Abstract
Enhancer of Zeste Homolog 2 (EZH2) is the core component of the polycomb repressive complex 2 (PRC2), possessing the enzymatic activity in generating di/tri-methylated lysine 27 in histone H3. EZH2 has important roles during early development, and its dysregulation is heavily linked to oncogenesis in various tissue types. Accumulating evidences suggest a remarkable therapeutic potential by targeting EZH2 in cancer cells. The first part reviews current strategies to target EZH2 in cancers, and evaluates the available compounds and agents used to disrupt EZH2 functions. Then we provide insight to the future direction of the research on targeting EZH2 in different cancer types. We comprehensively discuss the current understandings of the 1) structure and biological activity of EZH2, 2) its role during the assembling of PRC2 and recruitment of other protein components, 3) the molecular events directing EZH2 to target genomic regions, and 4) post-translational modification at EZH2 protein. The discussion provides the basis to inspire the development of novel strategies to abolish EZH2-related effects in cancer cells.
Keywords: Chemotherapy, DNA methylation, DZNep, EZH2, H3K27me3, LncRNA, PRC2, SET domain
1. Introduction
Enhancer of Zeste Homolog 2 (EZH2) is a histone methyltransferase (HMTase) methylating lysine 27 in histone H3 (H3K27). The crucial role of EZH2 during development is highlighted by early embryonic lethality observed in mice after the deletion of EZH2 gene [1]. EZH2 belongs to the catalytic subunit of the polycomb repressive complex 2 (PRC2). Together with the core members EED, SUZ12, and RbAp46/48 [2], they are required for the enzymatic activity of PRC2 in vitro. PRC2 mediates gene silencing mostly by modulating chromatin structure, and by such mean, it contributes to diverse biological functions via regulating gene expression. It was shown that PRC2 consists of several subunits including AEBP2, PCLs, and JARID2, and that it transiently interacts with other proteins such as DNA methyltransferases (DNMTs), histone deacetylase 1 (HDAC1), and SIRT1 [3-5]. These additional subunits determine the functions and activities of PRC2, which explain the diversity of its biochemical and functional properties.
Despite the discrepancy in the composition of different PRC2 complex, EZH2 is pivotal as the core catalytic unit, and via the recruitment of the subunits. When the PRC2 complex is recruited to chromatin, EZH2 induces trimethylation of Lys-27 in histone H3 (H3K27me3). H3K27me3 is frequently associated with gene repression, and it is a critical epigenetic event during tissue development and stem cell fate determination. Other than H3K27me3, the repressive effect of PRC2 involves multiple levels of mechanisms via the recruitment of DNMTs and polycomb repressive complex 1 (PRC1) with EZH2 playing a pivotal role in all events. DNMTs catalyze DNA methylation [3], while PRC1 has histone modification activity through the ubiquitylation of lysine 119 of histone H2A. The composition of PRC1 includes two core components, RING1A/B together with BMI1, MEL18 or NSPC1 [2], which are the major subunits contributed for its ubiquitylation ability. PRC1 induces the condensation of chromatin, and blocks the transcriptional elongation of polymerase II (POLII). The functional link of EZH2 with both DNA methylation and PRC1 are critical in the establishment of long term epigenetic memory.
2. Characteristics of EZH2
Human EZH2 gene is mapped to chromosome 7q35, and contains 20 exons encoding 746 amino acid residues. EZH2 is usually located in the nucleus due to its nuclear localization signal, but they are also found in the cytoplasm [6]. Sequence analysis shows that members of the EZH family [7] have four homologous domains, namely homologue domain I (H1 domain), homologue domain II (H2 domain), cysteine rich domain, and C-terminal SET domain.
Both the cysteine-rich region and the SET domain are required for its HMTase activity. The HMTase activity of SET domain was first observed in SUV39H1 and later several SET domains containing HMTases have been identified [8]. Although EZH2 contains the SET domain, it was shown that recombinant EZH2 has no methyltransferase activity [9] suggesting that EZH2 has to be integrated into a functional complex in order to possess the HMTase activity. Reconstitution or purification of the EZH2-containing complex indicated first that the EED-EZH2 complex possesses a HMTase activity, but afterwards shows that SUZ12 is also required for the H3K27 methylation activity [9]. In addition to its HMTase activity the SET domain can also bind to the long non-coding RNA (lncRNA) molecules HOTAIR and possibly XIST [10, 11]. The formation of the EZH2-RNA complex is essential to the target specificity of PRC2.
EZH2 and EZH1 are the two EZ homologs in vertebrates. Although both EZH members form a complex with polycomb group proteins, they have different expression patterns and their final products have different functional roles. EZH1 is present in both dividing and differentiated cells, whereas EZH2 is only found in actively dividing cells [7]. Similar to EZH2, EZH1 contains four homologous and also interacts with core PRC2 members EED, SUZ12, RbAP46/48, and other PRC members SirT1 and PHF1 that interact with the PRC2 complex containing EZH2 [12, 13]. Despite the similarity, it is assumed that EZH2 is the major contributor to establish cellular H3K27me2/3 pattern, as the PRC2 complex containing EZH1 has a low methyltransferase activity compared to PRC2 in complex with EZH2 [7]. Although EZH1 is 65% identical to EZH2 and has a SET domain, EZH1 lacks all the threonine residues of EZH2 that are phosphorylated by CDKs and p38. These residues are important for the binding of EZH2 to PRC2 recruiting either ncRNAs or YY1. The presence of threonine on EZH2 may contribute to the functional difference to its homolog EZH1 [13].
The N-terminal domains H1 and H2 are the protein interaction domains, which are required for the assembly of partner subunits for proper PRC2 functions. EZH2 binds to the WD40 domains of EED through H1 and H2 domains. EZH2 interacts also with diverse factors important for the regulation of gene expression, such as DNMTs and HDAC. The N-terminal H1 and H2 domains of EZH2 are required for establishing and maintaining the complexes. While EZH2 transfers methyl-groups to the histone H3 tail, it is also phosphorylated and ubiquitylated. These modifications regulate the activation of EZH2, its ability to transfer methyl-group, the stability, and the spatial and temporal distribution of EZH2 in various biological processes.
3. Physiological and cellular function of EZH2
Since the mammalian homolog of drosophila Ezh has been identified, a plethora of cellular processes regulated by EZH2 has been revealed. EZH2 is important for the regular mammalian development as it regulates genomic imprinting. In mice, EZH2 was required for genomic contraction to prevent access of POLII, which leads to imprinted silencing [14]. EZH2 maintained stem cell pluripotency and determined cell fate in the context of PRC2 mediated gene repression. It was suggested that EZH2 silenced developmental regulators that resulted in a loss of pluripotency, and promoted differentiation [15]. In the hematopoietic system, EZH2 plays a critical role to regulate cell proliferation [16] and T cell differentiation [17]. EZH2 also controls B cell development through the regulation of H3 methylation and immunoglobin heavy chain gene rearrangement [18]. Downregulation of EZH2 was observed in stressed and senescing cells in which the INK4A-ARF locus was re-expressed to induce cell senescence [19]. Besides its nuclear localization, EZH2 may also possesses important cellular functions by regulating actin polymerization in various cell types, such as fibroblasts and peripheral T-lymphocytes. It controlled cellular signaling via ligand-induced actin polymerization. This finding suggests that EZH2 is generally involved in lysine methylation and thus regulates both nuclear and extra nuclear signaling processes [6].
4. Roles of EZH2 in Cancer
Recent findings suggest that EZH2 enhances the development and progression of cancer, as it is overexpressed in diverse cancer types, such as prostate, breast, bladder, gastric, liver, lung, and pancreatic cancers [20-26], and a high EZH2 level is a good indicator of various pathological features and outcomes. In prostate and breast cancer, overexpression of EZH2 is associated with metastasis and poor prognosis [20, 21]. The oncogenic role of EZH2 has been well demonstrated as ectopic expression of EZH2 in cancer cells induced anchorage independent colony growth and promotion of cell invasion [21]. Depletion of EZH2 in cancer cells induces cell cycle arrest and inhibits cell proliferation [25, 26] EZH2 exhibits an anti-apoptotic effect in both p53 wildtype and deficient cancer cells [27, 28]. Inhibition of EZH2 enhanced E2F1-dependent Bim expression promoting cell apoptosis [27]. EZH2 is also a key regulator of tumor angiogenesis as it is directly induced by VEGF in endothelial cells and thus promotes tumor angiogenesis [29]. Besides cell growth, survival, and metastasis, some studies revealed the role of EZH2 to maintain self-renewal of adult and ES cells, which links EZH2 with poorly differentiated tumor cells and cancer stem cells. A genome wide integrated analysis revealed a common subset of genes in both aggressive prostate cancer cells and embryonic cells was targeted by PRC2 [30]. Moreover, EZH2 downregulated DNA damage repair in breast tumor-initiating cells that leads to the amplification of RAF1 gene and activation of B-catenin signaling [31]. These findings substantiated the function of EZH2 in maintaining the stemness features in a subpopulation of cancer cells. Furthermore, EZH2 may have a broad influence on transcriptome regulation. EZH2 is capable of repressing the transcription of RNA molecules other than mRNA, as we showed that EZH2 silenced tumor suppressor microRNA-218 in pancreatic cancer [32].
