Abstract
Regulation of protein expression by non-coding RNAs typically involves effects on mRNA degradation and/or ribosomal translation. The possibility of virus-host mRNA-mRNA antisense tethering interactions (ATI) as a gain-of-function strategy, via the capture of functional RNA motifs, has not been hitherto considered. We present evidence that ATIs may be exploited by certain RNA viruses in order to tether the mRNAs of host selenoproteins, potentially exploiting the proximity of a captured host selenocysteine insertion sequence (SECIS) element to enable the expression of virally-encoded selenoprotein modules, via translation of in-frame UGA stop codons as selenocysteine. Computational analysis predicts thermodynamically stable ATIs between several widely expressed mammalian selenoprotein mRNAs (e.g., isoforms of thioredoxin reductase) and specific Ebola virus mRNAs, and HIV-1 mRNA, which we demonstrate via DNA gel shift assays. The probable functional significance of these ATIs is further supported by the observation that, in both viruses, they are located in close proximity to highly conserved in-frame UGA stop codons at the 3′ end of open reading frames that encode essential viral proteins (the HIV-1 nef protein and the Ebola nucleoprotein). Significantly, in HIV/AIDS patients, an inverse correlation between serum selenium and mortality has been repeatedly documented, and clinical benefits of selenium in the context of multi-micronutrient supplementation have been demonstrated in several well-controlled clinical trials. Hence, in the light of our findings, the possibility of a similar role for selenium in Ebola pathogenesis and treatment merits serious investigation.
Keywords: Antisense, Ebola, mRNA, Selenium, Selenoprotein, HIV, Tethering, Thioredoxin reductase
Introduction
Regulation of mRNA levels and protein synthesis by other RNA species, such as microRNA and siRNA, typically involves either the degradation of target mRNAs, or the inhibition of protein translation in the absence of RNA degradation; these alternative outcomes depend primarily on the degree of complementarity between the two RNAs [1]. For cellular genes having natural antisense transcripts (NATs), protein synthesis can be either downregulated [2] or upregulated [3] by NAT binding to the target mRNA. However, aside from such effects on mRNA degradation vs. stability and translational repression vs. enhancement, the possibility of antisense-based mRNA tethering as a general gain-of-function strategy, via the capture of functional RNA motifs, has not been widely considered, if at all.
Particularly for a small virus with a highly constrained genome size, an ideally suitable function for this hypothetical mechanism would be the ability to gain additional protein coding potential via the capture of a selenocysteine insertion sequence (SECIS) element, which would confer the
ability for appropriately located UGA stop codons to be “recoded” and translated as selenocysteine (Sec). Since SECIS elements have been shown to function in trans, i.e., even when carried by a separate mRNA [4], it is highly probable that the presence of a SECIS in a tethered mRNA would also confer UGA-recoding ability on the tethering mRNA partner, i.e., in this case, the ability to express virally-encoded selenoprotein modules. There is no doubt that selenoprotein-encoding capability can serve a viral agenda, because a functional viral homologue of the prototypical selenoprotein, glutathione peroxidase (GPx), has been identified in the genome of Molluscum contagiosum, a large DNA pox virus [5]. Encoding such an antioxidant selenoprotein could enhance the ability of a virus to respond to and survive oxidant-based immune attacks, thereby increasing viral fitness.
Significantly, our research group has identified GPx-related sequences with in-frame UGA codons in various RNA viruses, including hepatitis C virus and human immunodeficiency virus type one (HIV-1); the UGA-containing active site regions of these GPx-like modules are typically encoded in an overlapping reading frame of another viral protein [6]. The putative HIV-1 GPx was cloned and found to encode functional GPx activity [7] and to exert anti-oxidant and anti-apoptotic effects [8], but only when the vi-ral protein was expressed as a selenoprotein via inclusion of a cellular SECIS element in the expression vector. A functional SECIS element encoded by an RNA virus has never been demonstrated. However, a virus would not need its own SECIS if it possessed a mechanism to hijack such a function from the host, which is not only a classic viral strategy, but arguably the essence of the viral life style.
