Abstract
Human coronaviruses (HCoVs) cause 15 to 30% of mild upper respiratory tract infections. However, no specific antiviral drugs are available to prevent or treat HCoV infections to date. Here, we developed four infectious recombinant HCoVs-OC43 (rHCoVs-OC43) which express the Renilla luciferase (Rluc) reporter gene. Among these four rHCoVs-OC43, rOC43-ns2DelRluc (generated by replacing ns2 with the Rluc gene) showed robust luciferase activity with only a slight impact on its growth characteristics. Additionally, this recombinant virus remained stable for at least 10 passages in BHK-21 cells. rOC43-ns2DelRluc was comparable to its parental wild-type virus (HCoV-OC43-WT) with respect to the quantity of the antiviral activity of chloroquine and ribavirin. We showed that chloroquine strongly inhibited HCoV-OC43 replication in vitro, with a 50% inhibitory concentration (IC50) of 0.33 μM. However, ribavirin showed inhibition of HCoV-OC43 replication only at high concentrations which may not be applicable to humans in clinical treatment, with an IC50 of 10 μM. Furthermore, using a luciferase-based small interfering RNA (siRNA) screening assay, we identified double-stranded-RNA-activated protein kinase (PKR) and DEAD box RNA helicases (DDX3X) that exhibited antiviral activities, which were further verified by the use of HCoV-OC43-WT. Therefore, rOC43-ns2DelRluc represents a promising safe and sensitive platform for high-throughput antiviral screening and quantitative analysis of viral replication.
INTRODUCTION
Coronaviruses (CoVs) belong to the family Coronaviridae in the order Nidovirales (1). They have a positive-sense RNA genome of ∼30 kb in length, the largest found in any RNA viruses. CoVs infect avian species and a wide range of mammals, including humans (2). Currently, six CoVs that are able to infect humans have been identified, i.e., the four circulating strains human CoV 229E (HCoV-229E), HCoV-OC43, HCoV-HKU1, and HCoV-NL63 and the two emergent strains severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Indeed, in 2003, an outbreak of SARS first demonstrated the potentially lethal consequences of zoonotic CoV infections in humans. In 2012, a similar, previously unknown CoV emerged, MERS-CoV, which has thus far caused over 1,650 laboratory-confirmed infections, with a mortality rate of about 30% (3, 4). However, to date, no effective drug has been identified for the treatment of HCoV infections, and few host factors have been identified that restrict the replication of HCoV. The emergence of these highly pathogenic HCoVs has reignited interest in studying HCoV biology and virus-host interactions. Therefore, a safe and sensitive screening model is required for rapid identification of potential drugs and screening of antiviral host factors capable of inhibiting HCoV infection.
The introduction of a reporter gene into the viral genome provides a powerful tool for initial rapid screening and evaluation of antiviral agents. The unique CoV transcription mechanism allows efficient expression of reporter genes by inserting reporter genes under the control of transcription regulatory sequence (TRS) elements. To date a number of reporter CoVs have been generated (5 – 11), and several reporter CoVs have been applied to antiviral screening assays (10 – 14), but most of them are animal CoVs which cause disease in only one animal species and generally do not do so in humans. Among these reporter CoVs, only one reporter CoV (SARS-CoV-green fluorescent protein [GFP]) was based on HCoV and applied to a small interfering RNA (siRNA) library screening (14). However, the SARS-CoV-GFP assay lacks sensitivity and requires a high infectious dose (multiplicity of infection [MOI] of 10) for quantitative screening. Moreover, experiments with this reporter virus require a biosafety level 3 (BSL3) facility, which is costly and labor-intensive. Thus, it is critical to generate a safe and sensitive reporter HCoV for high-throughput screening (HTS) assays. Moreover, generation of a reporter HCoV is more suitable to screen drugs for clinical treatment than the reporter animal CoVs. HCoV-OC43 shows promise as a reporter virus for screening anti-HCoV drugs or identifying host factors. HCoV-OC43 was first isolated from a patient with upper respiratory tract disease in the 1960s; together with severe beta-CoVs (SARS-CoV and MERS-CoV), it belongs to the Betacoronavirus genus (15, 16), and these three virus strains have a high level of conservation for some essential functional domains, especially within 3CLpro, RdRp, and the RNA helicase, which represent potential targets for broad-spectrum anti-HCoV drug design (17, 18). Moreover, unlike SARS-CoV or MERS-CoV, HCoV-OC43 usually causes a mild respiratory tract disease and can be used for screening antivirals in a BSL2 facility. Furthermore, a small animal model of HCoV-OC43 has been developed and used successfully for antiviral trials (18, 19).
HCoV-OC43 carries two accessory genes, ns2 and ns12.9 (20). The ns2 gene, located between the nsp13 and HE gene loci, encodes a protein of unknown function. The ns12.9 gene, located between the S and E structural genes, encodes a protein that was recently demonstrated as a viroporin involved in HCoV-OC43 morphogenesis and pathogenesis (21). In this study, four infectious recombinant HCoVs-OC43 (rHCoVs-OC43) were generated based on the ATCC VR-759 strain of HCoV-OC43 by genetic engineering of the two accessory genes. Successfully rescued viruses were characterized and subsequently investigated for genetic stability. One reporter virus, rOC43-ns2DelRluc, showed robust Rluc activity and had growth kinetics similar to those of the parental wild-type HCoV-OC43 (HCoV-OC43-WT). Furthermore, this reporter virus was used successfully to evaluate the antiviral activity of Food and Drug Administration (FDA)-approved drugs and siRNA screening assays. Our study indicated that the replacement of accessory gene ns2 represents a promising target for the generation of reporter HCoV-OC43 and provides a useful platform for identifying anti-HCoV drugs and host factors relevant to HCoV replication.
MATERIALS AND METHODS
Plasmid construction.
The infectious full-length cDNA clone pBAC-OC43FL (22), containing a full-length cDNA copy of HCoV-OC43, was used as the backbone to generate four rHCoVs-OC43 (Fig. 1). The Rluc gene was amplified from pGL4.75hRluc/CMV vector (Promega) and introduced into the plasmid pBAC-OC43FL by standard overlapping PCR. Modified fragments of HCoV-OC43 cDNA, for replacing the ns2 gene with Rluc gene (between 21,523 and 22,915 nucleotides, inclusively) or in-frame insertion of the Rluc gene into the ns2 gene (between nucleotides 21,517 and 21,518, inclusively), were generated by overlapping PCR and cloned into NarI/PmeI-digested pBAC-OC43FL to generate pBAC-OC43-ns2DelRluc or pBAC-OC43-ns2FusionRluc. The same strategies were employed for replacing the ns12.9 gene with Rluc gene or in-frame insertion of the Rluc gene into the ns12.9 gene, resulting in plasmids pBAC-OC43-ns12.9StopRluc and pBAC-OC43-ns12.9FusionRluc, respectively. Further details are available on request. All final constructs were verified by Sanger sequencing.
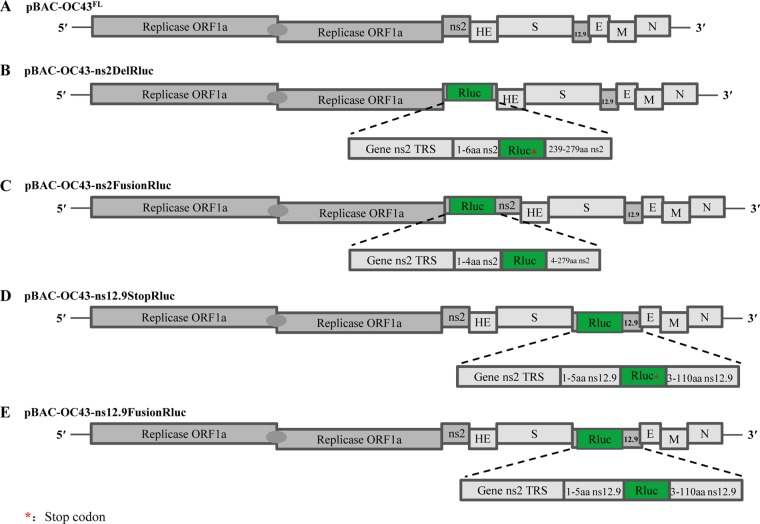
FIG 1.