Somatic mutations of EZH2 gene are commonly observed in myelodysplastic syndrome (MDS), T-cell acute lymphoblastic leukaemia (ALL), B-cell lymphoma, non-Hodgkin lymphoma (NHL), and follicular lymphoma (FL), in which they are responsible for the altered EZH2 activity [33-40]. A mutation of exon 15 of the EZH2 gene was found in some cases of germinal center B-cell-like (GCB) subtype of diffuse large B-cell lymphoma and FL [33]. This mutation leads to a single tyrosine substitution Tyr641 located at the SET domain of EZH2 protein, which is a key residue in the catalytic site of the SET. It was initially suggested that this mutant caused a reduction of H3K27 trimethylation activity in vitro [33], but a subsequent report suggested that while the transition from non-methylated to mono-methylated H3K27 was hampered, the mutant at Tyr641 increased the transition rate from mono-methyl to di- and tri-methyl H3K27 in GCB and NHL [34, 35]. Lymphoma cell lines and tissues bearing Tyr641 mutant exhibited higher levels of H3K27me3 than those having wild type EZH2 [36]. It illustrates a human disease that is dependent on the coordinated activities of normal and disease-associated mutant enzymatic functions. In addition to Tyr641, mutation of A677 and A687 in lymphoma promotes hypertrimethylation of H3K27 [37]. Treatment of EZH2-mutant B-cell leukemia cells by EZH2 inhibitor led to a global decrease of H3K27me3, robust gene activation, caspase activation, and decreased proliferation. This shows that cells harboring an EZH2 mutant are highly dependent on EZH2 activity for their survival [38]. However, the role of EZH2 and the effect of mutation to cancer development are still open to debate. Frequent mutations of EZH2 gene, that include deletion, missense, and frameshift mutations, are observed in MDS resulting in loss of EZH2 function [39]. These mutations of EZH2 are predictors of poor overall survival rates in MDS patients [40]. Furthermore, structural modeling predicted that somatic mutations of EZH2 in T-cell ALL were likely to disrupt the SET domain resulting in a loss of enzymatic function [41]. These findings extend the traditional understanding that EZH2 only has an oncogenic role in cancer.
More recently, a study demonstrated the PRC2-independent function of EZH2 in cells, which suggests that gene activation function of EZH2 is also important in cancer progression. Silencing of EZH2 significantly downregulated genes in prostate cancer cells. EZH2 occupied the promoters of the activated genes with a lack of H3K27me3 and the enrichment of active histone marks H3K4me2 and H3K4me3 [42]. Further study showed that genes activated by EZH2 in prostate cancer depend on cooperative recruitment of the androgen receptor, the catalytic site of EZH2, and the phosphorylation status of EZH2 Ser21 [42]. Nevertheless, this novel function of EZH2 provides an additional layer for the importance of EZH2 in cancer development and progression.
5. Targets of EZH2 in Cancer
The oncogenic role of EZH2 is largely contributed to its ability to repress the expression of tumor suppressor genes in cancer cells, which underlies the induction of various changes of the phenotype of cancer cells. EZH2 suppresses the INK4B-ARF-INK4A tumor suppressor locus to induce cell cycle progression and inhibit cell senescence. Repression of the locus also determines the balance between progenitor cells and cancer cells [43, 44]. EZH2 can inhibit cell differentiation and represses BMPR1B expression for BMPR1B-mediated differentiation signaling, which inhibits astroglial differentiation and promotes glioma tumorigenicity [45]. In hypoxic conditions, EZH2 was induced to repress RAD51, a protein important for the cellular response to DNA damage leading to genomic instability [46]. Studies have also identified various EZH2 targets in different cancer types that mediate cancer progression. EZH2 promoted cancer metastasis by suppressing E-cadherin, DAB2IP and ADRB2. E-cadherin was silenced by EZH2 in multiple cancer types promoting epithelial-mesenchymal transition of cells, which is a critical event in promoting cancer metastasis [47]. In prostate cancer, suppression of DAB2IP led to the activation of Ras and NF-kB to promote metastasis while inhibition of ADRB2 induced cell invasion [48, 49]. EZH2 also promotes angiogenesis of tumor in response to the activation of VEGF signaling by repressing VASH1, which is a negative regulator of angiogenesis [29]. In addition to protein coding genes, EZH2 can regulate non-coding RNA expression. MicroRNAs repressed by the PRC2 complex are responsible for the inhibition of cancer cell growth, invasiveness, and cancer stem cell properties [50]. In pancreatic cancer, inhibition of miR-218 by EZH2 could promote tumor growth and metastasis [27].
These evidences indicate EZH2 plays crucial roles during tumor growth and progression pointing to potential therapeutic approaches if we can attenuate the effect of EZH2 in cancer cells. In the following sections, we will first review the current approaches to inhibit EZH2 in cancer cells and describe the mechanism of the inhibition and the documented anticancer effects (Summarized in Table 1). Afterwards, we will provide insight to the future direction of EZH2 inhibition based on the molecular structure of EZH2, its role during the assembling of PRC2 and recruitment of cofactors, the molecular mechanism guiding EZH2 to its target genes, and the critical post-translational modification at EZH2. This review will inspire the development of novel approaches and strategies to target EZH2 in cancer cells.
Table 1.
Compounds and agents in use to target EZH2 in cancers.
| Name of Compound | Abbreviation/ Synonym | Mode of Inhibition | Anticancer Effects | Specificity to EZH2 | References |
|---|---|---|---|---|---|
| 3-Deazaneplanocin A | DZNep | Decreased EZH2, EED and SUZ12 protein level | Induced apoptosis in AML, breast cancer cell; impaired GBM cancer stem cell renewal in vitro and tumor initiation in vivo; reduced prostate cancer cell tumorigenicity; eradicated tumor initiating HCC cells; inhibited growth of non-small cell lung cancer cell; induced apoptosis in colon cancer cell | No | [75-78, 80, 103, 104] |
| 3-Deazaadenosine | DZA | Decreased EZH2 protein level | Inhibited growth in breast cancer cell | No | [80] |
| Diflourinated-curcumin | CDF | Induced microRNAs let-7, miR-26a and miR-101 targeting EZH2 | Inhibited tumor growth, cancer progression | No | [74] |
| Panobinostat | LBH589 | Promoted degradation of EZH2 | Induced apoptosis and differentiation of AML cells; selectively killed AML cells | No | [83-85] |
| Methotrexate | MTX | Reduced EZH2 expression | Derepressed E-cadherin in vitro | No | [70] |
| Bosutinib | SKI-606 | Inactivated EZH2 expression by inhibiting of Src-signalling | Derepressed E-cadherin and increased epithelial organization in vivo | No | [58] |
| GSK-A | - | SAM-Competitive inhibitor | No report | Yes | [89] |
| GSK126 | - | SAM-Competitive inhibitor | Inhibited cell proliferation and induced cell apoptosis in vitro; induced tumor regression and prolonged survival of mice | Yes | [88] |
| GSK343 | - | SAM-Competitive inhibitor | Inhibited epithelial ovarian cancer cells invasion; suppressed the growth and induced apoptosis of epithelial ovarian cancer cells in 3D culture | Yes | [90, 91] |
| GSK926 | - | SAM-Competitive inhibitor | Inhibited epithelial ovarian cancer cells invasion; suppressed the growth and induced apoptosis of epithelial ovarian cancer cells in 3D culture | Yes | [90, 91] |
| EPZ-6438 | E-7438 | SAM-Competitive inhibitor | Caused dose-dependent tumor growth inhibition in EZH2-mutant NHL xenograft-bearing mice | Yes | [92, 93] |
| EPZ005687 | - | Inhibited T641 and A677 mutant EZH2 methyltransferase activity | Induced apoptosis in heterozygous Tyr641 and Ala677 mutant lymphoma cells | Yes | [94] |
| EI1 | - | SAM-Competitive inhibitor | Selectively induced cell cycle arrest and apoptosis of DLBCL cells with Y641 mutations. | Yes | [95] |
| UNC1999 | - | SAM-Competitive inhibitor | Selectively killed diffused large B-cell lymphoma cell lines harboring EZH2 Y641 mutant; inhibited growth of mixed lineage leukemia (MLL)-rearranged leukemia cells | Specific to both EZH1/2 | [96, 97] |
6. Therapeutic strategies by targeting EZH2
In malignant cells, overexpression of EZH2 could result from genetic and epigenetic events. The EZH2 gene is amplified in 15% of primary human tumors [51], and has a strong correlation between its DNA copy number with transcription level in breast cancer [52]. Amplification of the EZH2 locus was observed in late-stage prostate cancer [53], which provides a clue to associate EZH2 with cancer progression. Overexpression of EZH2 can also be attributed to the increased transcription rate of EZH2 gene. EZH2 gene is directly modulated by E2F transcriptional factors [51] and high E2F3 levels are observed in a high proportion of prostate cancers, but is rare in non-neoplastic prostatic epithelial tissues [54]. Similarly, EZH2 is overexpressed in human papillomavirus-positive cancer cell in which the viral protein E7 induced the release of E2F from pocket proteins [55]. In epithelial ovarian cancer, increased levels of transcriptional factor NF-YA underlie the upregulation of EZH2 suggesting that EZH2 overexpression could be induced in different cancer types within a distinct gene context [56]. Other than DNA amplification and viral induction, upregulation of EZH2 is attributed to the hyperactivation of signaling pathway specific to the type of cancer. RAS signaling in pancreatic cancer and the hormonal signaling in breast cancer were identified as cause of enhanced EZH2 expression [57, 58].