In this study, we present both computational and experimental evidence for antisense tethering interactions (ATIs) between host selenoprotein mRNAs (specifically, thioredoxin reductase mRNAs) and certain mRNAs of RNA viruses. We focus here on the two most compelling examples we have identified, one involving the highly pathogenic Zaire strain of Ebola virus (EBOV), and another involving HIV-1. The first, shown as A in (Fig. 1), is between the mRNA of the human thioredoxin reductase 3 (TR3) and the mRNA encoding the nucleoprotein (NP) of EBOV; the second (B in Fig. 1) is between the mRNA of human thioredoxin reductase 1 (TR1) and a region of HIV-1 genomic mRNA, in the 3′-long terminal repeat (LTR).
Fig. (1).
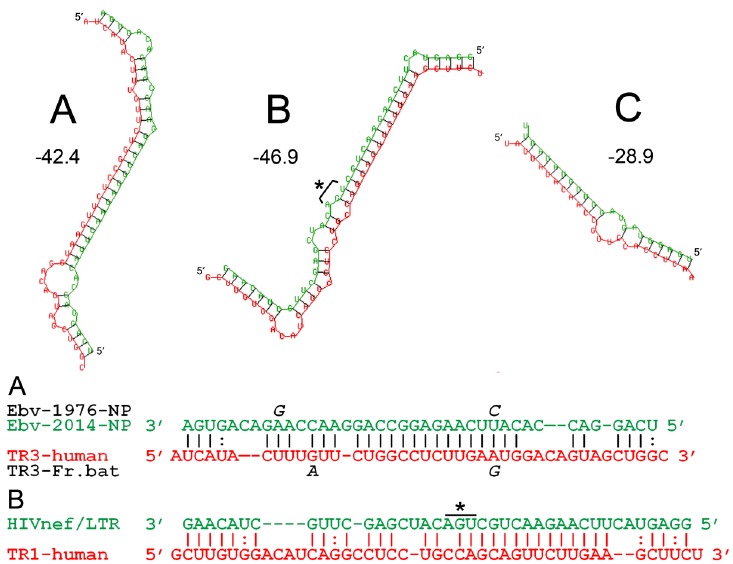
Predicted antisense interactions between regions of mRNAs of human thioredoxin reductases and viral mRNAs. These are shown as both RNA secondary structures and antisense sequence alignments. The RNA structures shown and computed interaction free energies in kcal/mol (numerals next to the structures) were generated using the RNAHybrid 2.2 program (see Methods). The antisense matches are: A. EBOV nucleoprotein mRNA (Ebv-2014-NP, green) vs. TR3 mRNA (TR3-human, red); B. HIV-1 (HIVnef/LTR, green) vs. TR1 (TR1-human, red); C. A typical imperfect microRNA interaction is included for comparison (Let-7 vs. a cellular target, the default example in RNAHybrid). The alignments A and B correspond exactly to the RNA secondary structures above; GU base pairs are indicated by a colon, and the highly conserved UGA stop codon of the HIV-1 nef gene is indicated by an asterisk. In alignment A, the letters in italics above and below the sequences correspond to genomic sequence variations between the 2014 EBOV (green) and the earliest 1976 EBOV isolates (black italics), and between human TR3 (red) and fruit bat TR3 (black italics); see text for a discussion of these mutations.
There are two major possibilities for how these antisense interactions would be likely to impact host selenium biochemistry during viral infection: either simply by antisense inhibition of cellular selenoprotein synthesis, or by direct competition for a limited pool of selenocysteine if the ATI with a selenoprotein mRNA can enable viral selenoprotein synthesis. We will present a model for the latter, and discuss the implications of these findings for well-established links between dietary selenium status and viral pathogenesis.