Development of human coronavirus OC43 (HCoV-OC43) reporter systems. A schematic representation of the cDNA clone pBAC-OC43FL and recombinant cDNA clones of HCoV-OC43 harboring the Renilla luciferase (Rluc) gene, which was introduced into the accessory genes by overlapping PCR as described in Materials and Methods, is shown. The Rluc gene (green) is depicted. Expanded regions show the transcription regulatory sequence (TRS) control of Rluc gene expression.
Cells and antibodies.
BHK-21, HEK-293T, and Huh7 cells were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 2 mM l-glutamine (Sigma-Aldrich) and were incubated at 37°C with 5% CO2.
The anti-Renilla luciferase (ab185925), anti-double-stranded-RNA-activated protein kinase (anti-PKR) (ab32052), and anti-phosphorylated PKR (ab81303) rabbit monoclonal antibodies were purchased from Abcam. The anti-Flag (F7425) rabbit polyclonal antibody was purchased from Sigma-Aldrich. The anti-eIF2α (D7D3), anti-phosphorylated-eIF2α (Ser51) (D9G8), anti-DDX3X (D19B4), and anti-β-actin (13E5) rabbit monoclonal antibodies were obtained from Cell Signaling Technology. The infrared IRDye 800CW-labeled goat anti-mouse IgG(H+L) and IRDye 680RD goat anti-rabbit IgG were purchased from Li-Cor Biosciences.
Generation and titration of recombinant viruses.
The reporter viruses rOC43-ns2DelRluc, rOC43-ns2FusionRluc, rOC43-ns12.9StopRluc, and rOC43-ns12.9FusionRluc were rescued from the infectious cDNA clones pBAC-OC43-ns2DelRluc, pBAC-OC43-ns2FusionRluc, pBAC-OC43-ns12.9StopRluc, and pBAC-OC43-ns12.9FusionRluc, respectively. In brief, BHK-21 cells grown to 80% confluence were transfected with 4 μg of pBAC-OC43FL, pBAC-OC43-ns2DelRluc, pBAC-OC43-ns2FusionRluc, pBAC-OC43-ns12.9StopRluc, or pBAC-OC43-ns12.9FusionRluc using the X-tremeGENE HP DNA transfection reagent (Roche) according to the manufacturer's instructions. After incubation for 6 h at 37°C in a humidified 5% CO2 incubator, the transfected cells were washed three times with DMEM and maintained in DMEM supplemented with 2% FBS for 72 h at 37°C and an additional 96 h at 33°C. Next, the rHCoVs-OC43 were harvested by three freeze-thaw cycles followed by centrifugation at 2,000 × g for 20 min at 4°C. HCoV-OC43-WT was obtained from the full-length cDNA clone pBAC-OC43FL. All viruses were propagated in BHK-21 cells in DMEM supplemented with 2% FBS.
The titers of rHCoVs-OC43 were determined by indirect immunofluorescence assay (IFA). Briefly, BHK-21 cells in 96-well plates were infected with 10-fold-diluted viruses. The viral titers were determined at 72 h postinfection (hpi) by IFA and expressed as median tissue culture infective dose (TCID50)/ml, according to the method of Reed and Münch (23).
Determination of viral growth kinetics.
BHK-21 cells seeded on 48-well plates were infected with HCoV-OC43-WT or rHCoVs-OC43 at an MOI of 0.01. After 2 h of incubation at 33°C, cells were washed with phosphate-buffered saline (PBS), and the medium was replaced with fresh medium before incubation at 33°C. The supernatants (150 μl) were harvested at 24, 48, 72, 96, 120, 144, and 168 hpi, and 150 μl of fresh medium was added to the cells. The titer for each virus at the indicated time point was determined by IFA, as described above.
Rluc activity assay.
Analysis of Rluc expression was performed in 48- or 96-well plates. Briefly, BHK-21 cells or HEK-293T cells in plates were infected with rHCoVs-OC43 at an MOI of 0.01. At various time points postinfection, the cells in each well were assayed for relative light units (RLU) using the Renilla-Glo luciferase assay system (Promega) according to the manufacturer's instructions.
Dual-luciferase reporter assay system.
HEK-293T cells were seeded in 24-well plates at a cell density of 2.5 × 105 cells per well. The next day, cells were transfected with plasmids expressing DDX3X or TBK1 (150 or 300 ng), along with IFN-β-Luc and Rluc internal reference reporter plasmids. At 24 h posttransfection, cells were lysed and analyzed with the dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Western blot analysis.
Infected or uninfected cells were washed twice with PBS, lysed with NP-40 buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% NP-40, and 0.5 mM EDTA) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mg/ml protease inhibitor cocktail (Roche) for 30 min at 4°C. An equal volume of each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Pall). The membranes were blocked with 5% skim milk in PBS containing 0.5% Tween (PBST) for 1 h at room temperature and incubated with primary antibody overnight at 4°C. After washes with PBST, the membranes were further incubated for 1 h with infrared IRDye 800CW-labeled goat anti-mouse IgG(H+L) (1:10,000) (Li-Cor) or IRDye 680RD goat anti-rabbit IgG(H+L) (1:10,000) (Li-Cor), and blots were scanned on the Odyssey Infrared Imaging System (Li-Cor).
RNA isolation and RT-PCR.
Total RNA was extracted from virus-infected BHK-21 cells using TRIzol reagent (Invitrogen) and treated with DNase I to remove potential genomic DNA. The RNA concentration was quantified using a NanoDrop 2000 series spectrophotometer (Thermo Scientific). For reverse transcription-PCR (RT-PCR), two sets of primer pairs flanking the inserted reporter gene were used: one pair for rOC43-ns2FusionRluc and rOC43-ns2DelRluc (5′-GTG TAA GCC CAA GGT TGA GAT AG-3′)/(5′-GTC GTT CAG ATT GTA ATC ATA TTG −3′) and another for rOC43-ns129FusionRluc and rOC43-ns129DelRluc (5′-CAT ATG AAT ATT ATG TAA AAT GGC −3′)/(5′-GCC ATA AAC ATT TAA CTC CTG TC −3′). The PCR products were subjected to electrophoresis on a 1% agarose gel.
Real-time PCR.
Semiquantitative PCR was performed using the one-step SYBR PrimeScript RT-PCR kit (TaKaRa) according to the manufacturer's instructions. Fold induction values were calculated using the 2−ΔΔCT method, and mRNA expression was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Genomic RNA copies of HCoV-OC43-WT or rOC43-ns2DelRluc were quantified using quantitative RT-PCR as described previously (24). All primer sequences are shown in Table S1 in the supplemental material.
Stability of rHCoVs-OC43.
To examine the stability of the inserted Rluc genes, rHCoVs-OC43 and their parental HCoV-OC43-WT were passaged 13 times in BHK-21 cells (see Fig. 3A). Briefly, cells in a 25-cm2 flask were infected with the rescued rHCoVs-OC43 and HCoV-OC43-WT (defined as P0) at an MOI of 0.01. At 120 hpi, 300 μl of cell culture supernatants from the passaged virus (P1) were added to naive cells to generate passage 2 virus (P2). After 13 rounds of serial passage, viral RNA was extracted from the supernatant of infected cells of each passage (P0 to P13), the stability of the inserted reporter genes was detected by RT-PCR as described above, and the genes were cloned into the pMD18-T vector (TaKaRa) for Sanger sequencing (four clones were sequenced for each passage). The titers of rHCoVs-OC43 from P1 to P13 were determined using IFA. In addition, BHK-21 cells in 48-well plates were infected with each passage of rHCoVs-OC43 at an MOI of 0.01, and the Rluc activity was measured at 72 hpi using the Renilla-Glo luciferase assay system.