Recent evidence demonstrates the role of microRNA in regulating EZH2 expression. In human prostate cancer, miR-101 expression is decreased in parallel with increased EZH2 expression. One or both of the two genomic loci encoding miR-101 were somatically lost in 37.5% of clinically localized prostate cancer cells and 66.7% of metastatic disease cells [59]. Loss of miR-101 expression is also observed in
gastric, pancreatic, and liver cancers in which EZH2 is overexpressed simultaneously [60-62]. The loss of other microRNAs was also identified as the cause of EZH2 overexpression in other cancer types, which included the loss of miR-26a in rhabdomyosarcoma and nasopharyngeal carcinoma [63, 64], miR-214 in breast cancer [65], miR-31 in melanoma [66] and Let-7 in high grade prostate cancer [67]. In the following paragraphs, we will discuss both the current and prospective strategies to target EZH2 in cancer cells (Fig. 1).
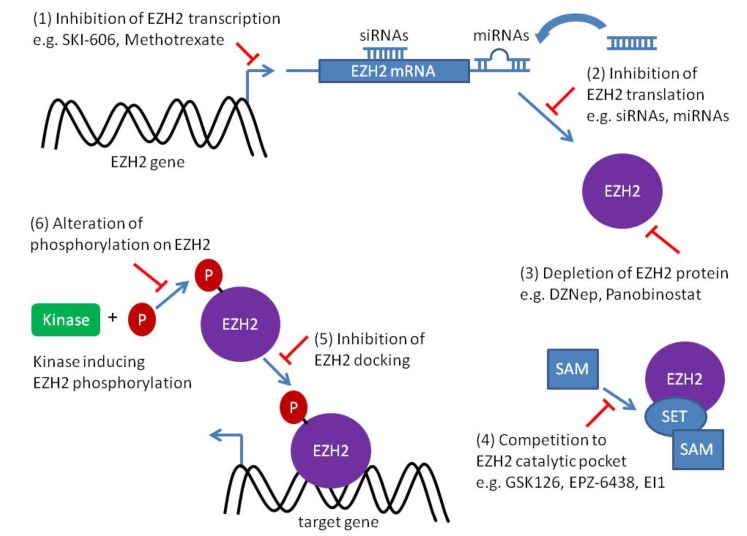
Fig. (1).
Summary of EZH2 targeting strategies. 1) Agents inhibiting the transcription of EZH2 (e.g. SKI-606 and methotrexate), which decrease subsequently the mRNA and protein levels of EZH2, are potential therapeutic drugs to treat EZH2 overexpressing cancer cells. 2) Degradation of EZH2 transcript via RNA interference (e.g. siRNAs or shRNAs) effectively inhibits translation of EZH2 mRNA and induces a significant anticancer effect. Restoring the regulation on EZH2 by miRNAs is another approach to reverse the epigenetic effects of ectopic EZH2. MiRNAs delivered to the cancer cells reduce the EZH2 protein level by either mRNA degradation or inhibiting the translation. 3) 3-Deazaneplanocin A (DZNep) and panobinostat (LBH589) provide a pharmacological approach to deplete the EZH2 protein directly in cancer cells. 4) Small molecule inhibitors (e.g. GSK-126, EPZ-6438, and EI1) specific to EZH2 compete with the methyl-donor S-adenosyl methionine (SAM) for the binding pocket of EZH2. They are highly selective to either wildtype or mutant EZH2 and effectively inhibit trimethylation of H3K27. 5) Silencing target recognition is a critical characteristic of EZH2 prior gene silencing. It was shown that lncRNAs guides EZH2 to dock to the right genomic region. EZH2 silencing can be abolished by abrogating the EZH2-cofactor RNA complex. 6) Phosphorylation of EZH2 protein determines EZH2 activity, stability, and proper targeting. Altering the phosphorylation status of EZH2 theoretically hampers the effect of EZH2 in cancer cells.
6.1. Inhibition of EZH2 at Transcription Level
In breast cancer, upregulation of EZH2 can be induced by hyperactivation of the Src signaling pathway. Src is a non-receptor tyrosine kinase activated by both steroid hormone receptors and ErbB family growth factor receptors, and it activates transcriptional factor such as STAT3, STAT5 and β-catenin by direct phosphorylation. EZH2 expression can be suppressed by Src inhibitor SKI-606 (bosutinib) [58], which decreased phosphorylation of Src in mammary tumors. Subsequently the mRNA and protein levels of EZH2 were reduced by approximately 50%. Rapid exposure of SKI-606 to tumors greatly diminished EZH2 and concurrently re-expressed EZH2-target E-cadherin, leading to a more organized epithelium [58]. Recently, SKI-606 was approved for the treatment of adult patients with Philadelphia chromosome positive chronic myelogenous leukemia (CML) resistant or intolerant to a previous therapy [68]. Moreover, SKI-606 is undergoing clinical trials on efficacy comparison to Imatinib in adult patients with newly diagnosed chronic phase CML [69].
Another study showed that the chemotherapeutic drug methotrexate exhibited EZH2-targeted anticancer effect, although it is known as an inhibitor of dyhydrofolate reductase. Methotrexate is widely used as an anticancer drug in different types of cancers, but it has been only recently reported that it down-regulates EZH2 expression in non-small cell lung cancer and consequently increases tumor suppressor gene and putative EZH2 target E-cadherin [70]. These examples illustrate that we can explore agents that target the upstream signaling to activate the expression of EZH2.
6.2. Inhibition of EZH2 at Translational Level
Depletion of EZH2 proteins in cancer cells is the most direct approach to attenuate its effect. Degradation of EZH2 transcript via RNA interference is an effective approach to reduce the EZH2 protein level in cancer cells. Transfection of small-interfering RNA (siRNA) duplexes targeting EZH2 inhibited proliferation of prostate cancer cells in vitro [20]. A lentivirus system expressing short hairpin RNA (shRNA) targeting EZH2 significantly diminished tumorigenicity of hepatocellular (HCC) cells [71]. Similar inhibition of EZH2 mediated by lentivirus reduced cell proliferation and delayed G(2)/M cell cycle transition, and significantly reduced breast xenograft growth and improved survival in vivo [72]. Knockdown of EZH2 by shRNA also delayed tumor growth and metastasis in orthotopic xenograft pancreatic cancer models [30]. RNAi-mediated approaches are effectively triggering tumor regression in HCC xenograft after intratumoral injection of either lentivirus-shRNA or siRNA targeting EZH2 [71], which provides a basis for an RNAi-mediated therapy.
