Abstract
A long-standing desire in biological and biomedical sciences is to be able to probe cellular chemistry as biological processes are happening inside living cells. Synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectral microscopy is a label-free and nondestructive analytical technique that can provide spatiotemporal distributions and relative abundances of biomolecules of a specimen by their characteristic vibrational modes. Despite great progress in recent years, SR-FTIR imaging of living biological systems remains challenging because of the demanding requirements on environmental control and strong infrared absorption of water. To meet this challenge, microfluidic devices have emerged as a method to control the water thickness while providing a hospitable environment to measure cellular processes and responses over many hours or days. This paper will provide an overview of microfluidic device development for SR-FTIR imaging of living biological systems, provide contrast between the various techniques including closed and open-channel designs, and discuss future directions of development within this area. Even as the fundamental science and technological demonstrations develop, other ongoing issues must be addressed; for example, choosing applications whose experimental requirements closely match device capabilities, and developing strategies to efficiently complete the cycle of development. These will require imagination, ingenuity and collaboration.
Keywords: FTIR, live cells, microfabrication, microfluidics, synchrotron radiation
1. Introduction
Research within the life sciences is at a defining moment. With an extraordinarily detailed genome-based understanding of cells, many researchers now seek to understand and define the principles guiding translation of the genetic code into the metabolic and regulatory networks underlying cellular function. The recent revolutions in single-cell genomic [1-4], proteomic [5-8], and transcriptomic [9-12] analysis have provided biomolecular details underscoring key processes in biological systems. Modern imaging techniques combining antibody staining or fluorescent markers with computation analysis have allowed researchers to study the spatiotemporal behavior of specific gene products in fixed cells, fixed tissue sections or living cells [13-15]. Complementary to these methods, Fourier transform infrared (FTIR) spectral microscopy (spectromicroscopy or microspectro-scopy), a label-free and non-destructive technique, enables real-time acquisition of broadband information on the cellular chemistry [16].
FTIR spectral microscopy uses a combination of visible light microscopy to examine the morphology of a biological specimen and infrared light illumination and interferometer to identify molecular composition. Illumination with infrared light promotes energy exchange between the inherent vibrational modes of molecular bonds and incident photons. These exchanges result in distinct, fingerprint-like spectral bands that appear in absorption spectrum measured as a function of wavelength of incident light (typically expressed in units of wavenumber, cm-1). Figure 1 highlights the origin of different stretching or bending vibrational bands commonly encountered in biological samples. The precise position, line shape, and intensity of these absorption bands depend on the molecular structure and conformation as well as intra- and inter- molecular interactions.
Figure 1.
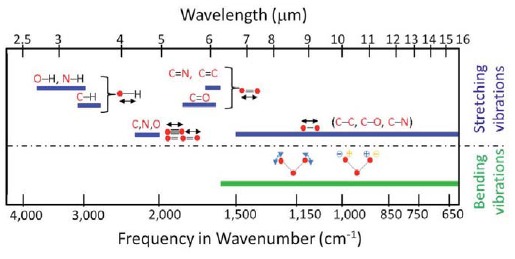
Vibrational modes of common biomolecular bonds - Inherent vibrational motion of molecules gives rise to distinct, fingerprint-like absorption bands in the mid-infrared region. Schematic shows different stretching or bending vibrational bands commonly encountered in biological samples. Infrared spectroscopy is sensitive to the presence of many chemical functional groups (structural fragments) in molecules, and taken together the set of vibration modes are unique for every molecular configuration.
The first experimental demonstration of infrared spectroscopy of biological samples was performed in 1949 using a thermal infrared light source and a dispersive infrared spectrometer approach [17]. In the 1950s, similar approaches were applied for the identification of different species and strains of bacteria [18], to study living muscle cells from insects and animals [19], and for the comparison of spectral features of normal and neoplastic tissues and the chemical constituents (e.g., nucleic acids, carbohydrates, fats and proteins) [20]. Because the measurement process was slow and data analysis was time-consuming, infrared spectral microscopy did not become a widely used tool for studying cellular or tissue systems until the 1990s as a result of three technological breakthroughs: (i) the application of the fast Fourier transform algorithm, (ii) the availability of inexpensive, fast digital computers which enabled the replacement of dispersive spectrometers with FTIR interferometers, and (iii) the introduction of fast-response, high-sensitivity photoconductive single-element mercury cadmium telluride (MCT) detectors. Chemical mapping could be performed by raster scanning the focused beam over the sample and collecting the data using a single point detector. In 1995, Lewis and Levin introduced focal plane array (FPA) detectors with a large number of small photovoltaic MCT detector elements [21] that could be used to image a larger sample region simultaneously. By the 2000s, most FTIR infrared microscopes were capable of using either a single-element detector for mapping or a FPA for imaging.
FTIR spectromicroscopy has emerged as a powerful tool for non-destructive, label-free chemical analysis of the structure and function of macromolecules in complex biological specimens such as cells and tissues. For example, FTIR spectral microscopy has been routinely applied to semi-quantitatively evaluate the relative abundance of carbonate, phosphate and collagen in mineralized bone and cartilage tissues [22], characterize the spatial distribution of main components (i.e., collagen and proteoglycans) in articular cartilage [23-26] and identify age-related structural changes in the DNA of prostate tissues and predict the metastatic state of tumors [27]. It has also been utilized for the detection of microcrystalline deposits of creatine in the brain tissues of post-mortem Alzheimer diseased humans as well as amyloid precursor protein (APP) transgenic mice [28] and for studying the effects of age and diet on the atherosclerotic lesion composition in rabbit aorta [29].
The introduction of synchrotron radiation as a brilliant and broadband infrared source (from far-IR to near-IR) made a significant contribution to modern FTIR spectral microscopy. According to the Rayleigh criterion, the theoretical spatial resolution limit of FTIR microscopes is ~λ, with microscope objectives having a typical numerical aperture (NA) of ~0.5 [30,31]. However, weak signal-to-noise ratio (S/N) provided by conventional thermal IR sources often limits the practical spatial resolution of FTIR microscopes to be ~20-50 μm. In contrast, the infrared light from synchrotron sources is 100-1000 times brighter than that from thermal sources [32], and allows for truly diffraction-limited spatial resolution with excellent S/N when used with a single point MCT detector. The diffraction-limited spot size of 2-10 μm allows mapping the composition and spatial variation of single (or clusters of) cells including microbial [33-36], fungal [37-41], algal [42-44], mammalian [45-51], and even subcellular components of certain mammalian cells [52-54]. Although SR-IR (or S-IR) is a tightly focused source, the low photon energy and low power (compared to IR laser sources) do not introduce detectable biochemical changes within a sample [55]. This non-invasive, label-free approach is greatly beneficial to experimenters that hope to perform further examination of the same sample using complementary methods such as staining [56], proteomic [57], genomic [58], or other –omics based analyses.
With the introduction of synchrotron illumination, interest in imaging live single cells and clusters of cells has increased dramatically. Imaging live cells can reduce some of the artifacts of fixation [59] and real-time FTIR measurements on living biological systems (Figure 2) are crucial to unravel how their chemical composition evolves with time [43]. This can give insights into many problems of biological or medical interest including metabolic processes [60,61], development [51], and real-time responses to treatment or changing environmental conditions [62,63]. Jamin et al. [46] performed the first synchrotron-based FTIR measurements of live cells in 1998. They measured the variation in lipid and protein distributions in mouse hybridoma B cells during cell division and necrosis using a humidified chamber. The use of humidified chambers has continued in a variety of applications including mammalian cell response to polychlorinated aromatic compounds [64], bacterial detoxification of chromium(VI) compounds [65], bacterial degradation of environmental carcinogens [60], oxygen-stress adaptive response in obligate anaerobes [35], and tracking differentiation process in neuron model cells [51]. These chambers, however, do not provide any way to replenish the media and only allow continuous measurement for a few hours before the biological system starts to degrade. Longer-term experiments require either repeated measurements of multiple samples at varying time points, sacrificing both temporal resolution and cell-cell variations within a population, or alternatively, the use of a microfluidic platform to maintain the biological system viability.
