bacterial kill, the selected combination should optimally suppress the development of resistance in order to preserve the utility of the antibiotic combination in the treatment of subsequent infection episodes (31).
To the best of our knowledge, ours is the first study to evaluate the activity of combinations in vitro against XDR E. cloacae using clinically relevant fluctuating drug concentrations. Unlike previous in vitro studies which had been conducted in KPC-producing carbapenem-resistant E. cloacae strains, our study focused on metallo-beta-lactamase (IMP and NDM)-harboring XDR E. cloacae strains, providing a fresh perspective compared to previous studies (8, 32). In addition, our study investigated the activity of polymyxin B in combination with various antibiotics, which is in contrast to most previous studies, which employed colistin in combination with other antibiotics against XDR E. cloacae (8, 9). Polymyxin B was employed in preference to colistin, as it has been shown to have superior clinical pharmacokinetics characteristics for infections (33). Unlike colistin, which may take several hours to reach desired peak concentrations even with loading doses, polymyxin B can rapidly achieve a desired plasma concentration that may then be maintained with a suitable daily dosage regimen (33). In addition, recent studies have suggested that polymyxin B is minimally eliminated renally; hence, adjustment of daily doses may not be required in patients with renal function (33, 34). As adequate dosing of antibiotics is pertinent in the treatment of CRE infections, we employed maximally clinically achievable free or unbound concentrations simulating maximally possible antibiotic doses for all tested antibiotics (Table 1). These concentrations were simulated based on parameters and results reported in previously published population pharmacokinetics studies and ensured that the killing effect observed in our TKS mimicked as closely as possible the killing that takes place in vivo (13 – 23).
In our TKS, we found that polymyxin B in combination with tigecycline is the most promising combination, demonstrating bactericidal activity against all four XDR E. cloacae strains at 24 h when exposed to static drug concentrations. As TKS poorly reflected antibiotic concentrations over time in vivo, we further validated the clinical applicability of the combinations in an HFIM. As expected, both tigecycline and polymyxin B monotherapies result in rapid regrowth for both strains, which was accompanied by the selective amplification of the resistant subpopulation over time, suggesting a strong selection pressure when nonoptimal antimicrobial regimens were used. The findings in our HFIM generally concurred with that of our TKS results, demonstrating bactericidal killing in both XDR E. cloacae strains in the HFIM at 24 h when polymyxin B was administered in combination with tigecycline. Interestingly, beyond 24 h, a slow and persistent regrowth was observed for ECL9800 but not ECL15118. This suggested the overall activity of antibiotic combinations was strain specific, and that prolonged and indiscriminate use of antibiotic combinations with initial bactericidal activity can result in the emergence of resistance in some XDR E. cloacae strains. The regrowth in ECL9800 was associated with the total bacterial population being replaced by mutants with reduced susceptibility to both polymyxin B and tigecycline over time, which ensued in the subsequent development of a polymyxin B- and tigecycline-resistant E. cloacae strain.
Upon further characterization of the resistant mutants, we found that the resistance phenotype was stable and did not revert to the susceptible phenotype despite repeated passages in drug-free media. This finding is similar to results published in a recent study by Lim et al. and is in contrast to previous findings in Pseudomonas aeruginosa, where susceptibility reversal was observed upon serial passage on drug-free medium (26, 35). Our findings suggest that the polymyxin B resistance in the XDR E. cloacae isolates is mutational and not adaptive in nature; however, this remains to be further studied. We observed a reduction in growth rate in most of the resistant mutants compared to the parent phenotype. This suggested that the development of several resistance mutations was associated with a corresponding biological fitness cost (36). Moving forward, the potential loss of virulence that may be associated with such loss of biofitness will be further explored.
To date, there have been a limited number of studies exploring antimicrobial combinations against XDR E. cloacae, and they were mainly conducted in KPC-harboring E. cloacae isolates. In a recent TKS study, polymyxin B plus carbapenems was found to be bactericidal against 6 KPC-producing CRE isolates, including two E. cloacae strains with high-level resistance to carbapenems (imipenem MIC, 64 mg/liter) (32). Polymyxin B plus tigecycline also showed bactericidal effect against both E. cloacae isolates; nonetheless, the reduction in colony count was lower than that of polymyxin B plus carbapenems (32). In another study, Betts et al. explored the activity of colistin plus tigecycline against CRE, including a tigecycline-resistant E. cloacae strain, by using standard checkerboard and time-kill assays and a simple invertebrate model (8). They found that colistin plus tigecycline was synergistic against the tigecycline-resistant E. cloacae strain in vitro, with significantly improved survival compared to either antibiotic when administered as a monotherapy. Interestingly, these were similar to our study findings, despite our XDR E. cloacae isolates having different underlying mechanisms of resistance (metallo-β-lactamases). The main advantages of our study were that, first, in addition to determining the activity of the potential combinations using static drug concentrations, we validated the combination in an in vitro HFIM system, subjecting the bacteria to humanized drug exposures. Second, we further characterized the resistant strains isolated during the course of testing, adding to the scarce literature on the properties of the resistant subpopulations in XDR E. cloacae that may develop during the course of treatment.