Restoring normal regulation on EZH2 is another feasible approach to reverse the epigenetic effects of ectopic EZH2. As mentioned, defects in microRNAs-mediated post-transcriptional regulation of EZH2 is one of the major reasons leading to EZH2 overexpression. Thus, miR-101 has a putative tumor suppressor function, and increasing miR-101 levels in bladder, gastric, and lung cancer cells modulated their epigenome by repressing EZH2 [60-62]. The reactivation of E-cadherin after depletion of EZH2 effectively inhibited cancer cell migration and invasion. Another study demonstrated that restoring miR-26a enhanced the expression of cyclin-dependent kinase inhibitor p14 and p21 in an EZH2-dependent manner, and suppressed cell growth and colony formation by inducing G(1)-phase cycle arrest in nasopharyngeal carcinoma cells. In primary breast cancer, allelic loss of miR-214 genes is observed that leads to overexpression of EZH2. Increase of miR-214 level reduced cell proliferation and inhibited the invasion potential of metastatic breast cancer cells [65]. Re-expression of miR-214 also suppressed the growth of HCC cell growth in vivo and inhibited tumorigenicity in vivo [73]. Furthermore, pharmacological agents that restore regulation on EZH2 can also effectively attenuate EZH2 oncogenic functions. Diflourinated-curcumin induced a panel of tumor suppressor microRNAs that included let-7, miR-26a, and miR-101, and simultaneously decreased EZH2 expression in pancreatic cancer cells. By targeting an EZH2-microRNA regulatory circuit this agent inhibited pancreatic cancer tumor growth and aggressiveness [74]. The microRNA approach allows us to develop therapeutic strategies based on the microRNA environment of different cancer types.
6.3. Inhibition of EZH2 by Depletion of EZH2 Protein
To date, the most commonly used inhibitor of EZH2 is 3-deazaneplanocin A (DZNep), which can effectively reduce the cellular EZH2 protein level. It provides a pharmacological approach to deplete EZH2 protein directly in cancer cells. DZNep inhibits S-adenosylhomocysteine hydrolase in cells leading to an accumulation of S-adenosylhomocysteine with knock-on disruption of methylation at H3K27. Although the effect of DZNep is non-specific to EZH2, as other PRC2 components EED and SUZ12 are also depleted simultaneously, this compound is potent to demethylate H3K27. Global reduction of the H3K27me3 level results upon DZNep treatment, which prompts the derepression of gene expression for induction of anticancer effects [75]. DZNep is suitable for therapeutic usage due to its selectivity to cancer cells. In breast cancer, it reactivated a specific set of genes that induced efficient cell apoptosis in cancer cells but not in normal cells [76]. DZNep strongly impairs glioblastoma cancer stem cell (CSC) self-renewal in vitro and tumor-initiating capacity in vivo [77]. DZNep also inhibits tumor-initiating cells in HCC as evidenced by reduced numbers of EpCAM(+) cells and reduced sphere formation [78]. In acute myeloid leukemia (AML), DZNep induced apoptosis by reactivating TXNIP, inhibiting thioredoxin activity, and in turn increasing reactive oxygen species [79]. The high effectiveness of DZNep observed by researchers prompted the characterization of DZNep variants for identifying EZH2 inhibitor and anti-cancer agent with higher potency. Currently, DZNep is tested in preclinical studies. Clinical trials are still missing, but studies investigated the biological effects of DZNep structural analogues such as 3-deazaadenosine. Similar to DZNep, 3-deazaadenosine is capable of depleting EZH2 protein in cancer cells and inducing similar biological activities, such as growth inhibition, cell cycle arrest, and cell differentiation in breast cancer cells [80].
Panobinostat (LBH589) is another agent that induces the depletion of EZH2 protein. The compound was first identified as a pan-HDAC inhibitor, but later its broad range of protein inhibition potency was shown. Panobinostat was tested in phase three clinical trials for several cancer types [81, 82]. Treatment of panobinostat inhibited the chaperone association of heat shock protein 90 with EZH2, which promoted the degradation of EZH2 in human acute leukemia cells [83]. Panobinostat also disrupted the interaction between EZH2 and DNMT1, which blocked the indirect silencing effect of EZH2 by inducing aberrant DNA methylation at target genes [63]. Synergistic effects between DZNep and panobinostat in targeting EZH2 was demonstrated for improving the drug efficacy and better selectivity in cancer cells initiating more depletion of EZH2 and re-expression of tumor suppressor genes including p16 and p21 in AML and mantle cell lymphoma cells [84, 85]. Increasing evidences suggest that the combinatorial pharmacologic approach is an effective way to target EZH2-mediated aberrant cancer epigenetics. Further reports demonstrate that DZNep combined with either HDAC or DNMT inhibitors re-expressed a specific set of genes silenced by EZH2 in breast cancer and leukemia [86, 87].
6.4. Selective Inhibitor of EZH2
To avoid interference to non-tumorigenic pathways, any approach or agent developed should possess a high specificity towards EZH2. Several research groups are actively searching for compounds selectively inhibiting different HMTases. Compound GSK126 is a potent, highly selective, small-molecule inhibitor of EZH2 [88], which inhibits the EZH2 methyltransferase activity without degradation of the PRC2 complex. Instead, it competes with the methyl-donor S-adenosyl methionine (SAM) for the binding pocket of EZH2. GSK126 decreased global H3K27me3 levels and reactivated silenced PRC2 target genes [88]. As a result of gene derepression, GSK126 inhibited the growth of various lymphoma cells containing both the wild type and mutant EZH2, inhibited xenograft tumor growth in mice, and prolonged their survival. Another EZH2 inhibitor GSK-A was highly selective for EZH2 when evaluated against a panel of methyltransferases. The compound is cell permeable and capable of inhibiting EZH2 in a cellular context, as breast cancer cells SK-Br-3 treated with GSK-A for 3 days reduced the global H3K27me3 level in a dose-dependent manner [89]. Furthermore, GSK926 and GSK343 are two more potent EZH2 inhibitors that reduce cellular H3K27 levels, are highly selective, SAM-competitive, and cell-active [90]. GSK343 decreased the EZH2 activity, was over 1000-fold selective for other histone methyltransferase and 60-fold to the closest homolog EZH1, and showed potent inhibition of H3K27 methylation in HCC1806 cells [90]. Both GSK343 and GSK926 suppress cell growth and induce apoptosis of epithelial ovarian cancer cells in 3-dimensional cultures and inhibit their invasiveness [91]. Another SAM competitive inhibitor EPZ-6438 is the first EZH2 inhibitor to enter human clinical development. i.e. a phase 1/2 study in patients with advanced solid tumors or will B cell lymphoma to examine the safety and efficacy of EPZ-6438 administration [92]. Treatment of EZH2-mutant NHL xenograft-bearing mice with EPZ-6438 inhibits tumor growth in a dose-dependent manner, including complete and sustained tumor regressions with correlative diminution of H3K27Me3 levels in tumors and selected normal tissues. Mice treated orally with EPZ-6438 for 28 days remained tumor free for up to 63 days afterwards in two EZH2-mutant xenograft models [92]. A synergistic anti-tumor activity was also observed using EPZ-6438 with glucocorticoid receptor agonists in models of germinal center non-Hodgkin lymphomas [93].
Inhibitors specific to mutated EZH2 products are ideal to eradicate mutants present in malignant cells. Mutation on EZH2 is frequently observed in hematopoietic malignancies with several inhibitors selectively killing these EZH2 mutant cells with high efficacy. The selective EZH2 inhibitor EPZ005687is highly potent in reducing H3K27 methylation in various lymphoma cells, and promotes cancer cell death in heterozygous Tyr641 and Ala677 mutant cells with minimum toxicity towards wild type cells [94]. It had a high selectivity when tested against 15 other protein methyltransferase and even a 50-fold selectivity against EZH1. EI1 is another EZH2 inhibitor with superior growth inhibition effects in mutant EZH2 cancer cells. This molecule induces a cell cycle arrest and apoptosis of DLBCL Cells with Y641 Mutations, but affects wild type EZH2 DLBCL cells only weakly. EI1 displays ∼90-fold selectivity for EZH2 over EZH1, and >10,000-fold selectivity over other HMTs [95]. UNC1999 is an orally bioavailable inhibitor of EZH2 and EZH1 that selectively killed diffused large B-cell lymphoma cell lines harboring EZH2 Y641 mutant [96] and inhibits growth of mixed lineage leukemia (MLL)-rearranged leukemia cells [97]. Oral administration of UNC1999 prolonged survival of a well-defined murine leukemia model bearing MLL-AF9 [97].
