Abstract
MicroRNAs (miRNAs), a recently discovered class of small non-coding RNAs, constitute a promising approach to anti-cancer treatments when they are used in combination with other agents. MiRNAs are evolutionarily conserved non-coding RNAs that negatively regulate gene expression by binding to the complementary sequence in the 3’-untranslated region (UTR) of target genes. MiRNAs typically suppress gene expression by direct association with target transcripts, thus decreasing the expression levels of target proteins. The delivery to cells of synthetic miRNAs that mimic endogenous miRNA targeting genes involved in the DNA-Damage Response (DDR) can perturb the process, making cells more sensitive to chemotherapy or radiotherapy. This review examines how cells respond to combined therapy and it provides insights into the role of miRNAs in targeting the DDR repair pathway when they are used in combination with chemical compounds or ionizing radiation to enhance cellular sensitivity to treatments.
Keywords: Apoptosis, Cell cycle checkpoints, DNA-Damage Response, DNA Repair, microRNAs, Tumor resistance
1. INTRODUCTION
Mammalian cells rely on a highly regulated and complex pathway to respond to DNA damage caused by genotoxic stress, which is commonly referred to as DNA-Damage Response (DDR), and includes damage sensors, mediators, signal transducers and effectors involved in cell cycle checkpoint activation, DNA repair, and apoptosis. Following genotoxic stress, the DDR pathway is post-transcriptionally regulated through selective mRNA stabilization or decay or translational control [1]; in this context, microRNAs (miRNAs) have emerged as important regulators of gene expression of key components of the DDR pathway. DNA damage can regulate miRNA expression at both the transcriptional and post-transcriptional levels as well as at miRNA biogenesis in an ataxia telangiectasia mutated (ATM)-dependent manner [2].
Alterations in the expression levels of DDR genes are frequently associated with resistance to conventional cancer therapies that rely on efficient DNA repair and/or alterations in cell cycle checkpoints and apoptosis regulation. The possibility of restoring the correct expression levels through miRNA targeting has recently been investigated as a potential therapeutic approach. The overexpression of tumor-suppressive miRNAs or inhibition of oncogenic miRNAs has been shown to have therapeutic potential in model systems [3]. Here we summarize recent findings linked to the emerging role of miRNAs in targeting the DDR pathway when they are used in combination with chemotherapic agents or ionizing radiation (IR) as a new strategy to enhance the cytotoxic effects of cancer treatments in tumor resistant cells.
1.1. DNA-Damage Response
To ensure genomic stability, the DDR must be able to recognize all types of DNA structural alterations, such as double-strand breaks (DSBs), stalled replication forks, nicks, and gaps. When cells are unable to properly repair DNA, apoptosis or senescence pathways may be triggered to eliminate the possibility of passing on damaged or unrepaired genetic material to its progeny.
DSBs represent the most deleterious DNA lesions to cells since they can lead to cell death if they are left unrepaired and can cause chromosomal translocations or genomic instability if they are repaired incorrectly. Upon DSBs are recognized, members of the phosphatidylinositol-3-OH kinase (PI(3)K)-like family, such as ATM (DNA-PKcs (DNA-dependent protein kinase catalytic subunit) and ATR (ataxia telangiectasia and Rad3 related) [4] relay and amplify the damage signal to effector proteins that in turn activate cell cycle checkpoints, regulate transcription, translation, and metabolism, and activate the appropriate DNA repair process, or seal the fate of the cells, such as apoptosis or senescence [5, 6]. Following DNA DSB induction, ATM undergoes spatial relocalization and catalytic activation. It is subsequently rapidly recruited to DNA damage sites; here ATM phosphorylates specific serines or threonines on many downstream protein substrates, including Ser-139 of the histone variant H2AX present in nucleosomes surrounding DSB sites, thereby regulating the DDR mechanism [7]. p53, which maintains genomic integrity by transactivating target genes involved in cell cycle arrest, DNA repair, apoptosis and senescence, is one of the proteins phosphorylated by ATM [8, 9]. In mammalian cells, the MRN complex (Mre11, Rad50, Nbs1) is one of the first complexes to be recruited to DSB sites where it acts as a damage sensor that can also form a physical bridge spanning the DSB ends [10]. The MRN complex recognizes DSBs and binds to the DNA surrounding the lesion, resecting the DNA around the break in a 5’–3’ dependent direction, and producing long 3′ ends that are recognized by the Replication Protein A (RPA) [10]. RPA is displaced by BRCA2, which binds to RAD51 and initiates repair by homologous recombination (HR) [11]. This pathway promotes accurate DNA repair by using an undamaged homologous sequence as its template. DSBs recognized and tethered by the MRN complex can also be repaired by the non-homologous end-joining pathway (NHEJ), which employs the products of the Ku70/80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4, DNA ligase IV and XLF (XRCC4-like factor, also called Cernunnos) [12]. NHEJ directly joins the two ends of a DSB regardless of the sequence template at the break’s exposed ends, making it error-prone but available at all times during the cell cycle. In contrast to NHEJ, HR is active only in the S and G2 phases of the cell cycle, when an intact sister chromatid sequence is available as a template for error-free DSB repair.