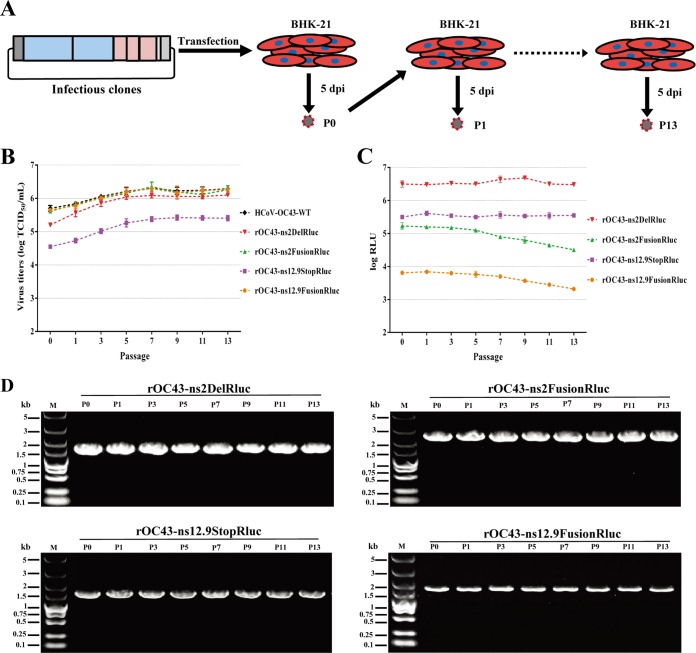
FIG 3.
Analysis of genetic stability of the reporter viruses. (A) Illustration of the virus passage procedure in BHK-21 cells. rHCoVs rescued from transfected cells were defined as P0. Culture supernatants from the transfected cells (P0) were added to naive cells to obtain passage 1 virus (P1). After 13 rounds of serial passages, the reporter viruses were passaged to P13. (B) Viral titers of reporter viruses during passages. Reporter viruses were passaged 13 times in BHK-21 cells, and the supernatants were collected from the virus-infected cells of each passage and titrated using the IFA-based viral titration assay. Data represent three independent experiments and are shown as means ± standard deviations. (C) Rluc activity of reporter viruses of each passage. BHK-21 cells were infected with reporter viruses (MOI = 0.01) of each passage in 48-well plates and assayed for the Rluc activity in RLU at 72 h postinfection. Data represent mean values from three independent experiments, with error bars representing the standard deviations of the means. (D) Analysis of genetic stability of the reporter viruses after several passages in BHK-21 cells. Viral RNA was extracted from culture supernatants of each passage, and RT-PCR was performed with a primer set flanking the Rluc gene. The resulting RT-PCR products were resolved by 1% agarose gel electrophoresis.
Cell viability assay.
The cell viability assay was performed using a Cell Titer-Glo luminescent cell viability assay kit (Promega). Briefly, cells were seeded in 96-well plates in triplicate. After 24 h, various concentrations of chloroquine (0 to 80 μM) and ribavirin (0 to 320 μM) (Sigma-Aldrich) were added to the medium. At 72 h, the plates were equilibrated at room temperature for 60 min, and 100 μl of Cell Titer-Glo reagent was added to the medium. The plates were subsequently shaken on a shaker for 2 min to induce cell lysis. After a final incubation for 10 min at room temperature, the luminescence was measured using a Glomax luminometer system (Promega).
Antiviral drug assay.
For the viral RNA load-based antiviral assay, confluent BHK-21 cells in 48-well culture plates were infected in triplicate with HCoV-OC43-WT or rOC43-ns2DelRluc at an MOI of 0.01. After 2 h of adsorption at 33°C, the inoculum was removed, and the cells were washed three times with DMEM. Subsequently, complete DMEM containing various concentrations of chloroquine (0 to 80 μM) or ribavirin (0 to 320 μM) was added to the cells. Cells were incubated for 72 h at 33°C in a humidified 5% CO2 incubator. The supernatants of cells infected with HCoV-OC43-WT or rOC43-ns2DelRluc were collected, and the viral RNA loads were determined using quantitative RT-PCR as described above. For the luciferase-based antiviral assay, BHK-21 cells in 96-well culture plates were infected with rOC43-ns2DelRluc, followed by incubation with chloroquine or ribavirin for 72 h; the Rluc activity was then measured as described above.
RNA interference (RNAi) screening.
We designed siRNA pools targeting eight potential host antiviral restriction factors for screening, and each individual siRNA pool consisted of three siRNAs targeting the same gene. A nontargeting siRNA having no matches to the viral or human genome served as a blank control. The specific siRNAs targeting antiviral host factors were synthesized by GenePharma (sequences will be provided upon request).
For testing of siRNA pools, HEK-293T cells plated in poly-l-lysine (PLL)-coated 48-well plates were transfected with siRNA pools using X-tremeGene siRNA transfection reagent (Roche) at a final concentration of 300 nM. After incubation for 24 h, cells were subsequently infected in triplicate with rOC43-ns2DelRluc at an MOI of 0.01. At 60 hpi, the Rluc activity was measured as described above.
Mice and infection.
Twelve-day-old female BALB/c mice (Animal Care Centre, Chinese Academy of Medical Science, Beijing, China) were randomly distributed into three groups. Two groups were intracerebrally inoculated with 20 μl of DMEM containing 100 TCID50 of HCoV-OC43-WT or rOC43-ns2DelRluc, and another group was intracerebrally inoculated with 20 μl of DMEM. The infected mice were monitored for survival. For the passages of rOC43-ns2DelRluc in BALB/c mice, 12-day-old mice were intracerebrally inoculated with 500 TCID50 of rOC43-ns2DelRluc (P0) in 20 μl of DMEM and sacrificed at 3 days postinoculation, and brains were homogenized in 500 μl of PBS containing 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 0.5 μg/ml amphotericin B. The brain homogenate was then clarified by low-speed centrifugation at 3,000 rpm for 12 min to obtain passage 1 (P1) virus. After 5 rounds of serial passages, the rOC43-ns2DelRluc was passaged to P5.
Statistical analysis.
Differences between groups were examined for statistical significance using Student's t test. Confidence levels are indicated in the figures as follows: *, P < 0.05; **, P < 0.01.
RESULTS
Characterization of rHCoVs-OC43 expressing Rluc.
The use of a reporter virus is a valuable screening tool for identifying novel antiviral drugs or host factors. To generate a high-expression reporter HCoV-OC43 and evaluate the roles of the ns2 and ns12.9 genes in viral production, four rHCoVs-OC43 were obtained following replacement of the ns2 or ns12.9 gene with Rluc (rOC43-ns2DelRluc and rOC43-ns12.9StopRluc) or in-frame insertion of the Rluc gene into the ns2 or ns12.9 gene (rOC43-ns2FusionRluc and rOC43-ns12.9FusionRluc) (Fig. 1).
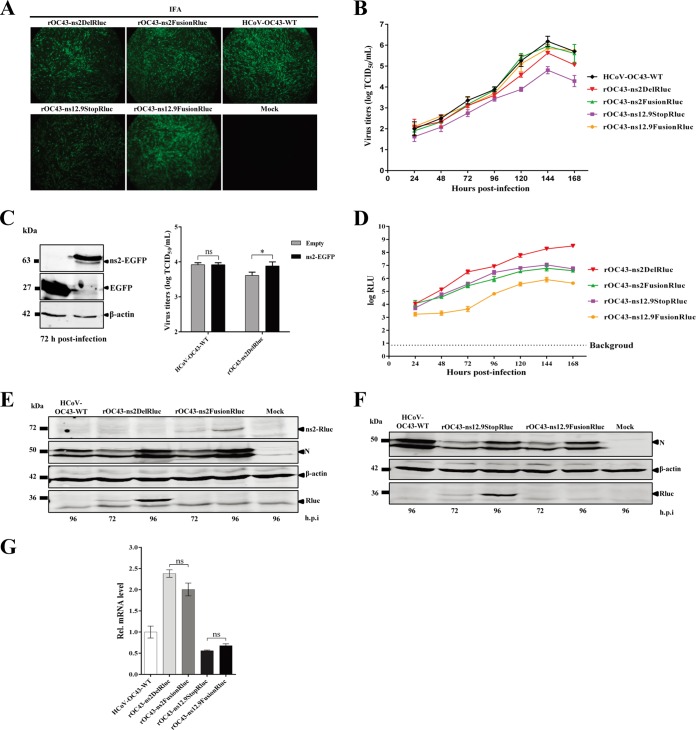
The in vitro growth characteristics of the reporter viruses were analyzed by growth kinetics in BHK-21 cells. rOC43-ns2FusionRluc and rOC43-ns12.9FusionRluc showed replication kinetics similar to that of HCoV-OC43-WT and reached a peak titer of 106 TCID50s/ml at 144 hpi (Fig. 2B), indicating that ns2-Rluc or ns12.9-Rluc fusion proteins were likely to retain their biological functions in the life cycle of HCoV-OC43. Moreover, the viral titer of rOC43-ns2DelRluc was only 4-fold lower than that of HCoV-OC43-WT at 144 hpi, indicating that the ns2 gene is nonessential for virus replication (Fig. 2B). In contrast, rOC43-ns12.9StopRluc showed impaired growth kinetics, with a peak titer of 104.8 TCID50s/ml at 144 hpi, which was ∼27-fold lower than that of the parental HCoV-OC43-WT (Fig. 2B). This result indicated that the ns12.9 viroporin is important for viral propagation in cell culture. To further explore whether the reduction in virus titers of rOC43-ns2DelRluc was due to the abolition of ns2 protein expression, we performed a transient-complementation assay. An ns2 protein expression vector was constructed, and its expression levels were detected by Western blotting (Fig. 2C, left). Compared with the empty-vector-transfected cells, ns2-expressing cells exhibited a slight increase in virus titers for HCoV-OC43-WT (Fig. 2C, right). These results confirmed that the loss of infectious virus production by deletion of the ns2 gene could be compensated for by transient expression of ns2 in BHK21 cells.