7. Perspective and future direction
Currently, DZNep is the most frequently used compound to abolish oncogenic effects of EZH2. Though it reduces H3K27me3 level efficiently and shows promising anticancer properties, it does not specifically inhibit EZH2 but the PRC2 complex as a whole, and as a homocysteine hydrolase inhibitor it undoubtedly interferes with additional cellular components. To avoid undesired interference, inhibitors specific to EZH2 are preferred, such as GSK-126 and GSK-A. Both substances nicely illustrate the approach to identify specific inhibitors, and the benefits of conducting robust screening. They were successfully identified in a high-throughput screening using the scintillation proximity assay [89] from a library containing around 20,000 small molecules [89]. The design of the screening, which is essential for the identification of agents, relied on the competition with methyl-donor SAM for the binding pocket of EZH2. Although all HMTases use the same methyl-donor SAM as cofactor to methylate different histone residues, there are structural difference in the methyl-donor pocket among the HMTases. These differences allow the development of agents that can distinguish one HMTase from others. However, the molecular function of EZH2 is not limited to trimethylation of H3K27 at target genes, as near complete demethylation of global H3K27 was unable to induce re-expression of genes in certain lymphoma cell lines [88]. Most likely other epigenetic mechanisms, such as histone 3 lysine 9 methylation, histone 2A ubiquitylation or DNA methylation, may exist and compensate for the suppression. EZH2 serves further purposes including assembly of PRC2 and recruitment of DNMTs or PRC1. These molecular functions of EZH2 may represent further targets for pharmaceutical intervention. In the following sections, we will discuss additional features of EZH2 which may shed light on the development of novel approaches to target EZH2.
7.1. Blockage of EZH2 Docking on Target Genes
Human do not have PRC2-responsive elements or any equivalent similar to drosophila for the docking of the complex. The processes by which PRC2 targets specific chromatin regions in cells are not fully revealed. Recently, it was shown that the SET domain of a variety of HMTs has direct RNA binding property, which prompted researchers to study the interaction of RNA molecules with EZH2. It was observed that lncRNA HOTAIR, and XIST bind to the SET domain of EZH2 [11, 98]. The lncRNAs function as a tag to the specific chromatin region to allow the docking of the PRC2 complex. The identification of HOTAIR-EZH2 interaction is important, as HOTAIR is overexpressed in breast, liver, colorectal, and pancreatic cancers. Overexpressed HOTAIR induces a genome-wide retargeting of PRC2 in colorectal cancer, which resulted in a significant increase of EZH2, SUZ12, and H3K27me3. Thus, the disruption of the EZH2-lncRNA binding in cancer cells can impair aberrant gene silencing. As shown in ChIP-on-chip assays, knockdown of HOTAIR by siRNA significantly altered the H3K27 methylation pattern and concordant PRC2 occupancy [58]. Depletion of HOTAIR leads to transcription derepression of target genes [58]. In pancreatic cancer cell lines, the knockdown of HOTAIR induced the re-expression of gene GDF15, which is suppressed by EZH2 [99], inhibited the growth of cancer cells, promoted cell cycle arrest at various phases, decreased cell invasion rate, and induced cell apoptosis. In mouse xenograft model, siHOTAIR treatment inhibited tumor growth, increased cell apoptotic rates, and decreased cell proliferation markers [99].
Decreasing the affinity of EZH2 to lncRNA is another way to affect its target specificity. EZH2 is phosphorylated by cyclin-dependent kinase 1 (CDK1) and cyclin-dependent kinase 2 (CDK2) at Tyr345 in a cell cycle dependent manner [98, 100]. A phospho-mimic at Tyr345 increased the binding of HOTAIR and the 5’ end of XIST. This domain of EZH2 is susceptible for phosphorylation and thus critical for assembly of the EZH2-lncRNA complex and recognition of target genes. Inhibition of phosphorylation of this EZH2 domain, significantly reduced the affinity of EZH2 to lncRNA and hence inhibited docking of EZH2 and silencing of target genes. All mentioned evidences suggest that the interaction between EZH2 and lncRNA is a potential target to attenuate EZH2-induced aberration.
7.2. Targeting on Phosphorylation of EZH2
EZH2 appears to be a nuclear phosphoprotein linking cell-cycle-intrinsic or extracellular signals to specific epigenetic signatures. Global proteomic profiling of phosphoproteins demonstrated that EZH2 is highly phosphorylated. Phosphorylation of EZH2 has already been linked to regulation of gene expression during cell cycle, tissue regeneration, embryonic stem cell maintenance, and cell lineage commitment. As mentioned previously, EZH1 lacks the threonine residues in the homologous regions of EZH2, which are susceptible to phosphorylation. Therefore, targeting the phosphorylation of EZH2 during therapy could be a more specific approach to attenuate EZH2-associated epigenetic aberration. Phosphorylation at Tyr345 favored the formation of the EZH2-lncRNA complex, which enhances binding of EZH2 to its targets. Additionally, phosphorylation of EZH2 at Tyr487 disrupted the binding of EZH2 with PRC2 components EED and SUZ12 [101] and thereby inhibited EZH2 methyltransferase activity. Phosphorylation of Tyr487 inhibits the migration and invasion of breast cancer cells [101]. Similarly, phosphorylation of EZH2 at Ser21 affected its methyltransferase activity, which is not achieved via the PRC2 complex assembly, but the substrate affinity of EZH2 [102]. As such, it impeded the binding of EZH2 to histone H3triggering a decrease of H3K27me3 and derepression of silenced genes [102]. Overall, to attenuate EZH2 based on phosphorylation, the effects of particular phosphorylation patterns in cancer cells have to be revealed first.
Besides, the linkage of CDK to EZH2 activation, stability, and the proper targeting of EZH2 emphasize the importance of cell cycle players during establishment of histone methylation memory during cell division. It ensures H3K27 methylation on de novo synthesized H3 incorporated in nascent nucleosomes before sister chromosomes are divided into two daughter cells [100]. Approaches to disrupt the establishment of H3K27me3 during cell division can prevent the driving epigenetic signals propagated to the daughter cells during expansion of cancer cells. By decoupling the functional linkage of the cell-cycle players and EZH2, H3K27me3 memory can be impaired in the cancer cells theoretically.
Additional strategies or inhibitors should be developed to attenuate the molecular function of EZH2 that contribute to cancer development. Other than the SAM binding pocket, we can target unique features of EZH2, such as 1) methyl-accepting amino acid pocket, 2) complex assembly domain, and 3) target specificity region. Another note is that certain EZH2 mutants display altered EZH2 biochemical and functional characters. Point mutations at Tyr641, Ala677, and Ala687 increased the hypertrimethylation activity of EZH2, possibly by increasing the size of the substrate recognition site. EZH2 protein is post-translationally modified and its structure is critical in complex assembly. Thus, it is not surprising that other mutations at EZH2 could alter its functions. Mutations can affect the assembly of PRC2, the integration of lncRNA to EZH2, the protein stability or target specificity of EZH2, and all together can attribute to oncogenic effects. As these mutants represent the drivers during cancer development, it is more logical to target them during therapy rather than wild type EZH2. Focusing on the mutants should possibly improve the effectiveness of any anticancer approaches.
ACKNOWLEDGEMENTS
The work described in this paper was supported by grants from the Research Grants Council-General Research Fund of Hong Kong Special Administrative Region, China (CUHK462211, CUHK462713 and 14102714), National Natural Science Foundation of China (81101888 and 8142730), Shenzhen Basic Research Program (JC20110-5201092A), and Direct Grant from CUHK to YC.