DNA structural alterations consisting in base and nucleotide damage are repaired by base excision repair (BER) and nucleotide excision repair (NER) pathways. The former repairs damaged bases, abasic sites, and single strand breaks (SSBs); the latter repairs DNA lesions that distort the DNA helical structure. The proteins of the NER/BER pathways seem to be connected to the DSB repair pathway; indeed ATM and ATR recruitment and their phosphorylation are negatively affected in cells defective in DDB2 or XPC functions, and DDB2 and XPC promote DNA repair through the BRCA1- and RAD51-dependent pathway [13]. While the mismatch repair (MMR) pathway is involved in the repair of small insertions, deletions and base-base mismatches caused by base deamination, oxidation, methylation, and replication errors [14, 15], the Fanconi Anaemia (FA) repair pathway is involved in repairing interstrand cross-links at replication forks, possibly by facilitating ATR- and ATM-dependent checkpoint signaling and activating HR repair [16].
2. MICRORNAS
MiRNAs are endogenous single stranded non-coding RNAs (18-24 nt), which act as post-transcriptional modulators of gene expression by interacting with 3’-untranslated regions (UTR) of target genes. Mature miRNAs recognize their target mRNAs by base-pairing interactions between nucleotides 2-8 of the miRNA (the seed region) and complementary nucleotides in the 3'UTR of mRNAs (referred to as miRNA response elements, MREs). MiRNAs typically repress translation and/or induce destabilization and decay of mRNAs [17]. Recruitment of the deadenylase and decapping complexes is required both for translational repression and mRNA degradation; the subsequent exonucleolytic digestion is required for mRNA decay [18].
MiRNAs play a physiological role in a variety of important biological and pathological processes, as well as in cellular stress. It is estimated that more than half of DNA damage checkpoint genes and DNA repair genes contain conserved miRNA target sites [19]. Genes that are key regulators or essential for cell function may have high rates of transcription and low rates of translation [20]. In this context, the negative translational modulation by miRNAs allows the cell to make many mRNA copies but to have a low and carefully controlled amount of protein [21].
MiRNA expression profiling has revealed drastic changes in the expression of multiple miRNAs in human cancers; indeed a variety of miRNAs are specifically overexpressed or lost in tumors compared to that in normal tissues. Oncogenic miRNAs, called oncomiRs, promote tumour development through the negative regulation of tumour suppressor genes, and tumour suppressor miRNAs, called anti-oncomiRs, are able to inhibit tumour growth by targeting oncogenes [22].
MiRNAs involved in the DDR pathway play a fundamental role in tumor formation and progression. Three members of the miR-34 family - miR-34a, miR-34b, and miR-34c - have been shown to inhibit genes involved in controlling cell cycle progression and apoptosis [23]. It has been demonstrated that miR-17/20 and miR-221/222 clusters target cell cycle regulators thus controlling cell cycle checkpoints and progression [24, 25]. To date, increasing numbers of miRNAs target genes involved in DDR pathways have been functionally validated (Table 1).
Table 1.
MiRNA-mRNA interactions in the DDR pathway functionally validated by luciferase assay.
| miRNA | Gene targeted | DDR pathway | Type of cells | Ref. |
|---|---|---|---|---|
| miR-100 miR-101 miR-27a miR-181a miR-26a miR-18a miR-203 miR-421 |
ATM | DNA damage signalling, DNA repair, cell cycle, apoptosis | Malignant glioma cells Lung cancer cells Gastric cancer cells Glioblastoma multiforme cells Breast cancer cells Colorectal cancer cells Neuroblastoma cells Neuroblastoma cells |
[26] [27] [28] [29] [30] [31] [32] [33] |
| miR-182 miR-1255b miR-9 miR-638 miR-146a/b-5p |
BRCA1 | DNA repair | Breast cancer cells Breast/ovarian cancer cells Ovarian cancer cells Breast cancer cells Breast cancer cells |
[34] [35] [36] [37] [38] |
| miR-1245 miR-148b* |
BRCA2 | DNA repair | Breast cancer cells Breast/ovarian cancer cells |
[39] [35] |
| miR-101 | DNA-PKcs | DNA repair | Malignant glioma cells | [27] |
| miR-424* | FANCF | DNA repair | Lung cancer cells | [40] |
| miR-383 | GADD45G | DNA repair, cell cycle, apoptosis | Breast cancer cells | [41] |
| miR-15b/16-2 miR-16 |
PPM1D | DNA Damage sensing | Bronchial epithelial cells Breast and osteosarcoma cancer cells |
[42] [43] |
| miR-155 miR-96-5p miR-96 miR-193b* miR-103/107 |
RAD51 | DNA repair | Breast cancer cells Not available Breast cancer cells Breast/ovarian cancer cells Osteosarcoma cells |
[44] [45] [46] [35] [47] |