The primary challenge in imaging living biological systems with SR-FTIR illumination is maintaining environmental conditions optimal for cell viability while minimizing the absorption signal from water. Strong IR absorption from the –OH stretching and HOH bending bands can quickly saturate the signal for transmission measurements with water thickness greater than about 10 micrometers [66]. Live cells, meanwhile, consume surrounding nutrients from the media and produce waste that must be exchanged periodically to maintain cell viability.
Despite great progress, SR-FTIR imaging of living biological systems remains challenging because of the demanding requirements on environmental control [16]. To meet this challenge, microfluidic systems have emerged in the last decade as a method to control the water thickness while providing a hospitable environment in order to measure cellular processes and responses over many hours or days. They can be broadly categorized by whether the channel used to confine the cells is closed, where upper and lower IR transparent windows are sandwiched between a spacer, or open, where surface effects are harnessed to maintain a thin water layer at an air-liquid interface. To date, the vast majority of papers on this topic have employed various iterations of closed channel devices while open-channel devices have emerged recently as a viable competitor, albeit with some limitations. This paper will provide an overview of microfluidic device development for FTIR imaging of living biological systems, provide contrast between the various techniques, and discuss future directions of development within this area.
2. Closed-channel Methods
Liquid flow cells (Figure 3) represent the foundation of the microfluidic closed-channel devices that comprise the majority of devices in use at various synchrotron facilities today. First employed by Wieliczka et al. [67] in 1989 to measure the absorption coefficients of water, their basic structure is a micron-scale thick gasket pressed between two infrared crystals and mechanically assembled as a stack in an external manifold that may also allow for sample injection and temperature control. This configuration is generally used for transmission experiments and allows for a sample to be maintained in relatively uniform, thin layer of water with absorption signal below saturation so that the water background can be subtracted later to obtain the sample spectra [66, 68-70]. This scheme is versatile in that a variety of different windows may be used and it is typically demountable, so that flow cells can be disassembled for cleaning and reuse. The main drawback of demountable flow cells is that the path length, which relies on mechanical pressure, is not easily reproducible between measurements, increasing bias when comparing data from different experiments. Poor sealing, leakage, and limited experimental complexity are two additional shortcomings when it is compared to approaches that use microfabrication.
Figure 3.
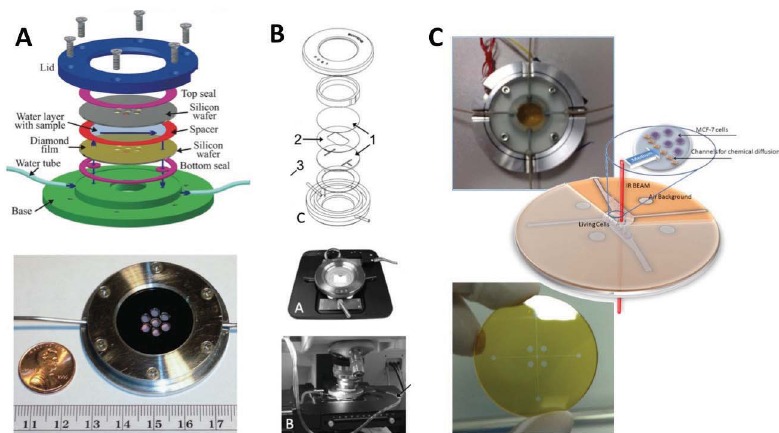
Closed-channel devices – A) Demountable stack-assembled flow cell (adapted from Nasse et al. [74]) to measure biological specimens with high spatial resolution. Cells are visualized through diamond films grown on silicon wafer and seals are maintained by mechanical pressure from assembled manifold. The flow cell can be disassembled after measurement to reuse parts or access cells. B) Demountable stack-assembled flow cell (adapted from Tobin et al. [73]) with machined features on separate layers. Based on a modified Bioptechs FCS3 cell, the channel and chamber layers are pressed between two CaF2 crystals for transmission measurement on a Bruker Hyperion microscope. C) Microfabrication allows more complex structures to be integrated into the flow cell and very precise control of water film thickness to be maintained uniformly over the entire measurement area. Device shown from Grenci et al. [87] has channels defined in a photosensitive polymer that covers the CaF2 window. The device is permanently sealed by thermomechanical bonding.
In 2005, Moss et al. [71] were the first to apply flow cell techniques to SR-FTIR imaging of single living cells. Human colorectal cancer cell lines (HT29, SW-480, WIDR, CaCO2), human fibroblasts (primary culture) and human umbilical vein endothelial cells from healthy controls, in both confluent and exponential cultures were plated onto one CaF2 window. The window was mounted in a standard liquid cell (SpectraTech EZ-Fill) with a 15 µm Teflon spacer and closed with another window to limit the water film thickness. Around the same time, Miljkovic et al. [72] also reported imaging live HeLa cells suspended in growth medium, acquired in both reflection and transmission modes, on different substrates (low-e slides and CaF2) and with spacers of different thicknesses using benchtop FTIR equipment. Heraud et al. [42] then built an in-house liquid cell based on the SpectraTech cell and demonstrated SR-FTIR mapping of the metabolite distribution in living algae Micrasterias hardyi. These proofs-of-principle experiments demonstrated the viability of microfluidic devices for SR-FTIR measurement of living cells.
The work that followed received significant interest from the IR community, particularly the IR synchrotron facilities [73-78]. Closed-channel configurations became a focal point and evolved into two main branches: One with demountable flow cell using plastic spacers and another micro-fabricated flow cells (both demountable and permanently sealed).
Demountable, stack-assembled cells inherited the basic layout of flow cells but added versatility for window selection and high-resolution imaging. The most common window material is CaF2 since it has good IR transparency, is relatively insoluble in water, and is cheap and robust compared to other materials such as BaF2 or ZnSe. In order to use high magnification objectives with small working distance, Nasse et al. [74] developed a demountable liquid cell with the viewports made of 0.4-0.8 µm diamond films grown by chemical vapor deposition on a silicon wafer. The total device thickness including the holder was only a few millimeters to take advantage of the high resolution imaging capabilities at the IRENI beamline and conform to the spatial constraints therein. Stack-assembled devices have been used to measure protein expression in live cells [79,80], DNA conformational changes [81], and monitor progression of the cell cycle [82]. This technique does not require microfabrication, and can mostly be implemented with off-the-shelf components outside of the custom manifold and machining through-holes in one of the windows. The drawback of this approach is the difficulty to implement sophisticated structures within the spacer and the path length reproducibility issues related to the mechanical clamping of the stack, giving variable compression of the spacer.
To cope with some of the limitations of stack-assembled cells, microfabrication has emerged as a method to implement more complex spacer designs and to build completely sealed, FTIR-compatible devices for live cell measurement. Hinsmann [83] performed the first use of microfluidic devices with infrared spectroscopy in 2001. With a mixer microstructure made between two calcium fluoride crystals, they were able to observe the real-time saponification of methyl monochloroacetate with sodium hydroxide. This approach was translated to living biological system imaging in 2010 by Birarda et al. [75] and Tobin et al. [73], where spacers were directly fabricated on top of IR-transparent windows using optical lithography or direct printing methods [84]. This method allowed microstructures like channels and reservoirs to be added to the cell chamber [85,86] or to change the material surface properties [87,88] while giving more precise control of the spacer thickness. The fabrication approach further allowed development of devices that were either thermo-mechanically or chemically sealed [59, 63] instead of being clamped together.