List of Abbreviations
- ALL
Acute Lymphoblastic Leukemia
- AML
Acute Myeloid Leukemia
- CDK
Cyclin-Dependent Kinase
- CSC
cancer stem cell
- DNMT
DNA Methyltransferase
- DZNep
3-Deazaneplanocin A
- EZH1
Enhancer of Zeste Homolog 1
- EZH2
Enhancer of Zeste Homolog 2
- FL
Follicular Lymphoma
- GCB
Germinal Center B-cell-like
- H1 domain
Homologue domain I
- H2 domain
Homologue domain II
- H3K27me3
Histone 3 Lysine 27 trimethylation
- HCC
Hepatocellular Carcinoma
- HDAC1
Histone deacetylase 1
- HMTase
Histone Methyltransferase
- MDS
Myelodysplastic Syndrome
- LncRNA
Long non-coding RNA
- NHL
Non-Hodgkin Lymphoma
- POLII
Polymerase II
- PRC1
Polycomb Repressive Complex 1
- PRC2
Polycomb Repressive Complex 2
- RNAi
RNA interference
- SAM
S-Adenosyl Methionine
- shRNA
short hairpin RNA
- siRNA
small-interfering RNA
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.O'Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21(13):4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 3.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 4.van der Vlag J., Otte A.P. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 1999;23(4):474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmichev A., Margueron R., Vaquero A., Preissner T.S., Scher M., Kirmizis A., Ouyang X., Brockdorff N., Abate-Shen C., Farnham P., Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA. 2005;102(6):1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su I.H., Dobenecker M.W., Dickinson E., Oser M., Basavaraj A., Marqueron R., Viale A., Reinberg D., Wülfing C., Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121(3):425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13(16):3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao R., Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margueron R.l. Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng X., Chen S., Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10(4):579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terranova R., Yokobayashi S., Stadler M.B., Otte A.P., van Lohuizen M., Orkin S.H., Peters A.H. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell. 2008;15(5):668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Bell G.W., Otte A.P., Vidal M., Gifford D.K., Young R.A., Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 16.Fukuyama T., Otsuka T., Shigematsu H., Uchida N., Arima F., Ohno Y., Iwasaki H., Fukuda T., Niho Y. Proliferative involvement of ENX-1, a putative human polycomb group gene, in haematopoietic cells. Br. J. Haematol. 2000;108(4):842–847. doi: 10.1046/j.1365-2141.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 17.Raaphorst F.M., Otte A.P., van Kemenade F.J., Blokzijl T., Fieret E., Hamer K.M., Satijn D.P., Meijer C.J. Distinct BMI-1 and EZH2 expression patterns in thymocytes and mature T cells suggest a role for Polycomb genes in human T cell differentiation. J. Immunol. 2001;166(10):5925–5934. doi: 10.4049/jimmunol.166.10.5925. [DOI] [PubMed] [Google Scholar]
- 18.Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 19.Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Mönch K., Minucci S., Porse B.T., Marine J.C., Hansen K.H., Helin K. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., Ghosh D., Pienta K.J., Sewalt R.G., Otte A.P., Rubin M.A., Chinnaiyan A.M. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Kleer C.G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S.A., Ghosh D., Sewalt R.G., Otte A.P., Hayes I.D., Sabel M.S., Livant D., Weiss S.J., Rubin M.A., Chinnaiyan A.M. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raman J.D., Mongan N.P., Tickoo S.K., Boorjian S.A., Scherr D.S., Gudas L.J. Increaserd expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin. Cancer Res. 2005;11(24 Pt 1):8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 23.Matsukawa Y., Semba S., Kato H., Ito A., Yanagihara K., Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97(6):484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudo T., Utsunomiya T., Mimori K., Nagahara H., Ogawa K., Inoue H., Wakiyama S., Fujita H., Shirouzu K., Mori M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br. J. Cancer. 2005;92(9):1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe H., Soejima K., Yasuda H., Kawada I., Nakachi I., Yoda S., Naoki K., Ishizaka A. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008;8:15. doi: 10.1186/1475-2867-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Xie D., Li Y.W., Cheung C.M., Yao H., Chan C.Y., Xu F.P., Liu Y.H., Sung J.J., Kung H.F. RNAi targeting EZH2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo. Cancer Lett. 2010;297(1):109–116. doi: 10.1016/j.canlet.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z.L., Zheng S.S., Li Z.M., Qiao Y.Y., Aau M.Y., Yu Q. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ. 2010;17(5):801–810. doi: 10.1038/cdd.2009.162. [DOI] [PubMed] [Google Scholar]
- 28.Wu Z., Lee S.T., Qiao Y., Li Z., Lee P.L., Lee Y.J., Jiang X., Tan J., Aau M., Lim C.Z., Yu Q. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011;18(11):1771–1779. doi: 10.1038/cdd.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C., Han H.D., Mangala L.S., Ali-Fehmi R., Newton C.S., Ozbun L., Armaiz-Pena G.N., Hu W., Stone R.L., Munkarah A., Ravoori M.K., Shahzad M.M., Lee J.W., Mora E., Langley R.R., Carroll A.R., Matsuo K., Spannuth W.A., Schmandt R., Jennings N.B., Goodman B.W., Jaffe R.B., Nick A.M., Kim H.S., Guven E.O., Chen Y.H., Li L.Y., Hsu M.C., Coleman R.L., Calin G.A., Denkbas E.B., Lim J.Y., Lee J.S., Kundra V., Birrer M.J., Hung M.C., Lopez-Berestein G., Sood A.K. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18(2):185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J., Cao Q., Mehra R., Laxman B., Tomlins S.A., Creighton C.J., Dhanasekaran S.M., Shen R., Chen G., Morris D.S., Marquez V.E., Shah R.B., Ghosh D., Varambally S., Chinnaiyan A.M. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Chang C.J., Yang J.Y., Xia W., Chen C.T., Xie X., Chao C.H., Woodward W.A., Hsu J.M., Hortobagyi G.N., Hung M.C. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C.H., To K.F., Xiao Z., Tong J.H., Zhu Y., Xia T., Lai P.B., Chan S.L., Marquez V.E., Chen Y.C. Enhancer of Zeste Homolog 2 Silences microRNA-218 in Human Pancreatic Ductal Adenocarcinoma Cells by Inducing Formation of Heterochromatin. Gastroenterology. 2013;144(5):1086–1097. doi: 10.1053/j.gastro.2013.01.058. [DOI] [PubMed] [Google Scholar]
- 33.Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F., Yap D., Humphries R.K., Griffith O.L., Shah S., Zhu H., Kimbara M., Shashkin P., Charlot J.F., Tcherpakov M., Corbett R., Tam A., Varhol R., Smailus D., Moksa M., Zhao Y., Delaney A., Qian H., Birol I., Schein J. Moore. R.; Holt, R.; Horsman, D.E.; Connors, J.M.; Jones, S.; Aparicio, S.; Hirst, M.; Gascoyne, R.D.; Marra, M.A. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sneeringer C.J., Scott M.P., Kuntz K.W., Knutson S.K., Pollock R.M., Richon V.M., Copeland R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wigle T.J., Knutson S.K., Jin L., Kuntz K.W., Pollock R.M., Richon V.M., Copeland R.A., Scott M.P. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 2011;585(19):3011–3014. doi: 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 36.Yap D.B., Chu J., Berg T., Schapira M., Cheng S.W., Moradian A., Morin R.D., Mungall A.J., Meissner B., Boyle M., Marquez V.E., Marra M.A., Gascoyne R.D., Humphries R.K., Arrowsmith C.H., Morin G.B., Aparicio S.A. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCabe M.T., Graves A.P., Ganji G., Diaz E., Halsey W.S., Jiang Y., Smitheman K.N., Ott H.M., Pappalardi M.B., Allen K.E., Chen S.B., Della Pietra A., III, Dul E., Hughes A.M., Gilbert S.A., Thrall S.H., Tummino P.J., Kruger R.G., Brandt M., Schwartz B., Creasy C.L. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA. 2012;109(8):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott H.M., Graves A.P., Pappalardi M.B., Huddleston M., Halsey W.S., Hughes A.M., Groy A., Dul E., Jiang Y., Bai Y., Annan R., Verma S.K., Knight S.D., Kruger R.G., Dhanak D., Schwartz B., Tummino P.J., Creasy C.L., McCabe M.T. A687V EZH2 Is a Driver of Histone H3 Lysine 27 (H3K27) Hypertrimethylation. Mol. Cancer Ther. 2014;13(12):3062–3073. doi: 10.1158/1535-7163.MCT-13-0876. [DOI] [PubMed] [Google Scholar]