| miR-7 | XRCC2 | DNA repair | Colorectal cancer cells | [48] |
| miR-125b miR-504 miR-25/miR-30d miR-19b miR-150 |
P53 | Cell cycle checkpoint, apoptosis | Neuroblastoma cells Colorectal carcinoma cells Lung adenocarcinoma cells Breast and cervical cancer cells Lung cancer cells |
[49] [50] [51] [52] [53] |
| miR-509-5p miR-660 |
MDM2 | p53 stabilization | Cervical/hepatoma cancer cells Lung cancer cells |
[54] [55] |
| miR-490-3p miR-545 |
CCND1 | Cell cycle | Lung cancer cells Lung cancer cells |
[56] [57] |
| miR-138 | CCND3 | Cell cycle | Hepatocellular carcinoma | [58] |
| miR-630 | CDC7 | Cell cycle | Lung cancer cells | [59] |
| miR-509-3p | CDK2 | Cell cycle | Lung cancer cells | [60] |
| miR-545 | CDK4 | Cell cycle | Lung cancer cells | [57] |
| miR-224 | CDKN1A (p21) | Cell cycle | Lung cancer cells | [61] |
| miR-451 | c-MYC | Cell cycle | Lung adenocarcinoma cells | [62] |
| miR-144 | BAX | Apoptosis | Lung cancer cells | [40] |
| miR-200bc/429 miR-181 miR-204 miR-24-2 miR-181a |
BCL2 | Apoptosis | Gastric cancer cells Chronic lymphocytic leukemia Neuroblastoma cells MCF-7 cells Leukemia cells |
[63] [64] [65] [66] [67] |
| miR-363 | MCL-1 | Apoptosis | Breast cancer cells | [68] |
| miR-181a | PRKCD | Apoptosis | Cervical cancer cells | [69] |
| miR-221 | PTEN | Apoptosis | Breast cancer cells | [70] |
| miR-222 | PUMA | Apoptosis | Oral squamous cell carcinoma | [71] |
| miR-424* | STAT5A | Apoptosis | Lung cancer cells | [40] |
| miR-155 | TP53INP1 | Apoptosis | Lung cancer cells | [53] |
| miR-200bc/429 miR-24 |
XIAP | Apoptosis | Gastric cancer cells Lung cancer cells |
[63] [72] |
3. Therapeutic resistance OF CANCER CELLS
Therapeutic resistance is the primary factor that limits the effectiveness of some cancer therapies such as chemotherapy and radiotherapy. Anticancer treatments target proliferating cancer cells by interfering with DNA replication, thereby generating lethal DNA damage. Some cancer treatments such as chemotherapeutic drugs, ionizing radiation (IR) and topoisomerase II poisons can induce DSBs both directly and indirectly via single-stranded DNA (ssDNA) lesions, which induce replication fork collapse leading to DSB formation. DNA damage caused by drugs or IR and the inherent DNA repair capacity of tumour cells are important factors that determine therapeutic outcome.
3.1. Tumour Resistance Linked to DDR Gene Alterations
Proficient DSB repair is required for cellular resistance to agents that induce DSBs in the S-phase, such as IR, topoisomerase poisons (e.g. camptothecin and etoposide), and poly ADP ribose (PARP) polymerase inhibitors. Several tumors have, however, a strong DSB repair capacity meaning that therapy has low efficacy against cancer cells. Cells that overexpress RAD51 exhibit resistance to radiotherapy and chemotherapy [73-75] and are correlated with poor clinical outcome in lung, prostate, and breast cancers [76-78]. RAD51 expression is increased during breast cancer progression and metastasis, and the subsequent knockdown of RAD51 has been found to repress cancer cell migration in vitro and to reduce primary tumor growth in a
syngeneic mouse model in vivo [79]. BRCA1 and BRCA2 are key components of the HR repair pathway and integral parts of the cellular DNA damage response. Mutations restoring BRCA1/BRCA2 function lead to a loss of sensitivity to cisplatin in human ovarian tumors [80]. Overexpression of DNA-PKcs, which is closely associated with tumor cell growth, poor prognosis, and the clinical therapeutic outcome, is frequently found in various cancers [81-83]. Increased expression of XRCC2 and XRCC4, involved respectively in HR and NHEJ repair, has been detected in lung cancer cells [84]. The down-regulation of the core MMR recognition protein complex, human mutS homolog 2 (hMSH2) and 6 (hMSH6) causes resistance to 5-FU [85]. Defects in MMR proteins have been associated with reduced or absent benefits from 5-FU adjuvant chemotherapy [86], topoisomerase inhibitors chemotherapy [87, 88], and alkylating agents [87; 89]. Examples exist of increased expression of genes associated with nucleotide excision repair (NER) in resistant cells and consequent increases in repair activity. The over-expression of excision repair cross-complementation group 1 ERCC1 protein has been linked to poor responses to chemotherapy in numerous cancer types, including non-small cell lung cancer, squamous cell carcinoma, and ovarian cancer [90-92]. Barckhausen et al. [93] demonstrated that drug-induced resistance in melanoma cells is a result of p53-dependent up-regulation of the NER genes XPC and DDB2, which stimulate the repair of DNA interstrand cross-links arising from O(6)-chloroethylguanine.