A few examples of the application of microfabricated devices include assessment of the effect of different fixatives and fixation protocols on living human monocytes [59], identification of the spectral markers of apoptotic and pre-apoptotic cells [89], monitoring the progression of cell cycle [90], and evaluating the chemical response of leukemia cells to drug treatments [91].
Some concerns have been raised with closed-channel devices in terms of the effects of both confinement and contact with non-conventional materials on the viability of cells over extended measurement periods. The use of deuterated water D2O [43,92] has been attempted to loosen the spatial constraint in the liquid film thickness to about 20 µm. However, it introduced other problems because cell viability cannot be sustained for long periods in D2O and isotopic exchange caused difficulties in water compensation during analysis. Birarda et al. [93] evaluated the effect of confinement on a circulating cell line. Monocyte cells (U937) were confined in devices having different thicknesses ranging from 9 (i.e. similar size of the cells) to 3 micrometers. Observation of the time evolution of DNA, protein and lipid bands showed that no cellular response was detectable for deformations lower than 60% for 100 minutes upon entering the device. This is an important consideration when working with large cells that may experience significant deformation to fit within 9-10 μm thick devices necessary for measurement.
Deformation is less of a concern for adherent cells but their interaction with the surface of infrared transparent materials may cause cytotoxicity problems. Wehbe et al. [94] found the infrared transparent Si, ZnS, and CaF2 did not impact the viability of adherent mammalian cell lines, but BaF2 and ZnSe did Mitri et al. [88] then demonstrated a nanometric layer of silicon dioxide can be used to protect cells from BaF2 and enable its exploitation of better transparency at lower frequencies. It remains an issue that prolonged exposure to shear stress during perfusion in closed-channel devices can damage cell viability [95] and has a limited measurement period of approximately 48 hours [87].
3. Open-channel Methods
A relatively new approach in microfluidic devices for live cell imaging is the application of open-channel devices (Figure 4). These are characterized by having one surface of the fluid exposed to atmosphere and the liquid thickness controlled by surface effects. In 2009, Holman et al. [96] used continuous flow of fluid through a microchannel etched in silicon, supplied by a combination of hydrostatic pressure and capillary forces. Hydrophobic treatment to other surfaces ensures water flows exclusively within the channel and forces were carefully balanced between the hydrostatic pressure of a feeder droplet, and capillary pull into a cleanroom tissue to maintain constant fluid thickness. This method was used to track the development and growth of an E. Coli biofilm over the course of two days using transmission measurement.
Figure 4.
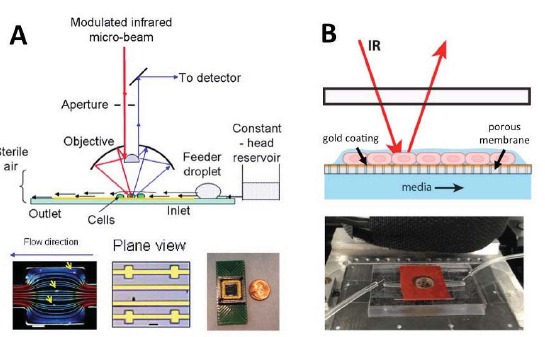
Open-channel devices – A) Open-channel device to study live bacteria in aqueous environments (adapted from Holman et al. [96]). The device is composed of channels etched into a silicon wafer. Flow of media is maintained in channels by hydrophobic treatment to non-channel surfaces and liquid flow is controlled by hydrostatic pressure in a feeder droplet. B) Membrane device to study cells that grow at the liquid air interface (adapted from Loutherback et al. [97]). The cells are maintained in a thin layer of water on top of a gold-coated porous membrane. Constant flow of media underneath the membrane allows for nutrients to be replenished by capillary action.
Loutherback et al. [97] recently demonstrated a second approach using an IR-reflective porous membrane to measure adherent mammalian cells for up to a week. Mammalian cells plated on top of the membrane were maintained in a thin layer of fluid above a larger flow channel from which they can draw fluid by capillary action. A thin (15 nm) gold layer deposited on top of the membrane allows FTIR measurement to be performed in a transflection mode. This device has allowed continuous measurements of live cells for up to seven days. The primary chemical observation showed an increase in carbohydrates associated with surfactants to manage evaporative stress by the cells, which suggests that a porous membrane-based approach is best suited for cells that naturally grow at air-liquid interfaces such as epithelial tissues of the skin, lung, eyes or microbial biofilms.
The advantage of the open-channel approach is that the fluid thickness can be maintained much thinner than that allowable by closed-channel devices, potentially reducing or removing the need to perform water background subtraction. These devices also reduce or avoid entirely the fringing effect from interference of multiple reflections at the infrared window surfaces by allowing the upper window to be placed much further from the lower window. A disadvantage of this approach is that environmental conditions must be maintained at high humidity to ensure that evaporation does not remove the media covering cell surface and the biological system must be chosen carefully to suit the environmental conditions.
4. Future directions
Figure 5 highlights future directions. Closed-channel devices using microfabrication can gain significant additional features with the incorporation of various well-developed microfluidic modalities [98]. The incorporation of cell traps [99,100] may be beneficial to place cells in well-registered locations for time-course measurements. Such structures may also help hold motile cells in place during measurement [43]. Water-in-oil droplets further may provide an alternative opportunity to encapsulate single cells [101] and monitor metabolic activity in well-characterized and isolated environments [102].
Figure 5.
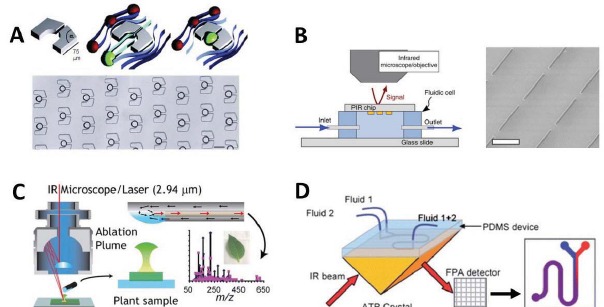
Future directions in microfluidic FTIR devices - A) Advanced microfluidic structures such as droplet generators and cell traps could be used for sample confinement and entrapment in an array format for measurement with SR-FTIR (adapted from Huebner et al.). B) Plasmonic microstructures can be used to increase the sensitivity of IR spectromicroscopy to extremely dilute analytes in solution (adapted from Adato et al. [112]). C) SR-FTIR in open-channel devices can be hyphenated with mass spectrometry for more detailed molecular identification as demonstrated by O’Brien et al. [57]. D) The coupling of SR illumination with large area focal plane array (FPA) imaging detectors can be used to employ SR for ATR imaging and fluidic micro incubators can be used for experiments on live cells (adapted from Chan et al. [108]).
One development that may prove particularly valuable is the ability to deposit smooth, IR-reflective layers of gold on polydimethylsiloxane (PDMS) [103]. While there remains concerns about electric field standing wave effects during transflection measurement [104], this material would allow cheap, rapid device fabrication and the inclusion of multilayer structures with a large suite of features including valves and mixers [105]. Mating through-holes in closed-channel devices to a PDMS-based, multiplexed in/out manifold may provide similar benefits while still allowing transmission measurements [106].
In addition to the open- and closed-channel devices, a third group of devices that are prominent in conventional FTIR imaging but have yet to see much use with synchrotron illumination sources are attenuated total reflection (ATR) cells or chambers. These ATR-based devices are functionally closer to environmental chambers than flow cells or microfluidic devices. ATR probes only a few microns at best at the crystal-liquid/cell interface, so the thickness of the total layer of water is not important, and it is possible to achieve petri dish-like condition for cell growth [107-111].