- 39.Nikoloski G., Langemeijer S.M., Kuiper R.P., Knops R., Massop M., Tönnissen E.R., van der Heijden A., Scheele T.N., Vandenberghe P., de Witte T., van der Reijden B.A., Jansen J.H. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 40.Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D., Ebert B.L. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., Lu C., Chen S.C., Wei L., Collins-Underwood J.R., Ma J., Roberts K.G., Pounds S.B., Ulyanov A., Becksfort J., Gupta P., Huether R., Kriwacki R.W., Parker M., McGoldrick D.J., Zhao D., Alford D., Espy S., Bobba K.C., Song G., Pei D., Cheng C., Roberts S., Barbato M.I., Campana D., Coustan-Smith E., Shurtleff S.A., Raimondi S.C., Kleppe M., Cools J., Shimano K.A., Hermiston M.L., Doulatov S., Eppert K., Laurenti E., Notta F., Dick J.E., Basso G., Hunger S.P., Loh M.L., Devidas M., Wood B., Winter S., Dunsmore K.P., Fulton R.S., Fulton L.L., Hong X., Harris C.C., Dooling D.J., Ochoa K., Johnson K.J., Obenauer J.C., Evans W.E., Pui C.H., Naeve C.W., Ley T.J., Mardis E.R., Wilson R.K., Downing J.R., Mullighan C.G. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu K., Wu Z.J., Groner A.C., He H.H., Cai C., Lis R.T., Wu X., Stack E.C., Loda M., Liu T., Xu H., Cato L., Thornton J.E., Gregory R.I., Morrissey C., Vessella R.L., Montironi R., Magi-Galluzzi C., Kantoff P.W., Balk S.P., Liu X.S., Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kheradmand Kia S., Solaimani Kartalaei P., Farahbakhshian E., Pourfarzad F., von Lindern M., Verrijzer C.P. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2(1):16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ezhkova E., Lien W.H., Stokes N., Pasolli H.A., Silva J.M., Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair folliclehomeostasis and wound repair. Genes Dev. 2011;25(5):485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Son M.J., Woolard K., Donin N.M., Li A., Cheng C.H., Kotliarova S., Kotliarov Y., Walling J., Ahn S., Kim M., Totonchy M., Cusack T., Ene C., Ma H., Su Q., Zenklusen J.C., Zhang W., Maric D., Fine H.A. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13(1):69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C.J., Yang J.Y., Xia W., Chen C.T., Xie X., Chao C.H., Woodward W.A., Hsu J.M., Hortobagyi G.N., Hung M.C. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Q., Yu J., Dhanasekaran S.M., Kim J.H., Mani R.S., Tomlins S.A., Mehra R., Laxman B., Cao X., Yu J., Kleer C.G., Varambally S., Chinnaiyan A.M. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H., Tu S.W., Hsieh J.T. Down-regulation of human DAB2IP gene expression mediated by polycombEzh2 complex and histone deacetylase in prostate cancer. J. Biol. Chem. 2005;280(23):22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 49.Yu J., Cao Q., Mehra R., Laxman B., Yu J., Tomlins S.A., Creighton C.J., Dhanasekaran S.M., Shen R., Chen G., Morris D.S., Marquez V.E., Shah R.B., Ghosh D., Varambally S., Chinnaiyan A.M. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12(5):419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Cao Q., Mani R.S., Ateeq B., Dhanasekaran S.M., Asangani I.A., Prensner J.R., Kim J.H., Brenner J.C., Jing X., Cao X., Wang R., Li Y., Dahiya A., Wang L., Pandhi M., Lonigro R.J., Wu Y.M., Tomlins S.A., Palanisamy N., Qin Z., Yu J., Maher C.A., Varambally S., Chinnaiyan A.M. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20(2):187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pollack J.R., Sørlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Børresen-Dale A.L., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saramäki O.R., Tammela T.L., Martikainen P.M., Vessella R.L., Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45(7):639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 54.Foster C.S., Falconer A., Dodson A.R., Norman A.R., Dennis N., Fletcher A., Southgate C., Dowe A., Dearnaley D., Jhavar S., Eeles R., Feber A., Cooper C.S. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23(35):5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 55.Holland D., Hoppe-Seyler K., Schuller B., Lohrey C., Maroldt J., Dürst M., Hoppe-Seyler F. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 2008;68(23):9964–9972. doi: 10.1158/0008-5472.CAN-08-1134. [DOI] [PubMed] [Google Scholar]
- 56.Garipov A., Li H., Bitler B.G., Thapa R.J., Balachandran S., Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol. Cancer Res. 2013;11(4):360–369. doi: 10.1158/1541-7786.MCR-12-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujii S., Fukamachi K., Tsuda H., Ito K., Ito Y., Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem. Biophys. Res. Commun. 2012;417(3):1074–1079. doi: 10.1016/j.bbrc.2011.12.099. [DOI] [PubMed] [Google Scholar]
- 58.Hebbard L., Cecena G., Golas J., Sawada J., Ellies L.G., Charbono A., Williams R., Jimenez R.E., Wankell M., Arndt K.T., DeJoy S.Q., Rollins R.A., Diesl V., Follettie M., Chen L., Rosfjord E., Cardiff R.D., Komatsu M., Boschelli F., Oshima R.G. Control of mammary tumor differentiation by SKI-606 (bosutinib). Oncogene. 2011;30(3):301–312. doi: 10.1038/onc.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varambally S., Cao Q., Mani R.S., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K., Brenner J.C., Yu J., Kim J.H., Han B., Tan P., Kumar-Sinha C., Lonigro R.J., Palanisamy N., Maher C.A., Chinnaiyan A.M. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman J.M., Liang G., Liu C.C., Wolff E.M., Tsai Y.C., Ye W., Zhou X., Jones P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69(6):2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 61.Wang H.J., Ruan H.J., He X.J., Ma Y.Y., Jiang X.T., Xia Y.J., Ye Z.Y., Tao H.Q. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur. J. Cancer. 2010;46(12):2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J.G., Guo J.F., Liu D.L., Liu Q., Wang J.J. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncol. 2011;6(4):671–678. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 63.Ciarapica R., Russo G., Verginelli F., Raimondi L., Donfrancesco A., Rota R., Giordano A. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 64.Lu J., He M.L., Wang L., Chen Y., Liu X., Dong Q., Chen Y.C., Peng Y., Yao K.T., Kung H.F., Li X.P. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 65.Derfoul A., Juan A.H., Difilippantonio M.J., Palanisamy N., Ried T., Sartorelli V. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis. 2011;32(11):1607–1614. doi: 10.1093/carcin/bgr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asangani I.A., Harms P.W., Dodson L., Pandhi M., Kunju L.P., Maher C.A., Fullen D.R., Johnson T.M., Giordano T.J., Palanisamy N., Chinnaiyan A.M. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3(9):1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong D., Heath E., Chen W., Cher M.L., Powell I., Heilbrun L., Li Y., Ali S., Sethi S., Hassan O., Hwang C., Gupta N., Chitale D., Sakr W.A., Menon M., Sarkar F.H. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 2012;7(3):e33729. doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gambacorti-Passerini C., Brümmendorf T.H., Kim D.W., Turkina A.G., Masszi T., Assouline S., Durrant S., Kantarjian H.M., Khoury H.J., Zaritskey A., Shen Z.X., Jin J., Vellenga E., Pasquini R., Mathews V., Cervantes F., Besson N., Turnbull K., Leip E., Kelly V., Cortes J.E. Bosutinib efficacy and safety in chronic phase chronic myeloid leukemia after imatinib resistance or intolerance: Minimum 24-month follow-up. Am. J. Hematol. 2014;89(7):732–742. doi: 10.1002/ajh.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gambacorti-Passerini C., Cortes J.E., Lipton J.H., Dmoszynska A., Wong R.S., Rossiev V., Pavlov D., Gogat Marchant K., Duvillié L., Khattry N., Kantarjian H.M., Brümmendorf T.H. Safety of bosutinib versus imatinib in the phase 3 BELA trial in newly diagnosed chronic phase chronic myeloid leukemia. Am. J. Hematol. 2014;89(10):947–953. doi: 10.1002/ajh.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang W.Y., Yang P.M., Chang Y.F., Marquez V.E., Chen C.C. Methotrexate induces apoptosis through p53/p21-dependent pathway and increases E-cadherin expression through downregulation of HDAC/EZH2. Biochem. Pharmacol. 2011;81(4):510–517. doi: 10.1016/j.bcp.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y., Lin M.C., Yao H., Wang H., Zhang A.Q., Yu J., Hui C.K., Lau G.K., He M.L., Sung J., Kung H.F. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46(1):200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez M.E., Li X., Toy K., DuPrie M., Ventura A.C., Banerjee M., Ljungman M., Merajver S.D., Kleer C.G. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28(6):843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia H., Ooi L.L., Hui K.M. MiR-214 Targets β-Catenin Pathway to Suppress Invasion, Stem-Like Traits and Recurrence of Human Hepatocellular Carcinoma. PLoS One. 2012;7(9):e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bao B., Ali S., Banerjee S., Wang Z., Logna F., Azmi A.S., Kong D., Ahmad A., Li Y., Padhye S., Sarkar F.H. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72(1):335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miranda T.B., Cortez C.C., Yoo C.B., Liang G., Abe M., Kelly T.K., Marquez V.E., Jones P.A. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009;8(6):1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan J., Yang X., Zhuang L., Jiang X., Chen W., Lee P.L., Karuturi R.K., Tan P.B., Liu E.T., Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suvà M.L., Riggi N., Janiszewska M., Radovanovic I., Provero P., Stehle J.C., Baumer K., Le Bitoux M.A., Marino D., Cironi L., Marquez V.E., Clément V., Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69(24):9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 78.Chiba T., Suzuki E., Negishi M., Saraya A., Miyagi S., Konuma T., Tanaka S., Tada M., Kanai F., Imazeki F., Iwama A., Yokosuka O. 3-Deazaneplanocin A is a promising therapeutic agent for the eradication of tumor-initiating hepatocellular carcinoma cells. Int. J. Cancer. 2012;130(11):2557–2567. doi: 10.1002/ijc.26264. [DOI] [PubMed] [Google Scholar]