As with all in vitro studies, the main limitation of our study is that the clinical relevance of our findings, including the virulence of the resistant subpopulation, will need to be further confirmed. In addition, the usefulness of the TKS and HFIM studies is limited by their labor-intensive nature, resulting in an inability to provide combination results in a clinically relevant time frame. Moving forward, a reliable in vitro methodology with a rapid turnaround time will be valuable in identifying effective combinations for the treatment of patients with XDR E. cloacae infections.
Conclusion.
In our study, we found that polymyxin B in combination with tigecycline is a promising treatment against XDR E. cloacae. However, it must be highlighted that prolonged and indiscriminate use, especially if employed at suboptimal doses, can result in the emergence of resistance strains. Future studies investigating polymyxin B combinations in animal models and in patients, with additional studies to characterize the virulence of the resistance strains, will be required.
ACKNOWLEDGMENTS
We thank the staff of Singapore General Hospital and Changi General Hospital for assisting in collection of the bacterial isolates.
A.L.K., T.-P.L., and W.L. have received funding for research from Pfizer Inc. This work was supported in part by a Singapore General Hospital Research Grant (SRG/C3/02/2014), a National Medical Research Council Centre Grant (NMRC/CG/016/2013), and unrestricted grant funding from Pfizer Inc. (WS 832447 and WS 776979).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflicts of interest to declare.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. [DOI] [PubMed] [Google Scholar]
- 2.Schultsz C, Geerlings S. 2012. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 72:1–16. doi: 10.2165/11597960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Kwa AL, Tam VH, Falagas ME. 2008. Polymyxins: a review of the current status including recent developments. Ann Acad Med Singapore 37:870–883. [PubMed] [Google Scholar]
- 6.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother 66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 8.Betts JW, Phee LM, Hornsey M, Woodford N, Wareham DW. 2014. In vitro and in vivo activities of tigecycline-colistin combination therapies against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 58:3541–3546. doi: 10.1128/AAC.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin KH, Chuang YC, Lee SH, Yu WL. 2010. In vitro synergistic antimicrobial effect of imipenem and colistin against an isolate of multidrug-resistant Enterobacter cloacae. J Microbiol Immunol Infect 43:317–322. doi: 10.1016/S1684-1182(10)60049-7. [DOI] [PubMed] [Google Scholar]
- 10.Voets GM, Fluit AC, Scharringa J, Cohen Stuart J, Leverstein-van Hall MA. 2011. A set of multiplex PCRs for genotypic detection of extended-spectrum beta-lactamases, carbapenemases, plasmid-mediated AmpC beta-lactamases and OXA beta-lactamases. Int J Antimicrob Agents 37:356–359. doi: 10.1016/j.ijantimicag.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 12.Davies TA, Marie Queenan A, Morrow BJ, Shang W, Amsler K, He W, Lynch AS, Pillar C, Flamm RK. 2011. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J Antimicrob Chemother 66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 13.Tod M, Lortholary O, Seytre D, Semaoun R, Uzzan B, Guillevin L, Casassus P, Petitjean O. 1998. Population pharmacokinetic study of amikacin administered once or twice daily to febrile, severely neutropenic adults. Antimicrob Agents Chemother 42:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam VH, McKinnon PS, Akins RL, Drusano GL, Rybak MJ. 2003. Pharmacokinetics and pharmacodynamics of cefepime in patients with various degrees of renal function. Antimicrob Agents Chemother 47:1853–1861. doi: 10.1128/AAC.47.6.1853-1861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebuck JA, Fish DN, Abraham E. 2002. Pharmacokinetics of intravenous and oral levofloxacin in critically ill adults in a medical intensive care unit. Pharmacotherapy 2002:1216–1225. [DOI] [PubMed] [Google Scholar]
- 16.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. 2007. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother 51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodvold KA, Gotfried MH, Cwik M, Korth-Bradley JM, Dukart G, Ellis-Grosse EJ. 2006. Serum, tissue and body fluid concentrations of tigecycline after a single 100 mg dose. J Antimicrob Chemother 58:1221–1229. doi: 10.1093/jac/dkl403. [DOI] [PubMed] [Google Scholar]