Another exciting area of future development is the integration of live-cell imaging with plasmonic nanoantennas for ultra-sensitive surface enhanced IR absorption spectroscopy. This could enable trace detection of physiologically significant yet elusive molecular species such as metabolites, cytokines, growth factors and antibodies. Towards this end, Adato et al. [112] have demonstrated an ultra-sensitive plasmonic internal reflection chip able to monitor monolayer protein binding events in real time with 10-fold signal enhancement over ATR.
Multimodal imaging in combination with mass spectrometry is also a promising area of future development. Infrared spectral data lacks sufficient chemical specificity for unique molecular identification. High-resolution mass spectrometry (MS) can make possible more complete identification of the full range of molecules involved in functional metabolism, including elemental composition obtained by accurate mass measurements and structural information gained from fragmentation products formed in tandem mass spectrometry measurements [113]. Combining these two techniques, using a demountable flow cell or open-channel device of certain kind, will allow the non-destructive, ambient chemical monitoring capabilities of SIR spectromicroscopy to be paired with efficient, spatially-resolved analysis by mass spectrometry.
5. Conclusion
Synchrotron-radiation Fourier transform infrared spectral microscopy has progressed rapidly in the last decade. Real-time measurements of biological processes in living cells have been and will continue to be one of the most exciting developments within this area. Various microfluidic devices have emerged as a platform for meeting the challenging requirement of providing both a hospitable environment where cellular processes and responses can be probed over many hours or days while maintaining water films thin enough to obtain high quality spectral information. Although it is in its infancy, and more work is needed before its full potential can be realized. The foundations of the field are already quite strong. The device development in this area is expected to proceed to improve environmental control and enable additional capabilities such as sequential chemical treatments, advanced microfluidic structures, and multimodal imaging. Other paradigms such as ATR or plasmonic antennas may allow circumvention of water thickness requirements and open whole new areas of application. Across all device designs, careful thought must be put into experimental pre-considerations for the biological system of interest, as each design has distinct strengths and weaknesses.
From infrared absorption data, spatially- and temporally-resolved chemical information, including the distributions and relative abundances of the classes of chemicals such as proteins, lipids, carbohydrates, or metabolites, is obtained. However, infrared data lack sufficient molecular or chemical specificity for unique identifications. The advent of genetically encoded labels enables the non-destructive fluorescence microscopy techniques [114,115] to provide highly selective and specific spatio-temporal information on the targeted cellular components or signalling molecules. Meanwhile, the destructive but high-chemical-resolution mass spectrometry (MS) can make possible more complete identification of the full range of molecules involved in functional metabolism, including elemental composition obtained by accurate mass measurements and structural information gained from fragmentation products formed in tandem mass spectrometry measurements [116-119]. Together, they will offer great, perhaps even revolutionary new capabilities for the future of SR-FTIR imaging of living biosystems.
Figure 2.
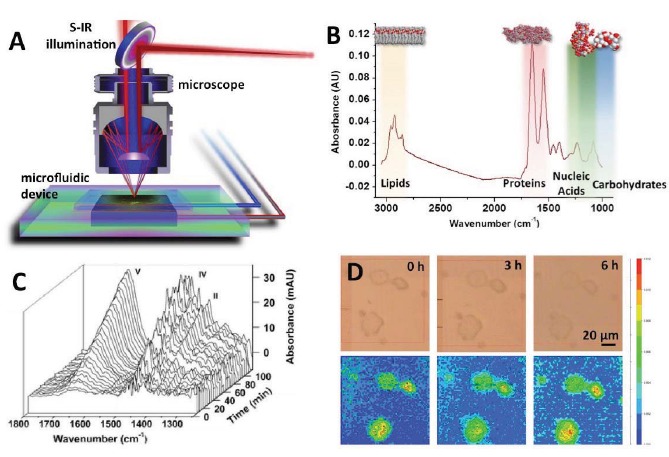
Synchrotron-radiation FTIR of live cells – A) Spatiotemporal chemical composition of live cells can be measured in microfluidic devices with an infrared microscope (adapted from Holman et al. [16]). Synchrotron infrared light is focused onto the sample by reflective optics and the reflected or transmitted light from the sample is directed to an infrared detector. Environmental conditions are maintained in the device through external fluidic connections. B) Infrared spectrum of a single human cell with color bands highlighting absorption peaks associated with constituent macromolecules. C) Repeated measurements of the same cells allow cellular processes to be recorded in real-time. Plot shows metabolite formation in single living Chlamydomonas reinhardtii (adapted from Goff et al. [43]). D) Chemical mapping shows how spatial distribution of chemical changes in several cells over time. Upper panels show visible images while lower panels show the lipid distribution in MCF-7 cells at time 0, 3, and 6 hours (adapted from Grenci et al. [87]).
ACKNOWLEDGEMENTS
This work was performed under the Berkeley Synchrotron Infrared Structural Biology (BSISB) Program funded by the US Department of Energy, Office of Science, and Office of Biological and Environmental Research. The Advanced Light Source is supported by the Director, Office of Science, and Office of Basic Energy Sciences. Both were supported through Contract DE-AC02-225 05CH11231.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Walker A., Parkhill J. Genome watch - Single-cell genomics. Nat. Rev. Microbiol. 2008;6(3):176–177. doi: 10.1038/nrmicro1862. [DOI] [PubMed] [Google Scholar]
- 2.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W.R., Hicks J. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalisky T., Quake S.R. Single-cell genomics. Nat. Methods. 2011;8(4):311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole Genome Amplification from a Single Cell - Implications for Genetic-Analysis. Proc. Natl. Acad. Sci. USA. 1992;89(13):5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman J.R., Ghaemmaghami S., Ihmels J., Breslow D.K., Noble M., DeRisi J.L., Weissman J.S. Single-cell proteomic analysis of S-cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 6.Shi Q.H., Qin L., Wei W., Geng F., Fan R., Shin Y.S., Guo D., Hood L., Mischel P.S., Heath J.R. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. USA. 2012;109(2):419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salehi-Reyhani A., Kaplinsky J., Burgin E., Novakova M., deMello A.J., Templer R.H., Parker P., Neil M.A., Ces O., French P., Willison K.R., Klug D. A first step towards practical single cell proteomics: a microfluidic antibody capture chip with TIRF detection. Lab Chip. 2011;11(7):1256–1261. doi: 10.1039/c0lc00613k. [DOI] [PubMed] [Google Scholar]