Many tumors display alterations in cell cycle progression that cause aberrant cell growth. Oncogenic alterations of cyclin-dependent serine/threonine kinases (CDKs), cyclins (CCNs) and inhibitors of cyclin-dependent kinases (CDKIs) have been reported in more than 90% of human cancers [94]. Tumor cell lines expressing higher levels of cyclin CCND1 have demonstrated greater resistance to cytotoxic drugs with respect to cells expressing lower levels [95, 96]. Overexpression of CCND1 in a human fibrosarcoma cell line has been shown to confer resistance to methotrexate [97]; conversely, suppression of CCND1 has been shown to potentiate the response of human pancreatic cancer cells to cisplatinum [98]. CDKN2A/2B deletion confers a poor prognosis in diffuse large B cell lymphoma under rituximab-CHOP (R-CHOP) chemotherapy [99]. MYC gene encodes for c-Myc, an oncogenic transcription factor involved in cell cycle progression, which is found to be deregulated in many human tumors, and frequently associated with tumor progression [100]. C-Myc is significantly upregulated in docetaxel-non-responding lung adenocarcinoma tissues in comparison with docetaxel-responding tissues [62].
Up-regulation of anti-apoptotic genes or down-regulation of pro-apoptotic genes is frequently observed in human tumors. Aberrant expression of members of the Bcl-2 (B-cell leukemia/lymphoma-2) family is, for example, strongly associated with resistance to chemotherapy and radiation [101-103]. Myeloid cell leukemia-1 (MCL-1) is an anti-apoptotic Bcl-2 family member that is often overexpressed in breast tumors and has been reported to play an important role in regulating drug resistance in various types of cancer [68]. X-linked inhibitors of apoptosis proteins (XIAP), which belong to the family of inhibitors of apoptosis proteins (IAPs), have been described as a chemoresistance factor in mammalian cancer [104].
4. MIRNAS targeting DDR genes to increase radio/chemo-sensitivity of tumor resistant cells
The disruption of DNA-damage response pathways via chemotherapeutic compounds used as monotherapy or in conjunction with radiotherapy has led to promising results in the clinical setting with regard to the treatment of various cancers. Inhibitors of the DDR pathway have been shown to have a great potential for chemo- and radio-sensitization of numerous cancers. Resistant cancers do not, however, respond to therapy as a consequence of the altered expression of genes that confer resistance to drugs or IR. Resistant cancer cells rely on efficient DNA repair, alterations in cell cycle checkpoints and apoptosis regulation, and no inhibitors have been successfully used until now.
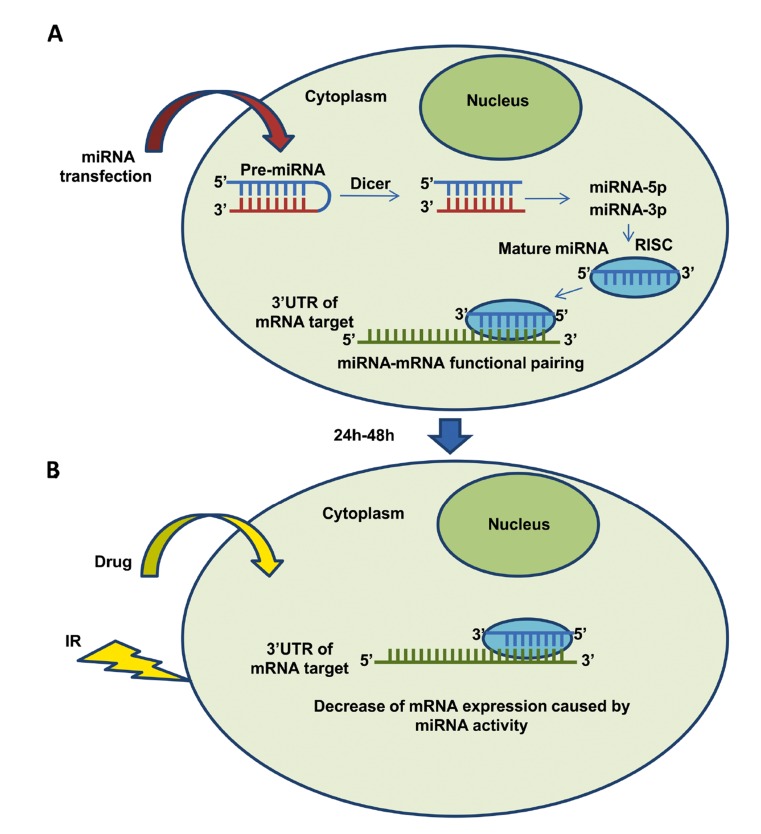
The approach that several current research studies are attempting is that of combining normal chemotherapy and radiotherapy using the RNA interference (RNAi) technique to specifically knock-down the expression of the drug- or IR-resistance genes. During gene silencing the resistant cells transiently become sensitized to the anti-cancer treatment. The therapeutic delivery to cells of synthetic miRNAs that mimic endogenous miRNAs that modulate genes involved in the DDR pathway has, therefore, been considered an appropriate approach to treating resistant cancer cells. MiRNA mimics, called miRNA precursor molecules (pre-miR), are, thus small, chemically modified double-stranded RNA molecules designed to mimic endogenous mature miRNA molecules, and they enable miRNA functional analysis by up-regulation of miRNA activity. Pre-miR molecules are carefully designed and modified to ensure that the correct strand, representing the desired mature miRNA, is taken up into the RNA-induced silencing-like complex (RISC) responsible for miRNA activity (Fig. 1). The delivery into cells of synthetic miRNAs that mimic endogenous miRNAs targeting genes involved in DDR can impair the process, making cells more sensitive to IR. An interval of 24-48h represents the time that is necessary for miRNA-transfected cells to show alterations in the expression level of target genes as a consequence of miRNA action. This time interval after transfection has been calculated on the basis of experiments described in