MATERIALS AND METHODS
Identification of Antisense Interactions via Computational Methods
To evaluate the hypothesis that certain viral mRNAs might engage in antisense interactions with host selenoprotein mRNAs, potential antisense matches were initially identified via nucleotide BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The HIV-1 genomic mRNA sequence, and individual mRNA sequences of Ebola virus, were used as probes (see below for specific Genbank accession numbers of reference sequences used). Nucleotide BLAST (blastn option) was used with default search parameters, against a search set of the Reference RNA sequence database (refseq.rna), restricted to either Homo sapiens (taxid:9606) or various bat taxa (e.g., Old World fruit bats, taxid:9398), since bats are now believed to be the most probable reservoir species for Ebola virus, and therefore may be hosts to which they have adapted in terms of potential mRNA interactions. The best candidates for ATI with selenoprotein-encoding mRNAs that were identified using BLAST and selected for further study were the HIV-1 nef region vs. TR1 and the Ebola NP vs. TR3, for both of which continuous 15 base pair antisense matches were identified via Blast, at high significance levels. The RNAHybrid program (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid; [9] was then used to establish the extent (in the 5’ and 3′ directions beyond the core match identified using BLAST) and computed binding energies of the potential RNA-RNA interactions; this generated the hybridizations shown in Fig. (1). The putative ATIs were further validated using another widely used method for accurate prediction of RNA-RNA interactions, IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp; [10-11]). This method uses a rigorous approach that considers not only the hybridization energy of the interacting pair of RNAs, but makes the assessment in the context of competing intramolecular mRNA secondary structures that must be unfolded in order for the intermolecular interaction to occur, using a sliding window to consider all possible competing intramolecular structures. For an interaction to be identified by the program, there must be a significant net interaction energy, after subtraction of the unfolding energies for each of the single strands. Using large input fragments of up to 1500 bases in length from the 3′ end of each of the cognate mRNAs as input (HIV-1 vs. TR1 and Ebola NP vs. TR3), with default parameters, IntaRNA identified exactly the same core antisense interactions as previously established using BLAST and RNAhybrid (see Results and Discussion section for details).
Reference Sequences Used in Computational Studies and Oligonucleotide Design
Genbank accession numbers for the viral and host gene reference sequences used, and the relevant sequence ranges shown in (Fig. 1), are: 1976 Zaire ebolavirus, NC_002549.1; 2014 Zaire ebolavirus, KJ660346.2 (nucleoprotein, 2350-2388); HIV-1, K02013.1 (nef/LTR region, 8989-9028); human TR1, NM_003330.3 (3612-3655); human TR3, NM_001173513.1 (1663-1703); fruit bat TR3, XM_006911434.
Demonstration of the Predicted Antisense Interactions at the DNA Level via Gel Shift Assays
Target-specific in vitro DNA hybridization of the cognate virus-selenoprotein pairs was demonstrated by electrophoretic mobility shift assays, using ~40mer synthetic single stranded ssDNA oligomers (Integrated DNA Technologies, Inc., Coralville, IA). Oligos (~1 μg each in 10 μl PBS), either singly or in cognate or mismatched pairs, were incubated at 37°C for 15 hours, except those in lanes 9 and 10 (Fig. 2), which were heated to 90°C for 10 minutes in the presence of 10 μg of sheered herring sperm DNA (Promega D1811, Madison, WI), followed by cooling to room temperature over 1 hour. The matched pairs of oligos from the respective viral and host mRNAs, corresponding to the sequence regions involved in the antisense interactions shown in Fig. 1, were as follows: EBOV nucleoprotein (Enp) vs. TR3, and HIV-1 nef/LTR region (Hnf) vs. TR1. Mismatched (non-cognate) mRNA pairs, e.g. Enp vs. TR1, were used as negative controls (see legend to Fig. 2 for details). Separation was on a 4% agarose gel with ethidium bromide visualization. GeneRuler 1 kb Plus DNA ladder (Thermo Scientific, Waltham, MA) was used as a guide.
Fig. (2).