- 8.Breker M., Gymrek M., Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 2013;200(6):839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D., Trombetta J.J., Gennert D., Gnirke A., Goren A., Hacohen N., Levin J.Z., Park H., Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam S., Zeisel A., Joost S., La Mannom G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11(2):163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 11.Grun D., Kester L., van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11(6):637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z.T., Wang D.C., Popescu L.M., Wang X. Single-cell transcriptome in the identification of disease biomarkers: opportunities and challenges. J. Transl. Med. 2014;12:212–214. doi: 10.1186/s12967-014-0212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi Y., Choi P.J., Li G.W., Chen H., Babu M., Hearn J., Emili A., Xie X.S. Quantifying E-coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberwine J., Yeh H., Miyashiro K., Cao Y., Nair S., Finnell R., Zettel M., Coleman P. Analysis of Gene-Expression in Single Live Neurons. Proc. Natl. Acad. Sci. USA. 1992;89(7):3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holman H.Y., Bechtel H.A., Hao Z., Martin M.C., Synchrotron I.R. Spectromicroscopy: Chemistry of Living Cells. Anal. Chem. 2010;82(21):8757–8765. doi: 10.1021/ac100991d. [DOI] [PubMed] [Google Scholar]
- 17.Blout E.R., Mellors R.C. Infrared Spectra of Tissues. Science. 1949;110(2849):137–138. doi: 10.1126/science.110.2849.137. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson H.J., Bolduan O.E. Infrared Spectrophotometry as a Means for Identification of Bacteria. Science. 1952;116(3005):111–113. doi: 10.1126/science.116.3005.111. [DOI] [PubMed] [Google Scholar]
- 19.Wood D.L. Infrared Microspectrum of Living Muscle Cells. Science. 1951;114(2950):36–38. doi: 10.1126/science.114.2950.36. [DOI] [PubMed] [Google Scholar]
- 20.Woernley D.L. Infrared Absorption Curves for Normal and Neoplastic Tissues and Related Biological Substances. Cancer Res. 1952;12(7):516–523. [PubMed] [Google Scholar]
- 21.Lewis E.N., Treado P.J., Reeder R.C., Story G.M., Dowrey A.E., Marcott C., Levin I.W. Fourier-Transform Spectroscopic Imaging Using an Infrared Focal-Plane Array Detector. Anal. Chem. 1995;67(19):3377–3381. doi: 10.1021/ac00115a003. [DOI] [PubMed] [Google Scholar]
- 22.Boskey A., Camacho N.P. FT-IR imaging of native and tissue-engineered bone and cartilage. Biomaterials. 2007;28(15):2465–2478. doi: 10.1016/j.biomaterials.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieppo L., Rieppo J., Jurvelin J.S., Saarakkala S. Fourier Transform Infrared Spectroscopic Imaging and Multivariate Regression for Prediction of Proteoglycan Content of Articular Cartilage. PLoS One. 2012;7(2):8. doi: 10.1371/journal.pone.0032344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieppo L., Saarakkala S., Närhi T., Holopainen J., Lammi M., Helminen H.J., Jurvelin J.S., Rieppo J. Quantitative Analysis of Spatial Proteoglycan Content in Articular Cartilage With Fourier Transform Infrared Imaging Spectroscopy: Critical Evaluation of Analysis Methods and Specificity of the Parameters. Microsc. Res. Tech. 2010;73(5):503–512. doi: 10.1002/jemt.20789. [DOI] [PubMed] [Google Scholar]
- 25.Potter K., Kidder L.H., Levin I.W., Lewis E.N., Spencer R.G. Imaging of collagen and proteoglycan in cartilage sections using Fourier transform infrared spectral imaging. Arthritis Rheum. 2001;44(4):846–855. doi: 10.1002/1529-0131(200104)44:4<846::AID-ANR141>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Camacho N.P., West P., Torzilli P.A., Mendelsohn R. FTIR microscopic imaging of collagen and proteoglycan in bovine cartilage. Biopolymers. 2001;62(1):1–8. doi: 10.1002/1097-0282(2001)62:1<1::AID-BIP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Malins D.C., Johnson P.M., Barker E.A., Polissar N.L., Wheeler T.M., Anderson K.M. Cancer-related changes in prostate DNA as men age and early identification of metastasis in primary prostate tumors. Proc. Natl. Acad. Sci. USA. 2003;100(9):5401–5406. doi: 10.1073/pnas.0931396100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallant M., Rak M., Szeghalmi A., Del Bigio M.R., Westaway D., Yang J., Julian R., Gough K.M. Focally elevated creatine detected in amyloid precursor protein (APP) transgenic mice and Alzheimer disease brain tissue. J. Biol. Chem. 2006;281(1):5–8. doi: 10.1074/jbc.C500244200. [DOI] [PubMed] [Google Scholar]
- 29.Palombo F., Cremers S.G., Weinberg P.D., Kazarian S.G. Application of Fourier transform infrared spectroscopic imaging to the study of effects of age and dietary L-arginine on aortic lesion composition in cholesterol-fed rabbits. J. R. Soc. Interface. 2009;6(37):669–680. doi: 10.1098/rsif.2008.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller L.M., Dumas P. Chemical imaging of biological tissue with synchrotron infrared light. Biochim. Biophys. Acta. 2006;1758(7):846–857. doi: 10.1016/j.bbamem.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Matthaus C. Chapter 10: Infrared and Raman microscopy in cell biology. Methods Cell Biol. 2008;89:275–308. doi: 10.1016/S0091-679X(08)00610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan W.D., Williams G.P. Infrared synchrotron radiation from electron storage rings. Appl. Opt. 1983;22(18):2914. doi: 10.1364/ao.22.002914. [DOI] [PubMed] [Google Scholar]
- 33.Probst A.J., Holman H.Y., DeSantis T.Z., Andersen G.L., Birarda G., Bechtel H.A., Piceno Y.M., Sonnleitner M., Venkateswaran K., Moissl-Eichinger C. Tackling the minority: sulfate-reducing bacteria in an archaea-dominated subsurface biofilm. ISME J. 2013;7(3):635–651. doi: 10.1038/ismej.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luef B., Frischkorn K.R., Wrighton K.C., Holman H.Y., Birarda G., Thomas B.C., Singh A., Williams K.H., Siegerist C.E., Tringe S.G. Downing KH5, Comolli LR5, Banfield JF6. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015:6. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 35.Holman H.Y., Wozei E., Lin Z., Comolli L.R., Ball D.A., Borglin S., Fields M.W., Hazen T.C., Downing K.H. Real-time molecular monitoring of chemical environment in obligate anaerobes during oxygen adaptive response. Proc. Natl. Acad. Sci. USA. 2009;106(31):12599–12604. doi: 10.1073/pnas.0902070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baelum J., Borglin S., Chakraborty R., Fortney J.L., Lamendella R., Mason O.U., Auer M., Zemla M., Bill M., Conrad M.E., Malfatti S.A., Tringe S.G., Holman H.Y., Hazen T.C., Jansson J.K. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ. Microbiol. 2012;14(9):2405–2416. doi: 10.1111/j.1462-2920.2012.02780.x. [DOI] [PubMed] [Google Scholar]
- 37.Kaminskyj S., Jilkine K., Szeghalmi A., Gough K. High spatial resolution analysis of fungal cell biochemistry - bridging the analytical gap using synchrotron FTIR spectromicroscopy. FEMS Microbiol. Lett. 2008;284(1):1–8. doi: 10.1111/j.1574-6968.2008.01162.x. [DOI] [PubMed] [Google Scholar]