the literature and of our own previous data [28, 40]. Pre-miRNAs rather than mature miRNAs are usually employed for therapeutical purposes. MiRNA precursors are double-stranded RNA molecules composed of a guide strand that is identical to the mature miRNA and a passenger strand that is partially or fully complementary to the guide strand [105]. Chorn et al. [106] recently demonstrated that modified single-stranded miRNA mimics also exhibit significant miRNA seed-based activity in cultured cells. The efficacy of single-stranded miRNA mimics was, however, reduced with respect to that of double-stranded ones, suggesting that further chemical modifications could be developed to enhance the targeting activity of single-stranded miRNA mimics. The experimental methods to examine the interaction between the microRNA and its targeting site(s) in the mRNA are important for understanding microRNA functions. Their interaction should be validated by luciferase assay to ensure that miRNA targets the exact mRNA. Luciferase reporter vectors, including recently introduced ones, are designed to test miRNA-mediated gene silencing. Cells are co-transfected with luciferase constructs containing the 3’UTRs of target genes (wild type/mutant) and with pre-miRTM miRNA precursor molecules. Cell lysates are collected 24h after transfection and the effect of miRNA on the luciferase gene expression can be seen as changes in the relative luciferase activity. Importantly, this assay does not reveal whether an interaction occurs endogenously, and unless care is taken with the miRNA concentration and promoter selection for the reporter, it does not reveal whether it could happen under physiological conditions. The functional interaction between miRNA and target transcript should, therefore, be validated under physiological conditions in miRNA-transfected cells by measuring the expression level of miRNA target genes using quantitative real-time PCR (qRT-PCR) and western blotting.
Fig. (1).
Therapeutical approach for resistant cancer cells. A) Cells are transfected with miRNA mimics targeting DDR genes. B) After transfection (24-48h) cells are treated with chemotherapic drugs or ionizing radiation (IR).
Transfecting mammalian cells with exogenous small interfering RNAs (siRNAs) has been adopted as a technology for targeted gene silencing. SiRNAs are double-strand RNAs of 21 to 23 nucleotides that can function in translational repression by binding to mRNAs with perfect sequence complementarity and causing cleavage of the mRNAs. One siRNA cleaving its mRNA target may provide robust gene silencing of target mRNA expression while a single miRNA leading to translational repression generally does not lead to robust gene silencing of its target. Indeed, the power of miRNA-based gene silencing may be due to the fact that one miRNA targets multiple genes, many of which participate in common signalling pathways. Furthermore, several different miRNAs may target a single mRNA [107]. One major concern linked to the therapeutical use of siRNAs is the potential for off-target effects, depending on the translational repression by siRNAs of unintended mRNA targets. MiRNA mimics carry the same sequence as naturally occurring endogenous miRNAs and are therefore expected to target the same set of genes, while siRNAs are exogenously introduced and no endogenous siRNAs have been identified in mammals. As cells already express the miRNA in question, the administration of a miRNA to cells can unlikely induce adverse events. Another concern linked to the therapeutic use of siRNAs is their potential to be immunogenic; they can indeed induce high levels of inflammatory cytokines and type I interferons, in particular interferon-alpha, after systemic administration in mammals and in primary human blood cell cultures [108].
The combination of miRNA delivery with chemotherapy or IR has recently been proposed as a strategy to improve therapy efficacy in patients with cancers refractory to standard therapies. To date, several studies have shown that miRNAs may be a useful modality to restore DDR gene networks in different cancer cell lines and in vivo models, rendering cells more sensitive to anti-cancer treatments.