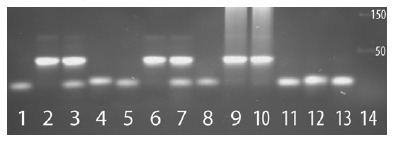
Virus vs. human selenoprotein antisense interactions demonstrated at the DNA level for the Ebola nucleoprotein and HIV nef regions shown in Fig. 1. Target-specific in vitro DNA hybridization of the cognate virus-selenoprotein pairs is shown by gel shift assay using ~40mer synthetic single stranded ssDNA oligos. The matched pairs of oligos from the respective viral and host mRNAs, corresponding to the sequence regions involved in the antisense interactions shown in Fig. 1, were as follows: EBOV nucleoprotein (Enp) vs. TR3, and HIV-1 nef/LTR region (Hnf) vs. TR1. Lanes have either a single oligo that runs as ssDNA (the lowest bands) or an incubated pair of oligos, as follows: 1. Enp; 2. 1:1 Enp+TR3; 3. 2:1 Enp+TR3; 4. TR3; 5. Hnf; 6. 1:1 Hnf+TR1; 7. 2:1 Hnf+TR1; 8. TR1; 9. 1:1 Enp+TR3 plus sheered herring sperm DNA; 10. 1:1 Hnf+TR1 plus herring DNA; 11. 1:1 Enp+TR1; 12. 1:1 Hnf+TR3; 13. 1:1 Hnf+Enp; 14. DNA size markers. The bright bands at the size of ~40mer double stranded dsDNA correspond to the expected hybridizations (Enp+TR3 or Hnf+TR1) at 1:1 (lanes 2,6,9,10) and 2:1 ratios (lanes 3,7). The faint bands above the 50 size marker (e.g. lanes 2,3,6,7) are possibly from trimer formation (e.g. TR3+Enp+TR3, etc.).
RESULTS AND DISCUSSION
Discovery of these antisense matches was initially guided via BLAST searches to identify potential core helical interactions of ~15 or more consecutive base pairs, which would correspond to a probability of < 5 x 10-9 in purely random DNA sequences. The best candidates identified using BLAST were the Ebola NP vs. TR3, and the HIV-1 nef region vs. TR1, which both had core antisense matches of 15 consecutive base pairs (Supplemental Material files S1 and S2), with highly significant BLAST “Expect” scores of 0.001 (Ebola NP vs. TR3) and 0.002 (HIV nef vs. TR1) vs. the human selenoprotein mRNA database. However, BLAST is not an ideal tool for discovering antisense interactions, because of the unique non-Watson-Crick base pairings available to RNA. To get a better sense of the significance and stability of these matches, the RNAHybrid program [9] was used to establish the extent and binding energies of the RNA:RNA interactions. The results are shown as A and B in (Fig. 1), rendered as both RNA secondary structures and antisense sequence alignments. Both of these predicted interactions are more energetically stable and extensive than typical microRNA binding interactions, such as that shown as structure C, which are inherently limited by the 22 nucleotide size of microRNA.
These results were further validated by the IntaRNA program [10-11], which output essentially identical RNA-RNA interactions as those shown in (Fig. 1) for the Ebola NP vs. TR3 and HIV-1 nef region vs. TR1 (for which the raw IntaRNA output is in Supplemental Material files S3 and S4 respectively). Relative to the RNAhybrid results shown in Fig. 1, the IntaRNA results for the same mRNA pairs, although slightly truncated at either one (HIV-TR1) or both ends (Ebola-TR3), are identical in their core hybridizations. The IntaRNA results extend the BLAST 15-bp core antisense matches to 17/18 consecutive base pairs for the HIV-1 nef region vs. TR1, and 21/22 base pairs, with one single-base insertion, for Ebola NP vs. TR3 (see files S3 and S4). In both cases, even after subtraction of computed unfolding energies for internal mRNA secondary structures, both interacting pairs still have net interaction energies of ~20 kcal/mol, and are the unique results predicted by IntaRNA for these mRNA pairs.
Alignment A in Fig. 1 also shows differences between 1976 and 2014 strains of EBOV, and between humans and fruit bats, in the antisense-tethered region of TR3. In the mutated positions (black italic letters above or below the alignment), the 2014 EBOV sequence is in all cases a better antisense match to the human TR3. In one location where the mutations align, a C base unique to 1976 EBOV gives a better match to the fruit bat TR3 (GC base pair), whereas the 2014 EBOV has mutated to a U, giving a better match to the human TR3 (AU base pair). This is consistent with the possibility that, in regard to this putative interaction with TR3 mRNA, EBOV may be gradually adapting from bats to humans and other primate hosts such as chimpanzees and gorillas, whose TR3 sequences are identical to humans in this region.