- 38.Jilkine K., Gough K.M., Julian R., Kaminskyj S.G. A sensitive method for examining whole-cell biochemical composition in single cells of filamentous fungi using synchrotron FTIR spectromicroscopy. J. Inorg. Biochem. 2008;102(3):540–546. doi: 10.1016/j.jinorgbio.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Jamme F., Vindigni J.D., Méchin V., Cherifi T., Chardot T., Froissard M. Single Cell Synchrotron FT-IR Microspectroscopy Reveals a Link between Neutral Lipid and Storage Carbohydrate Fluxes in S. cerevisiae. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szeghalmi A., Kaminskyj S., Gough K.M. A synchrotron FTIR microspectroscopy investigation of fungal hyphae grown under optimal and stressed conditions. Anal. Bioanal. Chem. 2007;387(5):1779–1789. doi: 10.1007/s00216-006-0850-2. [DOI] [PubMed] [Google Scholar]
- 41.Saulou C., Jamme F., Maranges C., Fourquaux I., Despax B., Raynaud P., Dumas P., Mercier-Bonin M. Synchrotron FTIR microspectroscopy of the yeast Saccharomyces cerevisiae after exposure to plasma-deposited nanosilver-containing coating. Anal. Bioanal. Chem. 2010;396(4):1441–1450. doi: 10.1007/s00216-009-3316-5. [DOI] [PubMed] [Google Scholar]
- 42.Heraud P., Wood B.R., Tobin M.J., Beardall J., McNaughton D. Mapping of nutrient-induced biochemical changes in living algal cells using synchrotron infrared microspectroscopy. FEMS Microbiol. Lett. 2005;249(2):219–225. doi: 10.1016/j.femsle.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Goff K.L., Quaroni L., Wilson K.E. Measurement of metabolite formation in single living cells of Chlamydomonas reinhardtii using synchrotron Fourier-Transform Infrared spectromicroscopy. Analyst (Lond.) 2009;134(11):2216–2219. doi: 10.1039/b915810c. [DOI] [PubMed] [Google Scholar]
- 44.Hirschmugl C.J., Bayarrib Z-E., Buntaa M., Holta J.B., Giordanoc M. Analysis of the nutritional status of algae by Fourier transform infrared chemical imaging. Infrared Phys. Technol. 2006;49(1-2):57–63. [Google Scholar]
- 45.Flower K.R., Khalifa I., Bassan P., Démoulin D., Jackson E., Lockyer N.P., McGown A.T., Miles P., Vaccari L., Gardner P. Synchrotron FTIR analysis of drug treated ovarian A2780 cells: an ability to differentiate cell response to different drugs? Analyst (Lond.) 2011;136(3):498–507. doi: 10.1039/c0an00564a. [DOI] [PubMed] [Google Scholar]
- 46.Jamin N., Dumas P., Moncuit J., Fridman W.H., Teillaud J.L., Carr G.L., Williams G.P. Highly resolved chemical imaging of living cells by using synchrotron infrared microspectrometry. Proc. Natl. Acad. Sci. USA. 1998;95(9):4837–4840. doi: 10.1073/pnas.95.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pijanka J., Dumas P., Moncuit J., Fridman W.H., Teillaud J.L., Carr G.L., Williams G.P. Synchrotron-based FTIR spectra of stained single cells. Towards a clinical application in pathology. Lab. Invest. 2010;90(5):797–807. doi: 10.1038/labinvest.2010.8. [DOI] [PubMed] [Google Scholar]
- 48.German M.J., Hammiche A., Ragavan N., Tobin M.J., Cooper L.J., Matanhelia S.S., Hindley A.C., Nicholson C.M., Fullwood N.J., Pollock H.M., Martin F.L. Infrared spectroscopy with multivariate analysis potentially facilitates the segregation of different types of prostate cell. Biophys. J. 2006;90(10):3783–3795. doi: 10.1529/biophysj.105.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krafft C., Salzer R., Seitz S., Ern C., Schieker M. Differentiation of individual human mesenchymal stem cells probed by FTIR microscopic imaging. Analyst (Lond.) 2007;132(7):647–653. doi: 10.1039/b700368d. [DOI] [PubMed] [Google Scholar]
- 50.Gazi E., Dwyer J., Lockyer N.P., Miyan J., Gardner P., Hart C.A., Brown M.D., Clarke N.W. A study of cytokinetic and motile prostate cancer cells using synchrotron-based FTIR micro spectroscopic imaging. Vib. Spectrosc. 2005;38(1-2):193–201. [Google Scholar]
- 51.Chen L., Holman H.Y., Hao Z., Bechtel H.A., Martin M.C., Wu C., Chu S. Synchrotron Infrared Measurements of Protein Phosphorylation in Living Single PC12 Cells during Neuronal Differentiation. Anal. Chem. 2012;84(9):4118–4125. doi: 10.1021/ac300308x. [DOI] [PubMed] [Google Scholar]
- 52.Pijanka J.K., Kohler A., Yang Y., Dumas P., Chio-Srichan S., Manfait M., Sockalingum G.D., Sulé-Suso J. Spectroscopic signatures of single, isolated cancer cell nuclei using synchrotron infrared microscopy. Analyst (Lond.) 2009;134(6):1176–1181. doi: 10.1039/b821112d. [DOI] [PubMed] [Google Scholar]
- 53.Gazi E., Dwyer J., Lockyer N.P., Miyan J., Gardner P., Hart C., Brown M., Clarke N.W. Fixation protocols for subcellular imaging by synchrotron-based Fourier transform infrared microspectroscopy. Biopolymers. 2005;77(1):18–30. doi: 10.1002/bip.20167. [DOI] [PubMed] [Google Scholar]
- 54.Kastyak-Ibrahim M.Z., Nasse M.J., Rak M., Hirschmugl C., Del Bigio M.R., Albensi B.C., Gough K.M. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage. 2012;60(1):376–383. doi: 10.1016/j.neuroimage.2011.11.069. [DOI] [PubMed] [Google Scholar]
- 55.Holman H.Y., Bjornstad K.A., McNamara M.P., Martin M.C., McKinney W.R., Blakely E.A. Synchrotron infrared spectromicroscopy as a novel bioanalytical microprobe for individual living cells: cytotoxicity considerations. J. Biomed. Opt. 2002;7(3):417–424. doi: 10.1117/1.1485299. [DOI] [PubMed] [Google Scholar]
- 56.Miller L.M., Miller L.M., Dumas P. Jamin, Teillaud, J-L.; Miklossy, J.; Forro, L. Combining IR spectroscopy with fluorescence imaging in a single microscope: Biomedical applications using a synchrotron infrared source. Rev. Sci. Instrum. 2002;73(3):1357–1360. [Google Scholar]
- 57.O'Brien J.T., Williams E.R., Holman H.Y. Ambient Infrared Laser Ablation Mass Spectrometry (AIRLAB-MS) of Live Plant Tissue with Plume Capture by Continuous Flow Solvent Probe. Anal. Chem. 2015;87(5):2631–2638. doi: 10.1021/ac503383p. [DOI] [PubMed] [Google Scholar]
- 58.Probst A.J., Birarda G., Holman H.Y., DeSantis T.Z., Wanner G., Andersen G.L., Perras A.K., Meck S., Völkel J., Bechtel H.A., Wirth R., Moissl-Eichinger C. Coupling genetic and chemical microbiome profiling reveals heterogeneity of archaeome and bacteriome in subsurface biofilms that are dominated by the same archaeal species. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0099801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaccari L., Birarda G., Businaro L., Pacor S., Grenci G. Infrared microspectroscopy of live cells in microfluidic devices (MD-IRMS): Toward a powerful label-free cell-based assay. Anal. Chem. 2012;84(11):4768–4775. doi: 10.1021/ac300313x. [DOI] [PubMed] [Google Scholar]
- 60.Holman H.Y., Nieman K., Sorensen D.L., Miller C.D., Martin M.C., Borch T., Mckinney W.R., Sims R.C. Catalysis of PAH biodegradation by humic acid shown in synchrotron infrared studies. Environ. Sci. Technol. 2002;36(6):1276–1280. doi: 10.1021/es0157200. [DOI] [PubMed] [Google Scholar]