FIG 2.
Characterization of reporter viruses using engineered accessary genes. (A) The N protein of recombinant HCoVs-OC43 (rHCoVs) examined by indirect immunofluorescence assay (IFA). At 72 h postinfection, virus-infected BHK-21 cells were incubated with anti-OC43-N mouse polyclonal antibodies and then stained with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG. Cells were analyzed under a fluorescence microscope. (B) Growth kinetics of rHCoVs. BHK-21 cells were infected with rHCoVs and HCoV-OC43-WT at a multiplicity of infection (MOI) of 0.01. Viral titers from culture supernatants at the indicated time points were determined by indirect IFA. Data represent three independent experiments and are shown as means ± standard deviations. (C) Complementation of rOC43-ns2DelRluc infection in BHK-21 cells expressing ns2. Cells were transfected with a plasmid expressing ns2-enhanced GFP (EGFP) or a control vector using the X-tremeGENE HP DNA transfection reagent, and the expression levels of ns2-EGFP were analyzed by Western blotting using anti-GFP antibody (left). After 24 h posttransfection, BHK-21 cells were infected with HCoV-OC43-WT or rOC43-ns2DelRluc at an MOI of 0.01. Cell supernatants were collected at 72 h postinfection, and the viral titers were determined by IFA (right). (D) Time course analysis of the reporter gene expression. The Rluc activity, represented as relative light units (RLU), was measured in BHK-21 cells infected with rHCoVs at the indicated time points (MOI = 0.01). Data represent three independent experiments and are shown as means ± standard deviations. (E and F) Western blot analysis of reporter gene expression. Proteins in cell lysates of BHK-21 cells infected with rHCoVs and HCoV-OC43-WT were analyzed by Western blotting using anti-OC43-N, anti-Rluc, and anti-β-actin antibodies. Cell lysates from uninfected cells (mock) served as a negative control. (G) Effect of the inserted reporter gene on subgenomic RNA (sgRNA) synthesis. At 72 h postinfection (MOI = 0.01), total cellular mRNAs were extracted and subjected to RT-PCR to determine the mRNA levels of ns2 (HCoV-OC43-WT), Rluc (rOC43-ns2DelRluc and rOC43-ns12.9StopRluc), ns2-Rluc (rOC43-ns2FusionRluc), and ns12.9-Rluc (rOC43-ns12.9FusionRluc). HCoV-OC43-WT was used as a control. Data were normalized to the levels of internal mouse GAPDH mRNA. Error bars indicate means and standard deviations from three independent experiments.
Rluc activity in cells infected with reporter viruses was also characterized. Surprisingly, the viral titer of rOC43-ns2DelRluc was 4-fold lower than that of the rOC43-ns2FusionRluc at 144 hpi but showed robust Rluc expression levels, with Rluc activity 18-fold higher than that of rOC43-ns2FusionRluc (Fig. 2D). rOC43-ns12.9StopRluc, although having impaired growth kinetics, showed relatively high Rluc activity, with 107 RLU at 144 hpi. However, rOC43-ns12.9FusionRluc showed faint Rluc activity even though it showed replication kinetics similar to those of HCoV-OC43-WT (Fig. 2D). Moreover, Western blotting was performed to confirm the Rluc expression levels of the reporter viruses at 72 and 96 hpi. The results showed similar expression levels of N proteins in rOC43-ns2FusionRluc and rOC43-ns2DelRluc, but the expression of the ns2-Rluc fusion protein of rOC43-ns2FusionRluc was significantly reduced compared with that of Rluc proteins of rOC43-ns2DelRluc (Fig. 2E). In addition, we observed high levels of Rluc proteins in the lysates of cells infected with rOC43-ns12.9StopRluc, but no ns12.9-Rluc fusion proteins were detected in the lysates of rOC43-ns12.9FusionRluc-infected cells, perhaps due to its low Rluc expression levels at 96 hpi (Fig. 2F). These results correlated with the Rluc activity detected at the corresponding hpi (Fig. 2D).
These observations prompted us to ascertain whether replacement of ns2 or ns12.9 with the Rluc gene could enhance the subgenomic mRNA (sgmRNA) transcription efficiency compared with that of ns2-Rluc or ns12.9-Rluc fusion genes. We detected the transcription levels of the ns2 (HCoV-OC43-WT), Rluc (rOC43-ns2DelRluc or rOC43-ns12.9StopRluc), ns2-Rluc fusion (rOC43-ns2FusionRluc), and ns12.9-Rluc fusion (rOC43-ns12.9FusionRluc) genes using semiquantitative PCR. The sgmRNA level of Rluc (rOC43-ns2DelRluc) was only 2.3-fold higher than that of ns2 of HCoV-OC43-WT and was similar to the sgmRNA level of the ns2-Rluc fusion gene (Fig. 2G). Moreover, replacement of the ns12.9 gene with the Rluc gene or insertion of the Rluc gene in frame into the ns12.9 coding region caused a slight reduction in the Rluc or ns12.9-Rluc sgmRNA level during infection (Fig. 2G). This result indicated that the Rluc activity differences in the two reporter viruses were not due to the transcription levels of sgmRNAs.
Collectively, these results suggested that the ns2 gene is not required for HCoV-OC43 replication and that high-expression reporter virus can be generated by replacing the ns2 gene with Rluc.
Stability of rHCoVs-OC43 after multiple passages.
To examine the in vitro stabilities of the four reporter viruses, rHCoVs-OC43 and HCoV-OC43-WT were passaged 13 times in BHK-21 cells as described above (Fig. 3A). As shown in Fig. 3B, titers for all viruses increased over the first four passages and became stable in subsequent passages. Moreover, the Rluc activities of rOC43-ns2DelRluc and rOC43-ns12.9StopRluc at each passage showed no significant fluctuations during the passages in BHK-21 cells. However, rOC43-ns2FusionRluc and rOC43-ns12.9FusionRluc showed 4- to 6-fold reductions in Rluc activity during the 13 passages (Fig. 3C). To investigate whether mutations were introduced during the passages, viral RNA was extracted from the supernatant of infected cells of each passage. The Rluc gene and its flanking sequences were detected by RT-PCR. Surprisingly, the Rluc gene remained intact in the genomes of all rHCoVs-OC43, as no smaller PCR product was detected over the 13 passages (Fig. 3D). However, sequence analysis of clones of RT-PCR products identified same mutations (a two-nucleotide insertion between positions 70 and 71), which resulted in a stop codon in the regions of the Rluc expression cassettes of three rHCoVs-OC43 (see Fig. S1 and Table S2 in the supplemental material). It is worth mentioning that the replacement of accessory genes with the Rluc gene resulted in rHCoVs-OC43 with higher genetic stability than for in-frame insertions of the Rluc gene (see Table S2 in the supplemental material). Because rOC43-ns2DelRluc showed robust Rluc activity, with little impact on its replication kinetics, and remained genetically stable during 10 passages in BHK-21 cells, we next evaluated the pathogenicity of rOC43-ns2DelRluc in the mouse model. Unlike the previously reported ns12.9 deletion mutant (21), the result showed that BALB/c mice inoculated with 100 TCID50s of either rOC43-ns2DelRluc or HCoV-OC43-WT showed a severe symptom of twitching limbs at 3 days postinoculation and 100% mortality at 4 days postinoculation, indicating that deletion of ns2 had no influence on the pathogenicity of rOC43-ns2DelRluc in BALB/c mice (see Fig. S2A and B in the supplemental material). Moreover, rOC43-ns2DelRluc remained genetically stable after 5 passages in mice, and the viral titers in brain tissues were 107.1 TCID50s/g at 3 days postinoculation, further confirming the applicability of rOC43-ns2DelRluc in vivo (see Fig. S2D and E in the supplemental material).