The predicted interactions are unambiguously supported by gel shift assays, which show strong in vitro DNA hybridization between the antisense partners. In Fig. 2, the matched pairs of oligos corresponding to A and B in Fig. 1 are labeled Enp/TR3 and Hnf/TR1 respectively. Combined at a 1:1 ratio, both cognate antisense pairs form predominantly double-stranded dsDNA, either in buffer solution (lanes 2 and 6), or in the presence of sheared cellular DNA (lanes 9 and 10); at a 2:1 ratio, both single-stranded ssDNA and dsDNA bands are observed (lanes 3 and 7). The interactions are specific, as no visible dsDNA is formed from mismatched pairs like Enp/TR1, Hnf/TR3, or Hnf/Enp under identical conditions (lanes 11-13).
The existence of ATI between the 3′ regions of viral mRNAs and host selenoprotein mRNAs suggests a new model for viral selenoprotein synthesis, as a variant of the known mechanism of eukaryotic cellular selenoprotein synthesis (Fig. 3). As shown schematically in Fig. 3B, the “tail-to-tail” antisense interaction between the 3′ ends of the TR1 and HIV-1 mRNAs spans the 3′ end of the viral nef coding sequence, which terminates in a UGA codon that is highly conserved in global HIV-1 isolates [12]. Significantly, the EBOV NP gene also terminates in a UGA codon, conserved in all 1976 through 2014 Zaire EBOV isolates, but not in the much less pathogenic Reston ebolavirus, in which the NP terminates in a UAA. In both HIV-1 nef and the EBOV NP mRNAs, recoding of the terminal UGA of these proteins as Sec via a tethered SECIS element (Fig. 3B) would produce a slightly extended isoform of the protein, containing a single selenium atom as Sec, plus a few residues encoded past the UGA. This is analogous to TR, where the UGA is also at the protein C-terminus; in HIV-1 nef, the sequence even mimics the TR redox center, with a conserved Cys immediately preceding the UGA [12].
Fig. (3).
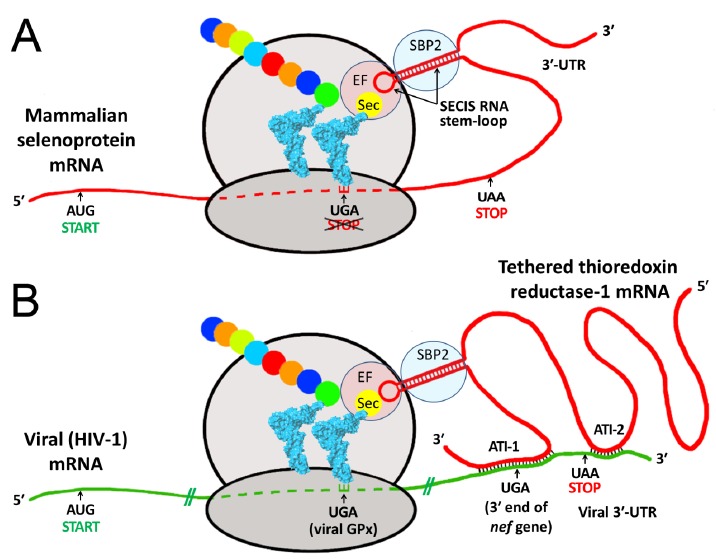
Proposed mechanism of selenocysteine (Sec) incorporation into viral proteins via hijacking of a SECIS element from a tethered host selenoprotein mRNA. Both panels show schematic ribosomes with bound tRNAs (blue), one carrying the rare selenium-containing amino acid Sec, the other a growing peptide chain (colored beads). A. During biosynthesis of mammalian selenoproteins, Sec is inserted at UGA codons, normally a STOP signal for protein synthesis. This mechanism requires a specialized RNA stem-loop structure, the SECIS element, generally located in the 3′ untranslated region (3′-UTR) of the selenoprotein mRNA [4]. By recruiting various protein factors, including SECIS binding protein 2 (SBP2) and elongation factor Sec (EF), the SECIS element enables delivery of tRNASec to the ribosome for Sec incorporation at the UGA codon, preventing it from acting as a stop signal. B. Using HIV-1 as an example, the lower panel shows how a viral mRNA, via antisense tethering interactions (ATI), could hijack a host SECIS element for decoding viral selenoprotein modules, such as the HIV-encoded glutathione peroxidase (viral GPx) [7-8, 13]. ATI-1 is the interaction shown as structure B in Fig. 1, and spans the highly conserved 3′-UGA codon of the nef gene; ATI-2 is a second shorter antisense region, consisting of 13 consecutive Watson-Crick base pairs near the end of the viral mRNA (bases 9111-9123, CAGCUGCUUUUUG). Similarly, in the Ebola nucleoprotein mRNA, less than 350 bases from the ATI shown as structure A in Fig. 1, there is also a secondary ATI region downstream of the conserved 3′-UGA stop codon (bases 2719-2734, CGACAAAUAGCUAACA), with only one mismatch to TR3 over 16 base pairs (A, opposite a G in TR3).