4.1. Sensitizing Tumor Resistant Cells Through miRNAs Targeting DNA Repair Genes
The inability of tumor cells to properly repair some types of DNA damage render them more sensitive than normal cells to DNA-damaging agents. Some cancer cells that rely on high repair efficiency due to the over-expression of DNA repair genes are nevertheless resistant to conventional therapy. There are several examples of miRNAs targeting DNA repair genes combined with cancer therapy described in the literature. MiR-26a enhanced the in vivo radiosensitivity of glioblastoma multiforme cells by targeting ATM and reducing their DNA repair ability [30]. The microRNA-mediated down-regulation of ATM by miR-421 leads to clinically manifest tumor radiosensitivity of oral carcinoma cells [109]. The down-regulation of ATM and DNA-PKcs by direct targeting of miR-101 sensitizes glioma cell lines to ionizing radiation [27]. Inhibition of H2AX expression by either miR-24 or miR-138 promotes cellular sensitivity to IR and/ or genotoxic agents [110, 111]. MiR-155 has been demonstrated to be a direct regulator of RAD51 and to enhance the radiosensitivity of breast cancer cells [44]. Chemosensitization to DNA damaging agents has been observed when miR-103/107 were overexpressed in several cell lines [47]. These miRNAs directly target and regulate RAD51 and RAD51D, which are essential proteins for the HR repair system. Overexpression of BRCA1 in breast cancer cells has been reported to lead to an increased resistance to cisplatin [112]. MiR-638 down-regulates BRCA1 expression sensitizing breast cancer cells to DNA-damaging agents [37]. MiR-9 mediates the down-regulation of BRCA1 and impedes DNA damage repair, increasing the sensitivity of ovarian cancer cells to cisplatin [36]. MiR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1 [113]. MiR-16 inhibits the DNA damage-mediated induction of Wip1 in human breast cancer cells (MCF-7), sensitizing them to doxorubicin [43]. MiR-21 has been demonstrated to negatively regulate mismatch repair genes MLH1 and MSH2, increasing the therapeutic efficacy of 5-FU in colorectal tumors [114].
4.2. Sensitizing Tumor Resistant Cells Through miRNAs Targeting Cell Cycle Genes
Tumor resistant cells commonly have cell cycle alterations as in the case of cisplatin-resistant cells in which cell-cycle kinases WEE1 and CHK1 are frequently overexpressed. It has been shown that miR-155 and the miR-15 family sensitized cisplatin-resistant cells to apoptosis by targeting WEE1 and CHK1 that regulate the G2/M checkpoint in the cell cycle [115]. MiR-122 sensitizes hepatocellular carcinoma cells to chemotherapeutic drugs by modulating the expression of the anti-apoptotic gene Bcl-w and the cell cycle related gene cyclin B1 [116]. MiR-133a sensitized colorectal cancer cells to chemotherapeutic drug treatment with doxorubicin or oxaliplatin by targeting the RFFL gene which regulates the p53/p21 signaling pathway [117]. MiR-224 targets p21Waf1/CIP1 [61], which is responsible for cell cycle control, blocking the transition from phase G1 to phase S. The down-regulation of miR-27a could sensitize gastric cancer cells to drugs by decreasing cyclin D1 expression and up-regulating p21expression [118]. Cells overexpressing miR-383, which targets the DDR gene GADD45G, exhibited a more severe cell death than control cells exposed to cisplatin treatment [41].
4.3. Sensitizing Tumor Resistant Cells Through miRNAs Targeting Apoptotic Genes
Tumor cells resistant to conventional therapy frequently show a decreased susceptibility to drug-induced apoptosis as a consequence of an up-regulation of anti-apoptotic proteins and a down-regulation of pro-apoptotic proteins. Several reports have shown that the miR-181b, miR-451, miR-497 and miR-200bc/429 cluster sensitize the cells to platinum induced cell death by down-regulating the expression of anti-apoptotic BCL-2 gene whose overexpression causes drug resistance [63, 64, 119]. MiR-129 triggers apoptosis by suppressing BCL2 and enhancing the cytotoxic effect of 5-fluorouracil in colorectal cancer cells [120]. MiR-204 significantly increased sensitivity to cisplatin and etoposide in neuroblastoma cells by targeting BCL-2 [65]. Xie et al. [72] demonstrated that miR-24 overexpression can overcome apoptosis resistance in cancer cells via down-regulation of XIAP expression in TRAIL-resistant lung cancer cells. In gastric and lung cell lines, the miRNA cluster miR-200bc/429 was shown to promote apoptosis by targeting BCL2 and XIAP, which sensitized resistant lines to vincristine as well as to cisplatin [63]. MiR-221 conferred resistance to trastuzumab in breast cancer cells by targeting PTEN, and suppression of miR-221 or restoration of PTEN expression reversed the malignant phenotypes of breast cancer cells [70]. MiR-181a negatively regulated the expression of PRKCD, a pro-apoptotic protein kinase, thereby inhibiting irradiation-induced apoptosis and decreasing G2/M block [69]. MiR-181a sensitizes, moreover, a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2 [67]. MiR-363 reversed the resistance of the MDA-MB-231 breast cancer cell line to cisplatin by targeting anti-apoptotic gene Mcl-1 [68]. MiR-222 targeted PUMA to sensitize human oral squamous cell carcinoma cells to cisplatin [71].