- 18.Jaruratanasirikul S, Sriwiriyajan S, Punyo J. 2005. Comparison of the pharmacodynamics of meropenem in patients with ventilator-associated pneumonia following administration by 3-hour infusion or bolus injection. Antimicrob Agents Chemother 49:1337–1339. doi: 10.1128/AAC.49.4.1337-1339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwa AL, Lim TP, Low JG, Hou J, Kurup A, Prince RA, Tam VH. 2008. Pharmacokinetics of polymyxin B1 in patients with multidrug-resistant Gram-negative bacterial infections. Diagn Microbiol Infect Dis 60:163–167. doi: 10.1016/j.diagmicrobio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Sakka SG, Glauner AK, Bulitta JB, Kinzig-Schippers M, Pfister W, Drusano GL, Sorgel F. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob Agents Chemother 51:3304–3310. doi: 10.1128/AAC.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaruratanasirikul S, Wongpoowarak W, Kositpantawong N, Aeinlang N, Jullangkoon M. 2012. Pharmacodynamics of doripenem in critically ill patients with ventilator-associated Gram-negative bacilli pneumonia. Int J Antimicrob Agents 40:434–439. doi: 10.1016/j.ijantimicag.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Burgess DS, Summers KK, Hardin TC. 1999. Pharmacokinetics and pharmacodynamics of aztreonam administered by continuous intravenous infusion. Clin Ther 21:1882–1889. doi: 10.1016/S0149-2918(00)86736-3. [DOI] [PubMed] [Google Scholar]
- 23.Shea KM, Cheatham SC, Wack MF, Smith DW, Sowinski KM, Kays MB. 2009. Steady-state pharmacokinetics and pharmacodynamics of piperacillin/tazobactam administered by prolonged infusion in hospitalised patients. Int J Antimicrob Agents 34:429–433. doi: 10.1016/j.ijantimicag.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Bilello JA, Bauer G, Dudley MN, Cole GA, Drusano GL. 1994. Effect of 2′,3′-didehydro-3′-deoxythymidine in an in vitro hollow-fiber pharmacodynamic model system correlates with results of dose-ranging clinical studies. Antimicrob Agents Chemother 38:1386–1391. doi: 10.1128/AAC.38.6.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 15(Suppl A):125–130. doi: 10.1093/jac/15.suppl_A.125. [DOI] [PubMed] [Google Scholar]
- 26.Lim TP, Cai Y, Hong Y, Chan EC, Suranthran S, Teo JQ, Lee WH, Tan TY, Hsu LY, Koh TH, Tan TT, Kwa AL. 2015. In vitro pharmacodynamics of various antibiotics in combination against extensively drug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 59:2515–2524. doi: 10.1128/AAC.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Argenio DZ, Schumitzky A (ed). 1997. ADAPT II user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, University of Southern California, Los Angeles, CA. [Google Scholar]
- 28.Tam VH, Schilling AN, Nikolaou M. 2005. Modelling time-kill studies to discern the pharmacodynamics of meropenem. J Antimicrob Chemother 55:699–706. doi: 10.1093/jac/dki086. [DOI] [PubMed] [Google Scholar]
- 29.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A, vol 19. NCCLS, Wayne, PA. [Google Scholar]
- 31.Zavascki AP, Bulitta JB, Landersdorfer CB. 2013. Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 11:1333–1353. doi: 10.1586/14787210.2013.845523. [DOI] [PubMed] [Google Scholar]
- 32.Barth N, Ribeiro VB, Zavascki AP. 2015. In vitro activity of polymyxin B plus imipenem, meropenem, or tigecycline against KPC-2-producing Enterobacteriaceae with high MICs for these antimicrobials. Antimicrob Agents Chemother 59:3596–3597. doi: 10.1128/AAC.00365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, Lee W, Kwa AL. 2015. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther 13:1481–1497. doi: 10.1586/14787210.2015.1093933. [DOI] [PubMed] [Google Scholar]
- 34.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 35.Tam VH, Schilling AN, Vo G, Kabbara S, Kwa AL, Wiederhold NP, Lewis RE. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3624–3630. doi: 10.1128/AAC.49.9.3624-3630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelraouf K, Kabbara S, Ledesma KR, Poole K, Tam VH. 2011. Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. J Antimicrob Chemother 66:1311–1317. doi: 10.1093/jac/dkr105. [DOI] [PubMed] [Google Scholar]