- 61.Carbone M., Zlateva T., Quaroni L. Monitoring and manipulation of the pH of single cells using infrared spectromicroscopy and a molecular switch. Biochim. Biophys. Acta. 2013;1830(4):2989–2993. doi: 10.1016/j.bbagen.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 62.Mitri E., Kenig S., Coceano G., Bedolla D.E., Tormen M., Grenci G., Vaccari L. Time-Resolved FT-IR Microspectroscopy of Protein Aggregation Induced by Heat-Shock in Live Cells. Anal. Chem. 2015;87(7):3670–3677. doi: 10.1021/ac5040659. [DOI] [PubMed] [Google Scholar]
- 63.Mitri E., Birarda G., Vaccari L., Kenig S., Tormen M., Grenci G. SU-8 bonding protocol for the fabrication of microfluidic devices dedicated to FTIR microspectroscopy of live cells. Lab Chip. 2014;14(1):210–218. doi: 10.1039/c3lc50878a. [DOI] [PubMed] [Google Scholar]
- 64.Holman H.Y., Goth-Goldstein R., Martin M.C., Russell M.L., McKinney W.R. Low-dose responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin in single living human cells measured by synchrotron infrared spectromicroscopy. Environ. Sci. Technol. 2000;34(12):2513–2517. [Google Scholar]
- 65.Holman H.Y., Perry D.L., Martin M.C., Lamble G.M., Mckinney W.R., Hunter-cevera J.C. Real-time characterization of biogeochemical reduction of Cr(VI) on basalt surfaces by SR-FTIR imaging. Geomicrobiol. J. 1999;16(4):307–324. [Google Scholar]
- 66.Rahmelow K., Hubner W. Infrared spectroscopy in aqueous solution: Difficulties and accuracy of water subtraction. Appl. Spectrosc. 1997;51(2):160–170. [Google Scholar]
- 67.Wieliczka D.M., Weng S.S., Querry M.R. Wedge Shaped Cell for Highly Absorbent Liquids - Infrared Optical-Constants of Water. Appl. Opt. 1989;28(9):1714–1719. doi: 10.1364/AO.28.001714. [DOI] [PubMed] [Google Scholar]
- 68.Venyaminov S.Y., Kalnin N.N., Quantitative I.R. Spectrophotometry of Peptide Compounds in Water (H2O) Solutions. 2. Amide Absorption-Bands of Polypeptides and Fibrous Proteins in Alpha-Coil, Beta-Coil, and Random Coil Conformations. Biopolymers. 1990;30(13-14):1259–1271. doi: 10.1002/bip.360301310. [DOI] [PubMed] [Google Scholar]
- 69.Kalnin N.N., Baikalov I.A., Venyaminov S.Y. Quantitative Ir Spectrophotometry of Peptide Compounds in Water (H2o) Solutions. 3. Estimation of the Protein Secondary Structure. Biopolymers. 1990;30(13-14):1273–1280. doi: 10.1002/bip.360301311. [DOI] [PubMed] [Google Scholar]
- 70.Venyaminov S.Y., Kalnin N.N., Quantitative I.R. Spectrophotometry of Peptide Compounds in Water (H2O) Solutions. 1. Spectral Parameters of Amino-Acid Residue Absorption-Bands. Biopolymers. 1990;30(13-14):1243–1257. doi: 10.1002/bip.360301309. [DOI] [PubMed] [Google Scholar]
- 71.Moss D.A., Keese M., Pepperkok R. IR micro spectroscopy of live cells. Vib. Spectrosc. 2005;38(1-2):185–191. [Google Scholar]
- 72.Miljkovic M., Romeo M., Matthäus C., Diem M. Infrared microspectroscopy of individual human cervical cancer (HeLa) cells suspended in growth medium. Biopolymers. 2004;74(1-2):172–175. doi: 10.1002/bip.20066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tobin M.J., Puskar L., Barber R.L., Harvey E.C., Heraud P., Wood B.R., Bambery K.R., Dillon C.T., Munro K.L. FTIR spectroscopy of single live cells in aqueous media by synchrotron IR microscopy using microfabricated sample holders. Vib. Spectrosc. 2010;53(1):34–38. [Google Scholar]
- 74.Nasse M.J., Ratti S., Giordano M., Hirschmugl C.J. Demountable Liquid/Flow Cell for in Vivo Infrared Microspectroscopy of Biological Specimens. Appl. Spectrosc. 2009;63(10):1181–1186. doi: 10.1366/000370209789553101. [DOI] [PubMed] [Google Scholar]
- 75.Birarda G., Grenci G., Businaro L., Marmiroli B., Pacor S., Vaccari L. Fabrication of a microfluidic platform for investigating dynamic biochemical processes in living samples by FTIR microspectroscopy. Microelectron. Eng. 2010;87(5-8):806–809. [Google Scholar]
- 76.Marcsisin E.J. Infrared microspectroscopy of live cells in aqueous media. Analyst (Lond.) 2010;135(12):3227–3232. doi: 10.1039/c0an00548g. [DOI] [PubMed] [Google Scholar]
- 77.Quaroni L., Zlateva T. Infrared spectromicroscopy of biochemistry in functional single cells. Analyst (Lond.) 2011;136(16):3219–3232. doi: 10.1039/c1an15060j. [DOI] [PubMed] [Google Scholar]
- 78.Zobi F., Quaroni L., Santoro G., Zlateva T., Blacque O., Sarafimov B., Schaub M.C., Bogdanova A.Y. Live-fibroblast IR imaging of a cytoprotective photoCORM activated with visible light. J. Med. Chem. 2013;56(17):6719–6731. doi: 10.1021/jm400527k. [DOI] [PubMed] [Google Scholar]
- 79.Miller L.M., Bourassa M.W., Smith R.J. FTIR spectroscopic imaging of protein aggregation in living cells. Biochim. Biophys. Acta. 2013;1828(10):2339–2346. doi: 10.1016/j.bbamem.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gelfand P., Smith R.J., Stavitski E., Borchelt D.R., Miller L.M. Characterization of Protein Structural Changes in Living Cells Using Time-Lapsed FTIR Imaging. Anal. Chem. 2015;87(12):6025–6031. doi: 10.1021/acs.analchem.5b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whelan D.R., Bambery K.R., McNaughton D., Puskar L., Wood B.R. Monitoring the Conformation and Concentration of DNA in Live Cells using Fourier Transform Infrared Spectroscopy. Biophys. J. 2014;106(2):206a–206a. [Google Scholar]
- 82.Whelan D.R., Bambery K.R., Puskar L., McNaughton D., Wood B.R. Synchrotron Fourier transform infrared (FTIR) analysis of single living cells progressing through the cell cycle. Analyst (Lond.) 2013;138(14):3891–3899. doi: 10.1039/c3an00316g. [DOI] [PubMed] [Google Scholar]
- 83.Hinsmann P. Design, simulation and application of a new micromixing device for time resolved infrared spectroscopy of chemical reactions in solution. Lab Chip. 2001;1(1):16–21. doi: 10.1039/b104391a. [DOI] [PubMed] [Google Scholar]
- 84.Chan K.L. Rapid prototyping of microfluidic devices for integrating with FT-IR spectroscopic imaging. Lab Chip. 2010;10(16):2170–2174. doi: 10.1039/c004246c. [DOI] [PubMed] [Google Scholar]
- 85.Vaccari L., Birarda G., Grenci G., Pacor S., Businaro L. Synchrotron radiation infrared microspectroscopy of single living cells in microfluidic devices: advantages, disadvantages and future perspectives. J. Phys. 2012;359(1):012007. [Google Scholar]
- 86.Chan K.L., Kazarian S.G. FT-IR Spectroscopic Imaging of Reactions in Multiphase Flow in Microfluidic Channels. Anal. Chem. 2012;84(9):4052–4056. doi: 10.1021/ac300019m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grenci G., Birarda G., Mitri E., Businaro L., Pacor S., Vaccari L., Tormena M. Optimization of microfluidic systems for IRMS long term measurement of living cells. Microelectron. Eng. 2012;98:698–702. [Google Scholar]
- 88.Mitri E., Pozzato A., Coceano G., Cojoc D., Vaccari L., Tormen M., Grenci G. Highly IR-transparent microfluidic chip with surface-modified BaF2 optical windows for Infrared Microspectroscopy of living cells. Microelectron. Eng. 2013;107:6–9. [Google Scholar]