5. Therapeutic delivery of MIRNAS TARGETING DDR GENES
The efficacy of the combined use of miRNAs targeting DDR genes together with chemo or radiotherapy has been demonstrated in mice. PEG-coated iron nanoparticles have been successfully used as delivery systems for miR-16 to reduce adriamycin resistance in a mouse gastric cancer model [121]. A xenograft of miR-101-overexpressing cells in mice yields tumors that shrink to a greater extent with respect to those in controls after IR [27]. Cortez et al. [122] recently demonstrated that the therapeutic delivery of a liposomal loaded with miR-200c mimics in combination with fractioned irradiation enhanced radiosensitivity in the lung cancer cells of xenograft mice by directly regulating the oxidative stress response genes PRDX2, GAPB/Nrf2, and SESN1 in ways that inhibited DNA double-strand break repair, increased the levels of reactive oxygen species, and up-regulated p21. Kasinski et al. [123] demonstrated that the systemic nanodelivery of miR-34 suppressed tumor growth leading to survival advantage. The therapeutic delivery of miR-205 mimics via nanoliposomes can sensitize the tumour to radiation by inhibiting DNA damage repair in a xenograft model [124]. Mir-205 combined with gemcitabine conjugated micelles in a pancreatic tumor model of gemcitabine resistant cells was found to produce significant inhibition of tumor growth [125]. MiR-34a, which inhibits tumor growth by regulating multiple tumor-related genes including p53 and cyclin D1, has been successfully incorporated in nanoparticle formulations to reduce cell proliferation and delay lung tumor growth in p53 deficient tumors [126]. MiR-34a incorporated together with paclitaxel into cationic solid lipid nanoparticles enhanced anti-cancer therapy in mice [127]. MiR-34a and doxorubicin can be efficiently encapsulated into hyaluronic acid-chitosan nanoparticles and delivered to breast tumor cells or tumor tissues and enhance the anti-tumor effects of the drug by suppressing the expression of non-pump resistance and anti-apoptosis proto-oncogene Bcl-2 [128]. MiR-129 enhances 5-FU cytotoxicity by suppressing BCL2 in a mouse colorectal tumor xenograft model [120].
Scarce data concerning the therapeutic delivery of miRNAs targeting genes involved in DDR are available with regard to humans. To date, MRX34 is the only miR-34a replacement therapy being assessed by a phase 1 clinical trial in patients with primary liver cancer or liver metastasis from other cancers using miR-34 mimic and the liposomal delivery technology [129]. The MRX34 phase 1 clinical study on solid tumors and hematological malignancies is expected to be completed shortly. The primary objectives of the clinical trial were to establish the maximum tolerated dose and the recommended phase 2 dose for future clinical trials. The secondary objectives were to assess the safety, tolerability and pharmacokinetic profile of MRX34 as well as to evaluate its biological activity and clinical outcomes. Further studies are critical to discover better delivery methods and improved oligonucleotide modification to enhance the performance of miRNAs as therapeutic agents in human patients.
CONCLUSION
MiRNAs which affect chemo- and radio-resistance could improve targeted miRNA-based therapeutic strategies and enhance the clinical outcome of conventional tumor therapy. MiRNAs as therapeutical agents have several advantages: they are, in fact, small, stable, can be delivered systemically and must enter into the cytoplasm of target cells to be active. As a result, the delivery of miRNA mimics seems to face less of a hurdle compared with protein-encoding DNA [130]. MiRNA mimics have the same sequence as naturally occurring miRNAs and are, therefore, expected to target the same set of mRNAs that are also regulated by natural miRNAs with little off-target effects. Moreover, since the genes targeted by a single miRNA are commonly related and function in a comparable cellular process, the targeting of a single miRNA can result in a combinatorial effect of gene expression changes in related target genes. Potential side effects of miRNA therapy must, nevertheless, be considered. Since miRNAs typically target hundreds of transcripts, there is a limit on the effect of a miRNA on an individual protein and miRNA modulation might have beneficial effects in one cell type but harmful ones in another. Moreover, in view of the fact that the majority of preclinical models have used mice with different degrees of immunosuppression, the interaction between the host immune system and miRNAs should be considered since it may improve or weaken the therapeutic effects of a particular miRNA [131]. Other limits associated to the use of miRNAs as therapeutic agents are their systemic delivery linked to degradation by nucleases, renal clearance, failure to cross the capillary endothelium, and cellular uptake of sufficient amounts of synthetic oligonucleotides to achieve sustained target inhibition [132-134]. In addition, the molecule's negative charge of miRNAs prevents their penetration through the cell membrane, although conjugation of miRNAs to cationic carriers should improve cell penetration and also prevent enzymatic cleavage of the miRNA, as has been reported for siRNAs [135]. Finally, the tumor microenvironment heterogeneity significantly enhances the efficacy of drug therapy, probably contributing to the development of drug resistance in some contexts [136].