The presence in a viral mRNA of an antisense region capable of tethering a host selenoprotein mRNA, located either immediately overlapping (HIV-1) or within 300 bases (Ebola NP) of a highly conserved in-frame UGA codon, is strong circumstantial evidence favoring the possibility that the UGA can be translated as Sec, even if inefficiently. Additionally, in both viral mRNAs, there is a shorter antisense binding region (labeled ATI-2 in Fig. 3) immediately downstream of the conserved UGA codon. Both viral mRNAs also have upstream -1 ribosomal frameshift sites: the viral GPx in HIV-1 [7, 13] and predicted sites in Ebola virus [14], that would enable access to other in-frame UGA codons during protein synthesis, providing a further rationale for a need to capture a host SECIS element. Because HIV-nef and Ebola-NP are produced in abundant quantities in infected cells, even if recoding of their terminal UGA codons as Sec was very inefficient, production of only a fraction of a percent of a selenium-containing isoform by this mechanism could perturb the synthesis of cellular selenoproteins, by depletion of a limited pool of Sec.
An alternate hypothesis to viral selenoprotein synthesis is that these antisense interactions may simply lead to downregulation of host selenoproteins via translational inhibition analogous to that by microRNA, without mRNA degradation [1]. In either case, levels of cellular selenoproteins would likely decrease, impairing host defenses in general and antioxidant defenses in particular, and/or helping to create conditions more favorable for viral replication and transfer to new hosts (e.g., coagulopathy in Ebola, because of the role of selenium in the regulation of blood clotting).
In the case of HIV-1, a negative correlation between selenium status and mortality has been firmly established [15-16], and significant clinical benefits of selenium supplementation have been demonstrated in various studies [17-19]. Nor is HIV an isolated example, because there are other established cases of chemoprotective/antiviral effects of dietary selenium, including mammary tumors caused by MMTV, a retrovirus [20], Keshan disease myocarditis, linked to coxsackie virus [21], liver cancer and hepatitis linked to hepatitis B virus [22], and even one example in which an Asian viral hemorrhagic fever was successfully treated with oral sodium selenite, giving an overall 80% reduction in mortality [23]. These results are not entirely unexpected, considering the many essential roles of selenium in the immune system [24].
Evidence for benefits of selenium supplementation in various RNA viral infections, and the host mechanisms involved, was recently reviewed by Steinbrenner et al., who pointed out that “Populations in several countries most afflicted by past and current outbreaks of Ebola fever (e.g., Liberia, Guinea, Democratic Republic of Congo) exhibit a high risk of selenium deficiency”, with Liberia being the lowest ranked African nation for dietary selenium supply [25].
Regarding Ebola, selenium is known to play a significant role in the regulation of blood clotting [26], and thus could play a role in the coagulopathy characteristic of Ebola hemorrhagic fever [14]. Intravenous selenite is also an effective treatment for septic shock [27], which has clinical similarities to hemorrhagic fever [28]. Such observations are consistent with the possibility of a virus-induced selenium defect, resulting from ATIs between Ebola mRNAs and host selenoprotein mRNAs, which could impair host selenoprotein synthesis by antisense translational inhibition, or by competition for Sec, or both. In either case, increasing selenium intake would be expected to reduce the detrimental effects of Ebola on host Se-dependent mechanisms, and selenium deficiency would be predicted to be a risk factor for increased mortality.