- 89.Birarda G., Bedolla D.E., Mitri E., Pacor S., Grenci G., Vaccari L. Apoptotic pathways of U937 leukemic monocytes investigated by infrared microspectroscopy and flow cytometry. Analyst (Lond.) 2014;139(12):3097–3106. doi: 10.1039/c4an00317a. [DOI] [PubMed] [Google Scholar]
- 90.Bedolla D.E., Kenig S., Mitri E., Ferraris P., Marcello A., Grenci G., Vaccari L. Determination of cell cycle phases in live B16 melanoma cells using IRMS. Analyst (Lond.) 2013;138(14):4015–4021. doi: 10.1039/c3an00318c. [DOI] [PubMed] [Google Scholar]
- 91.Munro K.L., Keith R.B., Elizabeth A.C., Ljiljana P., Mark J.T., Bayden R.W., Carolyn T.D. Synchrotron radiation infrared microspectroscopy of arsenic-induced changes to intracellular biomolecules in live leukemia cells. Vib. Spectrosc. 2010;53(1):39–44. [Google Scholar]
- 92.Quaroni L., Zlateva T., Sarafimov B., Kreuzer H.W., Wehbe K., Hegg E.L., Cinque G. Synchrotron based infrared imaging and spectroscopy via focal plane array on live fibroblasts in D2O enriched medium. Biophys. Chem. 2014;189:40–48. doi: 10.1016/j.bpc.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Birarda G., Gianluca G., Luca B., Benedetta M., Sabrina P., Federica P., Lisa V. Infrared microspectroscopy of biochemical response of living cells in microfabricated devices. Vib. Spectrosc. 2010;53(1):6–11. [Google Scholar]
- 94.Wehbe K., Filik J., Frogley M.D., Cinque G. The effect of optical substrates on micro-FTIR analysis of single mammalian cells. Anal. Bioanal. Chem. 2013;405(4):1311–1324. doi: 10.1007/s00216-012-6521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young E.W., Beebe D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010;39(3):1036–1048. doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holman H.Y., Miles R., Hao Z., Wozei E., Anderson L.M., Yang H. Real-Time Chemical Imaging of Bacterial Activity in Biofilms Using Open-Channel Microfluidics and Synchrotron FTIR Spectromicroscopy. Anal. Chem. 2009;81(20):8564–8570. doi: 10.1021/ac9015424. [DOI] [PubMed] [Google Scholar]
- 97.Loutherback K., Chen L., Holman H.Y. Open-Channel Microfluidic Membrane Device for Long-Term FT-IR Spectromicroscopy of Live Adherent Cells. Anal. Chem. 2015;87(9):4601–4606. doi: 10.1021/acs.analchem.5b00524. [DOI] [PubMed] [Google Scholar]
- 98.Squires T.M., Quake S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77(3):977–1026. [Google Scholar]
- 99.Di Carlo D., Wu L.Y., Lee L.P. Dynamic single cell culture array. Lab Chip. 2006;6(11):1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 100.Di Carlo D., Aghdam N., Lee L.P. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 2006;78(14):4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 101.Clausell-Tormos J., Lieber D., Baret J.C., El-Harrak A., Miller O.J., Frenz L., Blouwolff J., Humphry K.J., Köster S., Duan H., Holtze C., Weitz D.A., Griffiths A.D., Merten C.A. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol. 2008;15(5):427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 102.Huebner A., Bratton D., Whyte G., Yang M., Demello A.J., Abell C., Hollfelder F. Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab Chip. 2009;9(5):692–698. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 103.Graudejus O., Gorrn P., Wagner S. Controlling the Morphology of Gold Films on Poly(dimethylsiloxane). ACS Appl. Mater. Interfaces. 2010;2(7):1927–1933. doi: 10.1021/am1002537. [DOI] [PubMed] [Google Scholar]
- 104.Filik J., Frogley M.D., Pijanka J.K., Wehbe K., Cinque G. Electric field standing wave artefacts in FTIR micro-spectroscopy of biological materials. Analyst (Lond.) 2012;137(4):853–861. doi: 10.1039/c2an15995c. [DOI] [PubMed] [Google Scholar]
- 105.Unger M.A., Chou H.P., Thorsen T., Scherer A., Quake S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 106.Melin J., Quake S.R. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu. Rev. Biophys. Biomol. Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 107.Kazarian S.G., Chan K.L. Applications of ATR-FTIR spectroscopic imaging to biomedical samples. Biochim. Biophys. Acta. 2006;1758(7):858–867. doi: 10.1016/j.bbamem.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 108.Chan K.L., Gulati S., Edel J.B., de Mello A.J., Kazarian S.G. Chemical imaging of microfluidic flows using ATR-FTIR spectroscopy. Lab Chip. 2009;9(20):2909–2913. doi: 10.1039/b909573j. [DOI] [PubMed] [Google Scholar]
- 109.Kuimova M.K., Chan K.L., Kazarian S.G. Chemical Imaging of Live Cancer Cells in the Natural Aqueous Environment. Appl. Spectrosc. 2009;63(2):164–171. doi: 10.1366/000370209787391969. [DOI] [PubMed] [Google Scholar]
- 110.Chan K.L., Kazariana S.G., Vassoub D., Gionisb V., Chryssikosb G.D. In situ high-throughput study of drug polymorphism under controlled temperature and humidity using FT-IR spectroscopic imaging. Vib. Spectrosc. 2007;43:221–226. [Google Scholar]
- 111.Kazarian S.G. Enhancing high-throughput technology and microfluidics with FTIR spectroscopic imaging. Anal. Bioanal. Chem. 2007;388(3):2007–2007. doi: 10.1007/s00216-007-1193-3. [DOI] [PubMed] [Google Scholar]
- 112.Adato R., Altug H. In-situ ultra-sensitive infrared absorption spectroscopy of biomolecule interactions in real time with plasmonic nanoantennas. Nat. Commun. 2013;4:2154. doi: 10.1038/ncomms3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Junot C., Fenaille F., Colsch B., Bécher F. High Resolution Mass Spectrometry Based Techniques at the Crossroads of Metabolic Pathways. Mass Spectrom. Rev. 2014;33(6):471–500. doi: 10.1002/mas.21401. [DOI] [PubMed] [Google Scholar]
- 114.Lichtman J.W., Conchello J.A. Fluorescence microscopy. Nat. Methods. 2005;2(12):910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 115.Huang B., Bates M., Zhuang X.W. Super-Resolution Fluorescence Microscopy. Annu. Rev. Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X., Ser Z., Locasale J.W. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal. Chem. 2014;86(4):2175–2184. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Farre M., Pico Y., Barcelo D. Application of ultra-high pressure liquid chromatography linear ion-trap orbitrap to qualitative and quantitative assessment of pesticide residues. J. Chromatogr. A. 2014;1328:66–79. doi: 10.1016/j.chroma.2013.12.082. [DOI] [PubMed] [Google Scholar]
- 118.Ates E., Godula M., Stroka J., Senyuva H. Screening of plant and fungal metabolites in wheat, maize and animal feed using automated on-line clean-up coupled to high resolution mass spectrometry. Food Chem. 2014;142:276–284. doi: 10.1016/j.foodchem.2013.07.054. [DOI] [PubMed] [Google Scholar]
- 119.Junot C., Fenaille F., Colsch B., Bécher F. High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom. Rev. 2013;33(6):471–500. doi: 10.1002/mas.21401. [DOI] [PubMed] [Google Scholar]