- 79.Zhou J., Bi C., Cheong L.L., Mahara S., Liu S.C., Tay K.G., Koh T.L., Yu Q., Chng W.J. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood. 2011;118(10):2830–2839. doi: 10.1182/blood-2010-07-294827. [DOI] [PubMed] [Google Scholar]
- 80.Hayden A., Johnson P.W., Packham G., Crabb S.J. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast Cancer Res. Treat. 2011;127(1):109–119. doi: 10.1007/s10549-010-0982-0. [DOI] [PubMed] [Google Scholar]
- 81.Platzbecker U., Al-Ali H.K., Gattermann N., Haase D., Janzen V., Krauter J., Götze K., Schlenk R., Nolte F., Letsch A., Ottmann O.G., Kündgen A., Lübbert M., Germing U., Wermke M., Reinhard H., Weiss C., Lieder K., Ehninger G., Leismann O., Giagounidis A. Phase 2 study of oral panobinostat (LBH589) with or without erythropoietin in heavily transfusion-dependent IPSS low or int-1 MDS patients. Leukemia. 2014;28(3):696–698. doi: 10.1038/leu.2013.325. [DOI] [PubMed] [Google Scholar]
- 82.Berenson J.R., Hilger J.D., Yellin O., Boccia R.V., Matous J., Dressler K., Ghazal H.H., Jamshed S., Kingsley E.C., Harb W.A., Noga S.J., Nassir Y., Swift R.A., Vescio R. A phase 1/2 study of oral panobinostat combined with melphalan for patients with relapsed or refractory multiple myeloma. Ann. Hematol. 2014;93(1):89–98. doi: 10.1007/s00277-013-1910-2. [DOI] [PubMed] [Google Scholar]
- 83.Fiskus W., Wang Y., Sreekumar A., Buckley K.M., Shi H., Jillella A., Ustun C., Rao R., Fernandez P., Chen J., Balusu R., Koul S., Atadja P., Marquez V.E., Bhalla K.N. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114(13):2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiskus W., Buckley K., Rao R., Mandawat A., Yang Y., Joshi R., Wang Y., Balusu R., Chen J., Koul S., Joshi A., Upadhyay S., Atadja P., Bhalla K.N. Panobinostat treatment depletes EZH2 and DNMT1 levels and enhances decitabine mediated de-repression of JunB and loss of survival of human acute leukemia cells. Cancer Biol. Ther. 2009;8(10):939–950. doi: 10.4161/cbt.8.10.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fiskus W., Rao R., Balusu R., Ganguly S., Tao J., Sotomayor E., Mudunuru U., Smith J.E., Hembruff S.L., Atadja P., Marquez V.E., Bhalla K. Superior Efficacy of a Combined Epigenetic Therapy against Human Mantle Cell Lymphoma Cells. Clin. Cancer Res. 2012;18(22):6227–6238. doi: 10.1158/1078-0432.CCR-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun F., Chan E., Wu Z., Yang X., Marquez V.E., Yu Q. Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol. Cancer Ther. 2009;8(12):3191–3202. doi: 10.1158/1535-7163.MCT-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Momparler R.L., Idaghdour Y., Marquez V.E., Momparler L.F. Synergistic antileukemic action of a combination of inhibitors of DNA methylation and histone methylation. Leuk. Res. 2012;36(8):1049–1054. doi: 10.1016/j.leukres.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 88.McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Iii A.D., Diaz E., Lafrance L.V., Mellinger M., Duquenne C., Tian X., Kruger R.G., McHugh C.F., Brandt M., Miller W.H., Dhanak D., Verma S.K., Tummino P.J., Creasy C.L. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 89.Diaz E., Machutta C.A., Chen S., Jiang Y., Nixon C., Hofmann G., Key D., Sweitzer S., Patel M., Wu Z., Creasy C.L., Kruger R.G., Lafrance L., Verma S.K., Pappalardi M.B., Le B., Van Aller G.S., McCabe M.T., Tummino P.J., Pope A.J., Thrall S.H., Schwartz B., Brandt M. Development and Validation of Reagents and Assays for EZH2 Peptide and Nucleosome High-Throughput Screens. J. Biomol. Screen. 2012;17(10):1279–1292. doi: 10.1177/1087057112453765. [DOI] [PubMed] [Google Scholar]
- 90.Verma S.K., Tian X., LaFrance L.V., Duquenne C., Suarez D.P., Newlander K.A., Romeril S.P., Burgess J.L., Grant S.W., Brackley J.A., Graves A., Scherzer D.A., Shu A., Thompson C.S., Morgan-Ott H., Van Aller G.S., Machutta C.A., Diaz E., Jiang Y., Johnson N.W., Knight S., Kruger R.G., McCabe M.T., Dhanak D., Tummino P.J., Creasy C.L., Miller W.H. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med. Chem. Lett. 2012;3(12):1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Amatangelo M.D., Garipov A., Li H., Conejo-Garcia J.R., Speicher D.W., Zhang R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle. 2013;12(13):2113–2119. doi: 10.4161/cc.25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knutson S.K., Kawano S., Minoshima Y., Warholic N.M., Huang K.C., Xiao Y., Kadowaki T., Uesugi M., Kuznetsov G., Kumar N., Wigle T.J., Klaus C.R., Allain C.J., Raimondi A., Waters N.J., Smith J.J., Porter-Scott M., Chesworth R., Moyer M.P., Copeland R.A., Richon V.M., Uenaka T., Pollock R.M., Kuntz K.W., Yokoi A., Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 2014;13(4):842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 93.Knutson S.K., Warholic N.M., Johnston L.D., Klaus C.R., Wigle T.J., Iwanowicz D., Littlefield B.A., Porter-Scott M., Smith J.J., Moyer M.P., Copeland R.A., Pollock R.M., Kuntz K.W., Raimondi A., Keilhack H. Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas. PLoS One. 2014;9(12):e111840. doi: 10.1371/journal.pone.0111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J., Scott M.P., Jin L., Smith J.J., Olhava E.J., Chesworth R., Moyer M.P., Richon V.M., Copeland R.A., Keilhack H., Pollock R.M., Kuntz K.W. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 95.Qi W., Chan H., Teng L., Li L., Chuai S., Zhang R., Zeng J., Li M., Fan H., Lin Y., Gu J., Ardayfio O., Zhang J.H., Yan X., Fang J., Mi Y., Zhang M., Zhou T., Feng G., Chen Z., Li G., Yang T., Zhao K., Liu X., Yu Z., Lu C.X., Atadja P., Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.P., Kuznetsova E., Rougie M., Jiang A., Pattenden S.G., Norris J.L., James L.I., Roth B.L., Brown P.J., Frye S.V., Arrowsmith C.H., Hahn K.M., Wang G.G., Vedadi M., Jin J. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013;8(6):1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu B., On D.M., Ma A., Parton T., Konze K.D., Pattenden S.G., Allison D.F., Cai L., Rockowitz S., Liu S., Liu Y., Li F., Vedadi M., Frye S.V., Garcia B.A., Zheng D., Jin J., Wang G.G. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125(2):346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaneko S., Li G., Son J., Xu C.F., Margueron R., Neubert T.A., Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24(23):2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zeng X., Chen S., Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10(4):579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei Y., Chen Y.H., Li L.Y., Lang J., Yeh S.P., Shi B., Yang C.C., Yang J.Y., Lin C.Y., Lai C.C., Hung M.C. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 2011;13(1):87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cha T.L., Zhou B.P., Xia W., Wu Y., Yang C.C., Chen C.T., Ping B., Otte A.P., Hung M.C. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 103.Kikuchi J., Takashina T., Kinoshita I., Kikuchi E., Shimizu Y., Sakakibara-Konishi J., Oizumi S., Marquez V.E., Nishimura M., Dosaka-Akita H. Epigenetic therapy with 3-deazaneplanocin A, an inhibitor of the histone methyltransferase EZH2, inhibits growth of non-small cell lung cancer cells. Lung Cancer. 2012;78(2):138–143. doi: 10.1016/j.lungcan.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Benoit Y.D., Laursen K.B., Witherspoon M.S., Lipkin S.M., Gudas L.J. Inhibition of PRC2 histone methyltransferase activity increases TRAIL-mediated apoptosis sensitivity in human colon cancer cells. J. Cell. Physiol. 2013;228(4):764–772. doi: 10.1002/jcp.24224. [DOI] [PMC free article] [PubMed] [Google Scholar]