If these ATIs are found to be functionally significant at the mRNA level, and their effects confirmed via proteomics studies, it will necessitate a re-evaluation of the multifaceted role of selenium in virus-host interactions, and its clinical significance, particularly in Ebola infections.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Adam Hall for reviewing the manuscript and for providing helpful suggestions. This research was supported by a gift from the Dr. Arthur and Bonnie Ennis Foundation, Decatur, IL, to E.W.T.
SUPPLEMENTARY MATERIAL
The data files S1-S4 cited in the text can be obtained by email from the corresponding author, or downloaded as a merged PDF file from: http://www.researchgate.net/profile/ Ethan_Taylor2/publications.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Zeng Y., Yi R., Cullen B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khorkova O., Myers A.J., Hsiao J., Wahlestedt C. Natural antisense transcripts. Hum. Mol. Genet. 2014;23(R1):R54–R63. doi: 10.1093/hmg/ddu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahmoudi S., Henriksson S., Corcoran M., Mendez-Vidal C., Wiman K.G., Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Berry M.J., Banu L., Harney J.W., Larsen P.R. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shisler J.L., Senkevich T.G., Berry M.J., Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279:102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W., Ramanathan C.S., Nadimpalli R.G., Bhat A.A., Cox A.G., Taylor E.W. Selenium-dependent glutathione peroxidase modules encoded by RNA viruses. Biol. Trace Elem. Res. 1999;70:97–116. doi: 10.1007/BF02783852. [DOI] [PubMed] [Google Scholar]
- 7.Zhao L., Cox A.G., Ruzicka J.A., Bhat A.A., Zhang W., Taylor E.W. Molecular modeling and in vitro activity of an HIV-1-encoded glutathione peroxidase. Proc. Natl. Acad. Sci. USA. 2000;97:6356–6361. doi: 10.1073/pnas.97.12.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen I., Boya P., Zhao L., Metivier D., Andreau K., Perfettini J.L., Weaver J.G., Badley A., Taylor E.W., Kroemer G. Anti-apoptotic activity of the glutathione peroxidase homologue encoded by HIV-1. Apoptosis. 2004;9:181–192. doi: 10.1023/B:APPT.0000018800.87358.ba. [DOI] [PubMed] [Google Scholar]
- 9.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busch A., Richter A.S., Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright P. R., Georg J., Mann M., Sorescu D. A., Richter A. S., Lott S., Kleinkauf R., Hess W. R., Backofen R. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. 2014. [DOI] [PMC free article] [PubMed]
- 12.Taylor E.W., Cox A.G., Zhao L., Ruzicka J.A., Bhat A.A., Zhang W., Nadimpalli R.G., Dean R.G. Nutrition, HIV, and drug abuse: the molecular basis of a unique role for selenium. J. Acquir. Immune Defic. Syndr. 2000;25(Suppl. 1):S53–S61. doi: 10.1097/00042560-200010001-00009. [DOI] [PubMed] [Google Scholar]
- 13.Olubajo B., Taylor E.W. -1 frameshift in the HIV-1 env gene is enhanced by arginine deficiency via a hungry codon mechanism. Mutat. Res. 2005;579:125–132. doi: 10.1016/j.mrfmmm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Ramanathan C.S., Taylor E.W. Computational genomic analysis of hemorrhagic fever viruses. Viral selenoproteins as a potential factor in pathogenesis. Biol. Trace Elem. Res. 1997;56:93–106. doi: 10.1007/BF02778985. [DOI] [PubMed] [Google Scholar]
- 15.Baum M.K., Shor-Posner G., Lai S., Zhang G., Lai H., Fletcher M.A., Sauberlich H., Page J.B. High risk of HIV-related mortality is associated with selenium deficiency. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1997;15:370–374. doi: 10.1097/00042560-199708150-00007. [DOI] [PubMed] [Google Scholar]