Suitability of rOC43-ns2DelRluc for high-throughput antiviral drug screening.
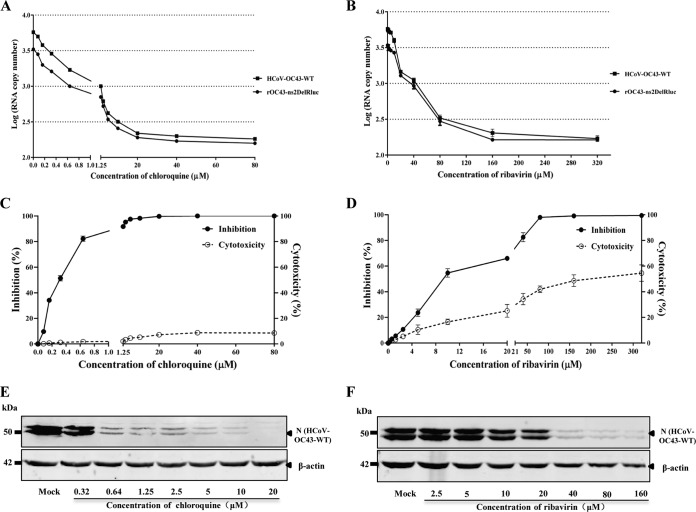
To verify whether rOC43-ns2DelRluc displayed sensitivity similar to that of the parental HCoV-OC43-WT under antiviral drug treatment, the reporter virus was used to evaluate of the antiviral activities of chloroquine and ribavirin in parallel. As shown in Fig. 4A, chloroquine treatment had a significant inhibitory effect on HCoV-OC43-WT or rOC43-ns2DelRluc replication at low-micromolar concentrations, while ribavirin showed no inhibitory effect at the same concentrations (Fig. 4B). Moreover, a similar decrease in viral copy numbers was observed for the two viruses in the presence of increasing levels of chloroquine or ribavirin, indicating that deletion of the ns2 gene had no effect on the sensitivity of rOC43-ns2DelRluc to the antiviral drugs.
FIG 4.
Replication of HCoV-OC43 in response to drug treatment in BHK-21 cells. (A and B) Effect of chloroquine or ribavirin on the replication of HCoV-OC43-WT or rOC43-ns2DelRluc. BHK-21 cells seeded in 48-well plates were infected with HCoV-OC43-WT or rOC43-ns2DelRluc at an MOI of 0.01 for 2 h and subsequently treated with chloroquine or ribavirin at the indicated concentrations. At 72 h postinfection, supernatants were removed and subsequently analyzed for viral load by real-time quantitative RT-PCR. Error bars indicate means and standard deviations from three independent experiments. (C and D) Chloroquine or ribavirin inhibition of Rluc activity of rOC43-ns2DelRluc and cytotoxic effects. The inhibition assay was performed as described in Materials and Methods. Rluc activity of chloroquine- or ribavirin-treated cells was normalized to that of dimethyl sulfoxide (DMSO)-treated control cells and measured relative to that of DMSO-treated cells. Viable cell numbers were used to determine the percent cytotoxic effect in drug-treated cells relative to that in DMSO-treated cells. Error bars indicate means and standard deviations from three independent experiments. (E and F) Antiviral effect of chloroquine or ribavirin on HCoV-OC43-WT N protein synthesis.
To verify whether the Rluc activity of rOC43-ns2DelRluc could be used for antiviral drug screening, we analyzed the antiviral activities of chloroquine and ribavirin against rOC43-ns2DelRluc in parallel using luciferase-based reporter assays in BHK-21 cells. As expected, Rluc activity was reduced in the presence of increasing levels of chloroquine or ribavirin in a dose-dependent manner (Fig. 4C and D). For chloroquine, a 50% inhibitory concentration (IC50) of 0.33 μM and a 50% cytotoxic concentration (CC50) of 397.54 μM were observed (mean values from three independent experiments), which is in line with a previous report (18). In contrast, ribavirin exhibited an inhibitory effect at concentrations of 3 μM or higher, with an IC50 of 10.00 μM and a CC50 of 156.16 μM. Our validation experiments suggested that the rOC43-ns2DelRluc-based Rluc assay allows more sensitive and more rapid quantification of viral replication than the traditional quantitative RT-PCR assay, with 106.2 RLU in dimethyl sulfoxide (DMSO)-treated cells at 72 hpi (data not shown) and the Rluc activity could be detected without extracting viral RNA, suggesting its utility for HTS of antiviral drugs.
Screening for potential host factors that inhibit HCoV-OC43 replication.
To further evaluate the applicability of rOC43-ns2DelRluc for antiviral screening, this reporter virus was employed to screen host factors that inhibit HCoV-OC43 replication. Here, we selected eight previously reported potential antiviral host factors against flaviviruses and tested them in RNAi screening. Among these eight host factors, tripartite motif protein 56 (TRIM56) served as a positive control as it belongs to a new class of host antiviral restriction factors that confer resistance to HCoV-OC43 (25). The effect of knockdown of the individual gene on rOC43-ns2DelRluc replication was expressed as relative luciferase activity (RLA), which is the ratio of RLU obtained from cells treated with targeting siRNA pools to that obtained from cells that were treated with a control siRNA (26).
As expected, compared with HEK-293T cells transfected with a control siRNA, knockdown TRIM56 (with mRNA levels reduced to 37.2% compared to those for control cells) increased Rluc activity ∼1.62-fold (Fig. 5A). In addition, we showed that knockdown double-stranded-RNA-activated protein kinase (PKR) or DEAD box RNA helicases (DDX3X) could significantly enhance Rluc activity, indicating that PKR and DDX3X are potential anti-HCoV-OC43 host factors (Fig. 5A). Moreover, the cell viability assay showed no significant differences between cells transfected with host factor siRNA pools and control cells transfected with scrambled siRNA, and the efficiency of RNAi-mediated knockdown was assessed using quantitative RT-PCR (Fig. 5B), demonstrating the validity of these host factors in the replication of the HCoV-OC43.
FIG 5.
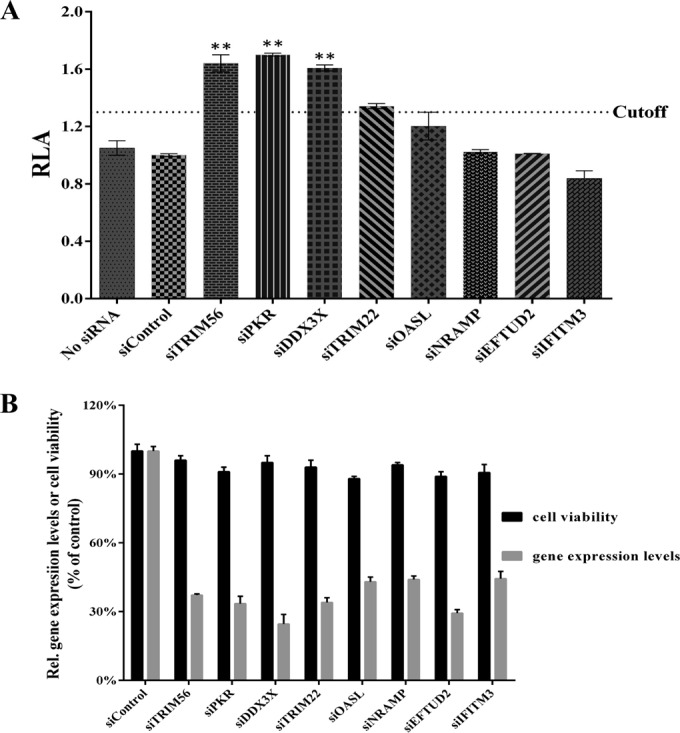
Screening of host factors influencing HCoV-OC43 replication using rOC43-ns2DelRluc. (A) HEK-293T cells were transfected with various small interfering RNA (siRNA) pools, followed by infection with rOC43-ns2DelRluc at an MOI of 0.01 in 48-well plates, and assayed for Rluc activity. The relative luciferase activity (RLA) represents the mean ± standard deviation (n = 3) of the ratio of relative light units (RLU) obtained from cells treated with targeting siRNAs to the RLU obtained from cells that were treated with a nontargeting siRNA (siControl), which had no adverse effect on the viruses and cells. (B) Real-time RT-PCR was used to quantitate the knockdown effect of the indicated siRNA pools at 36 h posttransfection (gray bars), and the effect of siRNA transfection on cell viability was analyzed in parallel (black bars). Values were normalized to those for nontargeting siRNA-transfected cells (100%). Error bars indicate means and standard deviations from three independent experiments. *, P < 0.05; **, P < 0.01.
Validation of PKR and DDX3X as antiviral factors in HCoV-OC43 replication.
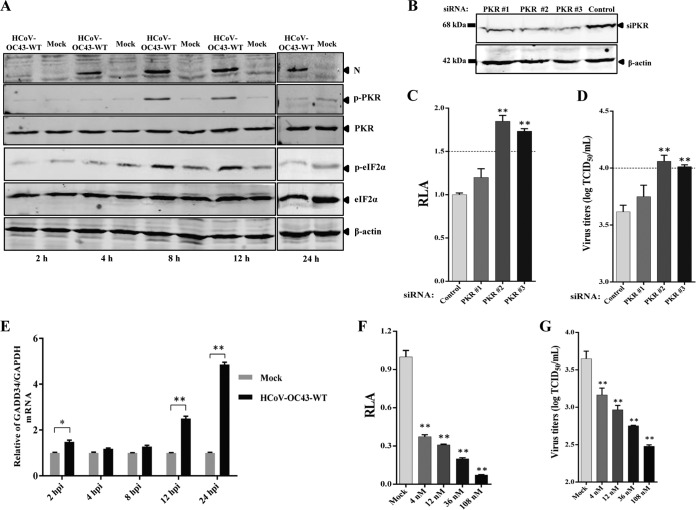
PKR is the strongest antiviral host factor identified in the primary siRNA screening assay. To further validate the antiviral role of PKR in HCoV-OC43 replication, Huh7 cells were infected with HCoV-OC43-WT at an MOI of 0.05. Cell pellets were collected at 2, 4, 8, 12, and 24 hpi. As shown in Fig. 6A, cells infected with HCoV-OC43-WT strongly induced PKR activation at 8 and 12 hpi, which decreased dramatically after 24 hpi. This observation was supported by the detection of phosphorylation of eIF2α, the substrate of phosphorylated PKR, which showed high basal levels at 8 and 12 hpi and became barely detectable at 24 h (Fig. 6A). These results suggested that phosphorylation of PKR and eIF2α was increased at the early stage of infection but quickly suppressed at 24 hpi. To determine the role of PKR in HCoV-OC43 replication, Huh7 cells were transfected with PKR-specific siRNAs to knock down PKR or nontargeting siRNA as a negative control. The results showed that two siRNAs (PKR 2 and PKR 3) efficiently reduced endogenous PKR levels compared with those in control cells (Fig. 6B). The reduction of endogenous PKR (PKR 2 and PKR 3) resulted in an obvious increase in both HCoV-OC43-WT and rOC43-ns2DelRluc replication, with a 1.83-fold increase in Rluc activity or virus titer (Fig. 6C and D), indicating that PKR plays an antiviral role in HCoV-OC43-infected cells. The observation of rapid dephosphorylation of eIF-2α in HCoV-OC43-infected cells prompted us to examine the expression of GADD34, which is a component of the protein phosphatase 1 (PP1) complex, which dephosphorylates eIF-2α. The mRNA level of GADD34 showed a 5-fold increase in HCoV-OC43-infected Huh7 cells at 24 hpi, which served as a feedback loop to mediate eIF-2α dephosphorylation at the corresponding time point (Fig. 6E). Interestingly, we also detected a 1.7-fold induction of the GADD34 mRNA level at 2 hpi, and this slight increase of the GADD34 mRNA level may play an important role in facilitating HCoV-OC43 replication during its invasion period. Okadaic acid (OA) was defined as a protein phosphatase inhibitor, promoting PKR and eIF2α phosphorylation. To further confirm the effect of PP1 activity on HCoV-OC43 replication, cells were incubated in the presence of OA or DMSO and then infected with HCoV-OC43-WT or rOC43-ns2DelRluc, respectively. As shown in Fig. 6F, in contrast to the case for non-DMSO-treated Huh7 cells in the presence of different concentrations of OA, the Rluc activity of rOC43-ns2DelRluc was significantly decreased at a concentration of 4 nM, with 10-fold inhibition observed at a concentration of 108 nM. This result was further confirmed by HCoV-OC43-WT, which showed obviously reduced virus titers (∼11-fold) at a concentration of 108 nM (Fig. 6G). Taken together, these results indicated that PKR and eIF2α phosphorylation induce an antiviral effect in HCoV-OC43-infected cells, and this inhibition was blocked by HCoV-OC43-induced GADD34 expression.
FIG 6.
Validation of PKR as an antiviral factor in HCoV-OC43 replication. (A) HCoV-OC43 infection induces the phosphorylation of PKR and eIF2α. Huh7 cells were infected with HCoV-OC43 or mock infected at an MOI of 0.05 and harvested at 2, 4, 8, 12, and 24 hpi. The cell lysates were collected and analyzed by Western blotting with anti-PKR, anti-p-PKR, anti-eIF2α, and anti-p-eIF2α (S51) antibodies. β-Actin was used as a protein loading control. (B) Knockdown of PKR expression at 48 h posttransfection. (C and D) Knockdown PKR induces the replication of HCoV-OC43 in Huh7 cells. The Rluc activity of rOC43-ns2DelRluc and titers of HCoV-OC43-WT were determined at 72 hpi. Data represent three independent experiments and are shown as means ± standard deviations. (E) Induction of GADD34 expression in HCoV-OC43-infected Huh7 cells at 12 h postinfection. (F and G) Reduction of HCoV-OC43 replication by inhibition of PP1 activity with okadaic acid (OA) in HCoV-OC43-infected Huh7 cells. Huh7 cells were treated with OA or DMSO after being infected with rOC43-ns2DelRluc or HCoV-OC43-WT. The Rluc activity of rOC43-ns2DelRluc and titers of HCoV-OC43-WT were determined at 72 hpi. Data represent three independent experiments and are shown as means ± standard deviations. *, P < 0.05; **, P < 0.01.
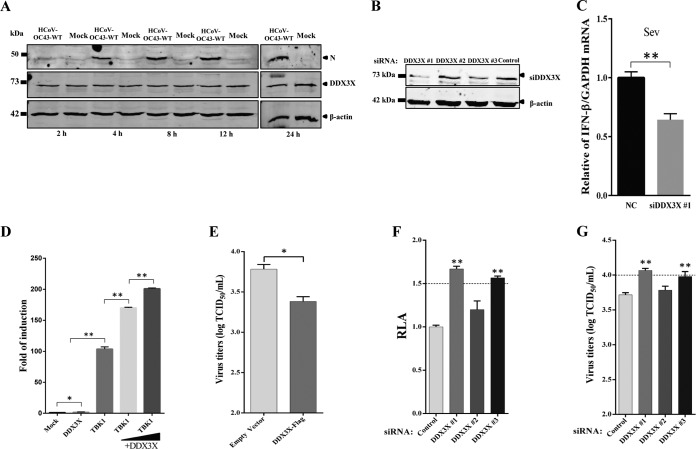
DDX3X is another potent antiviral host factor identified in the siRNA screening assay. Human DDX3X is a newly discovered DEAD box RNA helicase. In addition to its involvement in protein translation, the cell cycle, apoptosis, nuclear export, and eukaryotic gene regulation, human DDX3X is a critical molecule in innate immune signaling pathways and contributes to type I interferon (IFN) induction. A previous report showed that DDX3X is upregulated upon dengue virus (DENV) or porcine reproductive and respiratory syndrome virus (PRRSV) infection (27, 28). However, contrary to our expectations, Western blotting showed no change in DDX3X protein levels in Huh7 cells upon HCoV-OC43-WT infection (Fig. 7A). This result was further confirmed by semiquantitative PCR in HCoV-OC43-WT-infected Huh7 or HEK-293T cells (data not shown). A previous study demonstrated that coexpression of TBK1 with DDX3X rather than overexpression of DDX3X itself led to IFN promoter activation, because overexpression of TBK1 causes DDX3X activation (29). Our result showed that silencing of endogenous DDX3X expression using RNAi would significantly affect the transcription level of IFN-β; however, overexpression of DDX3X alone activated the IFN promoter only 2-fold (Fig. 7C and D). Moreover, the result showed that the Rluc activity of rOC43-ns2DelRluc or virus titers of HCoV-OC43-WT increased by 1.7-fold in DDX3X-silenced cells (DDX3X 1 or DDX3X 3) compared with control cells at 72 hpi (Fig. 7F and G). Furthermore, we performed an overexpression assay and demonstrated that overexpression of DDX3X showed antiviral activity against HCoV-OC43 infection (Fig. 7E). Thus, DDX3X may play an antiviral role during HCoV-OC43 infection through positive regulation of innate immune-signaling processes.