- 16.Constans J., Pellegrin J.L., Sergeant C., Simonoff M., Pellegrin I., Fleury H., Leng B., Conri C. Serum selenium predicts outcome in HIV infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;10:392. doi: 10.1097/00042560-199511000-00015. [DOI] [PubMed] [Google Scholar]
- 17.Baum M.K., Campa A., Lai S., Sales Martinez S., Tsalaile L., Burns P., Farahani M., Li Y., van Widenfelt E., Page J.B., Bussmann H., Fawzi W.W., Moyo S., Makhema J., Thior I., Essex M., Marlink R. Effect of micronutrient supplementation on disease progression in asymptomatic, antiretroviral-naive, HIV-infected adults in Botswana: a randomized clinical trial. JAMA. 2013;310:2154–2163. doi: 10.1001/jama.2013.280923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurwitz B. E., Klaus J. R., Llabre M. M., Gonzalez A., Lawrence P. J., Maher K. J., Greeson J. M., Baum M. K., Shor-Posner G., Skyler J. S., Schneiderman N. Suppression of human immunodeficiency virus type 1 viral load with selenium supplementation: a randomized controlled trial. 2007. [DOI] [PubMed]
- 19.Jiamton S., Pepin J., Suttent R., Filteau S., Mahakkanukrauh B., Hanshaoworakul W., Chaisilwattana P., Suthipinittharm P., Shetty P., Jaffar S. A randomized trial of the impact of multiple micronutrient supplementation on mortality among HIV-infected individuals living in Bangkok. AIDS. 2003;17:2461–2469. doi: 10.1097/00002030-200311210-00008. [DOI] [PubMed] [Google Scholar]
- 20.Schrauzer G.N., Molenaar T., Kuehn K., Waller D. Effect of simulated American, Bulgarian, and Japanese human diets and of selenium supplementation on the incidence of virally induced mammary tumors in female mice. Biol. Trace Elem. Res. 1989;20:169–178. doi: 10.1007/BF02919109. [DOI] [PubMed] [Google Scholar]
- 21.Beck M.A., Kolbeck P.C., Shi Q., Rohr L.H., Morris V.C., Levander O.A. Increased virulence of a human enterovirus (coxsackievirus B3) in selenium-deficient mice. J. Infect. Dis. 1994;170:351–357. doi: 10.1093/infdis/170.2.351. [DOI] [PubMed] [Google Scholar]
- 22.Yu S.Y., Zhu Y.J., Li W.G. Protective role of selenium against hepatitis B virus and primary liver cancer in Qidong. Biol. Trace Elem. Res. 1997;56:117–124. doi: 10.1007/BF02778987. [DOI] [PubMed] [Google Scholar]
- 23.Hou J.C. Inhibitory effect of selenite and other antioxidants on complement-mediated tissue injury in patients with epidemic hemorrhagic fever. Biol. Trace Elem. Res. 1997;56:125–130. doi: 10.1007/BF02778988. [DOI] [PubMed] [Google Scholar]
- 24.McKenzie R.C., Rafferty T.S., Beckett G.J. Selenium: an essential element for immune function. Immunol. Today. 1998;19:342–345. doi: 10.1016/s0167-5699(98)01294-8. [DOI] [PubMed] [Google Scholar]
- 25.Steinbrenner H., Al-Quraishy S., Dkhil M.A., Wunderlich F., Sies H. Dietary Selenium in Adjuvant Therapy of Viral and Bacterial Infections. Adv. Nutr. 2015;6:73–82. doi: 10.3945/an.114.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meydani M. Modulation of the platelet thromboxane A2 and aortic prostacyclin synthesis by dietary selenium and vitamin E. Biol. Trace Elem. Res. 1992;33:79–86. doi: 10.1007/BF02783995. [DOI] [PubMed] [Google Scholar]
- 27.Huang T.S., Shyu Y.C., Chen H.Y., Lin L.M., Lo C.Y., Yuan S.S., Chen P.J. Effect of parenteral selenium supplementation in critically ill patients: a systematic review and meta-analysis. PLoS One. 2013;8:e54431. doi: 10.1371/journal.pone.0054431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray M., Mahanty S. Ebola hemorrhagic fever and septic shock. J. Infect. Dis. 2003;188:1613–1617. doi: 10.1086/379727. [DOI] [PubMed] [Google Scholar]