FIG 7.
Validation of DDX3X as an antiviral factor in HCoV-OC43 replication. (A) The expression level of DDX3X is unchanged in HCoV-OC43-infected Huh7 cells. Huh7 cells were infected with HCoV-OC43 or mock infected at an MOI of 0.05 and harvested at 2, 4, 8, 12, and 24 hpi. Cell lysates were collected and analyzed by Western blotting with anti-DDX3X antibody. β-Actin was used as a protein loading control. (B and C) DDX3X is required for IFN-β induction. siRNA-treated HEK-293T cells were infected with Sendai virus (Sev). Induction of IFN-β mRNA was measured by semiquantitative PCR. (D) Overexpression of DDX3X alone is insufficient to activate the IFN promoter. HEK-293T cells were transfected with 500 ng of plasmids encoding DDX3X or TBKI or cotransfected with plasmids TBK1 (500 ng) and DDX3X (100 or 300 ng), together with 500 ng of IFN-β-Luc reporter plasmid and internal control plasmid pRL-TK (20 ng) as indicated. (E) Overexpression of DDX3X shows weak antiviral activity against HCoV-OC43. Huh7 cells were transfected with pFlag-DDX3X or empty vector. At 24 h posttransfection, cells were infected with HCoV-OC43 at an MOI of 0.05, and titers of HCoV-OC43-WT were determined at 72 hpi. (F and G) siRNA-mediated DDX3X silencing induces HCoV-OC43 replication. The Rluc activity of rOC43-ns2DelRluc and titers of HCoV-OC43-WT were determined at 72 hpi. Data represent three independent experiments and are shown as means ± standard deviations. *, P < 0.05; **, P < 0.01.
These data demonstrated the feasibility of using rOC43-ns2DelRluc for drug screening and identifying antiviral host factors.
DISCUSSION
Rapid identification of therapeutics is a high priority, as there is currently no specific therapy to treat novel Betacoronavirus (SARS-CoV and MERS-CoV) infections, which can cause high case-fatality rates (30). A marker virus with the introduction of a reporter gene into the viral genome provides a powerful tool to address this problem. To date, only one reporter HCoV (SARS-CoV-GFP) has been generated and applied to an siRNA library screening assay (14). However, SARS-CoV-GFP lacks sensitivity, as it requires a high infectious dose (MOI of 10) for quantitative screening. Moreover, this reporter virus assay must be performed in a BSL3 facility, which is costly and labor-intensive. Thus, the generation of a safe and sensitive reporter HCoV for HTS assays is urgent. Here, we report a sensitive antiviral screening platform based on recombinant HCoV-OC43 (rOC43-ns2DelRluc) that expresses Rluc as a reporter gene. Furthermore, using a luciferase-based siRNA screening assay, we identified two host factors (PKR and DDX3X) that exhibit antiviral effects.
HCoV-OC43 carries two accessory genes, ns2 and ns12.9; however, the biological functions of these HCoV-OC43 accessory genes remain poorly understood. In this study, we generated a variety of luciferase-based rHCoVs-OC43 by genetic engineering of the two accessory genes. Among the rHCoVs-OC43, rOC43-ns12.9StopRluc led to a lower virus yield in BHK-21 cells, suggesting that the ion channel activity of ns12.9 is important for the production of infectious virions. A recent study by Freeman et al. showed that in-frame insertion of reporter gene into replicase genes (ns2 or ns3) of murine hepatitis virus (MHV) was tolerated and resulted in replication kinetics similar to those for MHV-WT (10). These results are consistent with the results obtained for the two Rluc fusion reporter viruses (rOC43-ns2FusionRluc and rOC43-ns12.9FusionRluc), which showed replication kinetics similar to those of HCoV-OC43-WT. However, our study demonstrated that the use of Rluc fused with the accessory genes of HCoV-OC43 was an ineffective way to generate high-expressing reporter HCoV, because the two Rluc fusion reporter viruses showed impaired Rluc activity and genetic instability during passages in vitro. One reporter virus, rOC43-ns2DelRluc, had replication kinetics similar to those of the parent virus HCoV-OC43-WT, showing robust Rluc activity during infection of BHK-21 cells. Moreover, deletion of the ns2 gene had no influence on the pathogenicity of rOC43-ns2DelRluc in mice, and the inserted Rluc gene remained stable both in vitro and in vivo. Thus, rOC43-ns2DelRluc might be a superior reporter virus for screening antivirals in terms of growth characteristics, Rluc expression levels, and genetic stability.
Recently, examination of a library of FDA-approved drugs used for anti-MERS-CoV screening in cell culture successfully identified four potent inhibitors. Intriguingly, all the screened compounds were broad-spectrum anti-HCoV drugs that also inhibited the replication of SARS-CoV and HCoV-229E (31). However, traditional cytopathic effect (CPE)-based viral titration assays are ill-suited for HTS assays, as more and more novel antiviral drugs are developed every year. Thus, rOC43-ns2DelRluc would provide a powerful tool for rapid and quantitative screening of broad-spectrum anti-HCoV drugs. Chloroquine, a clinically approved drug, appeared to be a broad-spectrum CoVs drug, as it blocks the replication of SARS-CoV, HCoV-OC43, MERS-CoV, and HCV-229E in vitro (32, 33). Additionally, clinical experience gained from treating SARS and MERS suggested the effectiveness of a number of interventions, including ribavirin, interferon (alfacon-1), corticosteroids, or a combination of these interventions (34, 35). In our study, rOC43-ns2DelRluc was used to evaluate the antiviral activity of chloroquine and ribavirin in 96-well plates. rOC43-ns2DelRluc with deletion of the ns2 gene showed no impairment in response to drug treatment compared with HCoV-OC43-WT, showing a similar decrease in viral copy numbers in the presence of increasing concentrations of chloroquine or ribavirin (Fig. 4A and B). Moreover, the Rluc activity of rOC43-ns2DelRluc was reduced in the presence of increasing levels of chloroquine or ribavirin in a dose-dependent manner, with IC50s similar to those with HCoV-OC43-WT. It is worth mentioning that in our study, we demonstrated that ribavirin exhibited an inhibitory effect against HCoV-OC43 only at high concentrations and showed a significant cytotoxicity in BHK-21 cells. These data suggest that rOC43-ns2DelRluc represents a superior model for screening broad-spectrum HCoV drugs without the requirement of BSL3 confinement.
In the past decade, reporter viruses have been used widely for screening pooled siRNAs to discover host factors that can influence the replication of diverse positive-strand RNA viruses. Such reporter viruses have allowed sensitive and quantitative evaluation of antiviral or proviral effects (36 – 40). However, few host factors that can restrict the replication of CoV have been identified. Here, eight potential antiviral host factors in flavivirus infection were selected for RNAi screening using the reporter rOC43-ns2DelRluc, leading to the identification of two anti-HCoV-OC43 host factors (PKR and DDX3X).
Many viral families have evolved various regulatory mechanisms that modulate host protein synthesis to maximize the production of progeny viruses. In infection with the CoV infectious bronchitis virus (IBV), overexpression of a dominant negative kinase-defective PKR mutant enhanced IBV replication by almost 2-fold (41). In this study, we showed that the basal levels of phosphorylated PKR and eIF-2α were unregulated in cells infected with HCoV-OC43 at the early stage of infection. Intriguingly, phosphorylated eIF-2α decreased rapidly via induction of GADD34 expression. Upregulation of eIF-2α phosphorylation using OA significantly reduced HCoV-OC43 replication. These results indicated that PKR plays an antiviral role in HCoV-OC43-infected cells.
DDX3X (DDX3 represented on the X chromosome) belongs to the DEAD box family of ATP-dependent RNA helicases. It is a multiple-function protein involved in protein translation, cell cycle, apoptosis, nuclear export, translation, and assembly of stress granules. There is growing evidence that DDX3X is a component of the innate immune response against viral infections (42). In our study, using RNA interference and an overexpression approach, we first described the antiviral role of DDX3X during HCoV-OC43 infection via regulation of the type I IFN pathway. Other studies have suggested that DDX3X is an important host factor required for hepatitis C virus (HCV) and HIV infection (43, 44). The core protein of HCV interacts with DDX3X to manipulate splicing and regulation of transcription or translation, and the helicase activity of DDX3X was required for HIV RNA export. Therefore, DDX3X plays distinct roles in virus-specific situations.
In summary, we generated a robust and stable luciferase-based recombinant HCoV-OC43 by replacement of the ns2 gene. This reporter virus can be used for screening anti-HCoV drugs and host factors. To the best of our knowledge, this is the first construction of a luciferase-based HCoV-OC43 for quantitative antiviral assays. The reporter virus will contribute to future work focused on screening wide-spectrum drugs or host factors influencing HCoV replication.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Megaproject for Infectious Disease Research of China (2014ZX10004001 and 2013ZX10004601) and National Key Plan for Scientific Research and Development of China (2016YFD0500301).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that no competing interests exist.
Funding Statement
This work, including the efforts of Wenjie Tan, was funded by Megaproject for Infectious Disease Research of China (2014ZX10004001 and 2013ZX10004601) and National Key Plan for Scientific Research and Development of China (2016YFD0500301).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00814-16.
REFERENCES
- 1.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. 2014. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol 159:2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 2.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gralinski LE, Baric RS. 2015. Molecular pathology of emerging coronavirus infections. J Pathol 235:185–195. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desforges M, Le Coupanec A, Stodola JK, Meessen-Pinard M, Talbot PJ. 2014. Human coronaviruses: viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res 194:145–158. doi: 10.1016/j.virusres.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan CA, Haijema BJ, Boss D, Heuts FW, Rottier PJ. 2005. Coronaviruses as vectors: stability of foreign gene expression. J Virol 79:12742–12751. doi: 10.1128/JVI.79.20.12742-12751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stirrups K, Shaw K, Evans S, Dalton K, Casais R, Cavanagh D, Britton P. 2000. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J Gen Virol 81:1687–1698. doi: 10.1099/0022-1317-81-7-1687. [DOI] [PubMed] [Google Scholar]
- 7.Shen H, Fang SG, Chen B, Chen G, Tay FP, Liu DX. 2009. Towards construction of viral vectors based on avian coronavirus infectious bronchitis virus for gene delivery and vaccine development. J Virol Methods 160:48–56. doi: 10.1016/j.jviromet.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch BJ, de Haan CA, Rottier PJ. 2004. Coronavirus spike glycoprotein, extended at the carboxy terminus with green fluorescent protein, is assembly competent. J Virol 78:7369–7378. doi: 10.1128/JVI.78.14.7369-7378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becares M, Sanchez CM, Sola I, Enjuanes L, Zuñiga S. 2014. Antigenic structures stably expressed by recombinant TGEV-derived vectors. Virology 464–465:274–286. doi: 10.1016/j.virol.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman MC, Graham RL, Lu X, Peek CT, Denison MR. 2014. Coronavirus replicase-reporter fusions provide quantitative analysis of replication and replication complex formation. J Virol 88:5319–5327. doi: 10.1128/JVI.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts RS, Yount BL, Sims AC, Baker S, Baric RS. 2006. Renilla luciferase as a reporter to assess SARS-CoV mRNA transcription regulation and efficacy of anti-SARS-CoV agents. Adv Exp Med Biol 581:597–600. doi: 10.1007/978-0-387-33012-9_108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao G, Du L, Ma C, Li Y, Li L, Poon VK, Wang L, Yu F, Zheng BJ, Jiang S, Zhou Y. 2013. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS–CoV. Virol J 10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Forrest JC, Zhang X. 2015. A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res 114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wilde AH, Wannee KF, Scholte FE, Goeman JJ, Ten Dijke P, Snijder EJ, Kikkert M, van Hemert MJ. 2015. A kinome-wide small interfering RNA screen identifies proviral and antiviral host factors in severe acute respiratory syndrome coronavirus replication, including double-stranded RNA-activated protein kinase and early secretory pathway proteins. J Virol 89:8318–8333. doi: 10.1128/JVI.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntosh K, Becker WB, Chanock RM. 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U S A 58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. 2004. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol 78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Chen C, Tan W, Yang K, Yang H. 2016. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci Rep 6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keyaerts E, Li S, Vijgen L, Rysman E, Verbeeck J, Van Ranst M, Maes P. 2009. Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice. Antimicrob Agents Chemother 53:3416–3421. doi: 10.1128/AAC.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacomy H, Fragoso G, Almazan G, Mushynski WE, Talbot PJ. 2006. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology 349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mounir S, Labonte P, Talbot PJ. 1993. Characterization of the nonstructural and spike proteins of the human respiratory coronavirus OC43: comparison with bovine enteric coronavirus. Adv Exp Med Biol 342:61–67. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R, Wang K, Ping X, Yu W, Qian Z, Xiong S, Sun B. 2015. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J Virol 89:11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.St-Jean JR, Desforges M, Almazán F, Jacomy H, Enjuanes L, Talbot PJ. 2006. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J Virol 80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed LJ, Münch HA. 1938. Simple method of estimating fifty percent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 24.Hu Q, Lu R, Peng K, Duan X, Wang Y, Zhao Y, Wang W, Lou Y, Tan W. 2014. Prevalence and genetic diversity analysis of human coronavirus OC43 among adult patients with acute respiratory infections in Beijing. PLoS One 9:e100781. doi: 10.1371/journal.pone.0100781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Li NL, Wang J, Shi PY, Wang T, Miller MA, Li K. 2014. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J Virol 88:13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Feng T, Pan W, Shi X, Dai J. 2015. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem Biophys Res Commun 456:327–332. doi: 10.1016/j.bbrc.2014.11.080. [DOI] [PubMed] [Google Scholar]
- 28.Chen Q, Liu Q, Liu D, Wang D, Chen H, Xiao S, Fang L. 2014. Molecular cloning, functional characterization and antiviral activity of porcine DDX3X. Biochem Biophys Res Commun 443:1169–1175. [DOI] [PubMed] [Google Scholar]
- 29.Soulat D, Bürckstümmer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T, Superti-Furga G. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J 27:2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. 2013. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep 3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Wilde AH, Jochmans D, Posthuma CC, Zevenhoven-Dobbe JC, van Nieuwkoop S, Bestebroer TM, van den Hoogen BG, Neyts J, Snijder EJ. 2014. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother 58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. 2004. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun 323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kono M, Tatsumi K, Imai AM, Saito K, Kuriyama T, Shirasawa H. 2008. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antiviral Res 77:150–152. doi: 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross AE, Bryson ML. 2015. Oral ribavirin for the treatment of noninfluenza respiratory viral infections: a systematic review. Ann Pharmacother 49:1125–1135. doi: 10.1177/1060028015597449. [DOI] [PubMed] [Google Scholar]
- 35.Groneberg DA, Poutanen SM, Low DE, Lode H, Welte T, Zabel P. 2005. Treatment and vaccines for severe acute respiratory syndrome. Lancet Infect Dis 5:147–155. doi: 10.1016/S1473-3099(05)70022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 37.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J Virol 83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schröder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J 27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pène V, Li Q, Sodroski C, Hsu CS, Liang TJ. 2015. Dynamic interaction of stress granules, DDX3X, and IKK-α mediates multiple functions in hepatitis C virus infection. J Virol 89:5462–5477. doi: 10.1128/JVI.03197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.