ACKNOWLEDGEMENTS
This work was supported by grant 60A06-3595/12 of University of Padova to M.M. We apologize to the authors whose works we were unable to cite due to space limitations.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Boucas J., Riabinska A., Jokic M., Herter-Sprie G.S., Chen S., Höpker K., Reinhardt H.C. Posttranscriptional regulation of gene expression-adding another layer of complexity to the DNA damage response. Front. Genet. 2012;3:159. doi: 10.3389/fgene.2012.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X., Wan G., Berger F.G., He X., Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell. 2011;41(4):371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasinski A.L., Slack F.J. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinner A., Wu W., Staudt C., Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper J.W., Elledge S.J. The DNA damage response: Ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Stracker T.H., Roig I., Knobel P.A., Marjanović M. The ATM signaling network in development and disease. Front. Genet. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 8.Oren M. Decision making by p53: life, death and cancer. Cell Death. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 9.Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 10.Stracker T.H., Petrini J.H. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Doty T., Gibson B., Heyer W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieber M.R. The mechanism of human non-homologous DNA end joining. J. Biol. Chem. 2008;283(1):1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 13.Ray A., Milum K., Battu A., Wani G., Wani A.A. NER initiation factors, DDB2 and XPC, regulate UV radiation response by recruiting ATR and ATM kinases to DNA damage sites. DNA Repair (Amst.) 2013;12(4):273–283. doi: 10.1016/j.dnarep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christmann M., Tomicic M.T., Roos W.P., Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193(1-2):3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 15.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 16.Calderón-Montaño J.M., Burgos-Morón E., Orta M.L., López-Lázaro M. Effect of DNA repair deficiencies on the cytotoxicity of drugs used in cancer therapy - a review. Curr. Med. Chem. 2014;21(30):3419–3454. doi: 10.2174/0929867321666140601203816. [DOI] [PubMed] [Google Scholar]
- 17.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15(1):21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wouters M.D., van Gent D.C., Hoeijmakers J.H., Pothof J. microRNAs, the DNA damage response and cancer. Mutat. Res. 2011;717:54–66. doi: 10.1016/j.mrfmmm.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Fraser G.B., Hirsh A.E., Giaever G., Kumm J., Eisen M.B. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:2137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller H.A., Forman J.J., Legesse-Miller A. ‘Myc’ed messages’: Myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:3146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenouda S.K., Alahari S.K. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3-4):369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer. 2012;12(9):613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 24.Yu Z., Willmarth N.E., Zhou J., Katiyar S., Wang M., Liu Y., McCue P.A., Quong A.A., Lisanti M.P., Pestell R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. USA. 2010;107(18):8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Liang C., Ma H., Zhao Q., Lu Y., Xiang Z., Li L., Qin J., Chen Y., Cho W.C., Pestell R.G., Liang L., Yu Z. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19(6):7122–7137. doi: 10.3390/molecules19067122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng W.L., Yan D., Zhang X., Mo Y.Y., Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst.) 2010;9(11):1170–1175. doi: 10.1016/j.dnarep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Yan D., Ng W.L., Zhang X., Wang P., Zhang Z., Mo Y.Y., Mao H., Hao C., Olson J.J., Curran W.J., et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Francesco A., De Pittà C., Moret F., Barbieri V., Celotti L., Mognato M. The DNA-Damage Response to γ-radiation is affected by miR-27a in A549 cells. Int. J. Mol. Sci. 2013;14:17881–17896. doi: 10.3390/ijms140917881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Nie Y., Li X., Wu G., Huang Q., Cao J., Du Y., Li J., Deng R., Huang D., Chen B., Li S., Wei B. MicroRNA-181a Functions as an Oncomir in Gastric Cancer by Targeting the Tumour Suppressor Gene ATM. Pathol. Oncol. Res. 2014;20(2):381–389. doi: 10.1007/s12253-013-9707-0. [DOI] [PubMed] [Google Scholar]
- 30.Guo P., Lan J., Ge J., Nie Q., Guo L., Qiu Y., Mao Q. MiR-26a enhances the radiosensitivity of glioblastoma multiforme cells through targeting of ataxia-telangiectasia mutated. Exp. Cell Res. 2014;320(2):200–208. doi: 10.1016/j.yexcr.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Song L., Lin C., Wu Z., Gong H., Zeng Y., Wu J., Li M., Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One. 2011;6(9):e25454. doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Wan G., Spizzo R., Ivan C., Mathur R., Hu X., Ye X., Lu J., Fan F., Xia L., Calin G.A., Ellis L.M., Lu X. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol. Oncol. 2014;8(1):83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu W., Chan C.S., Wu R., Zhang C., Sun Y., Song J.S., et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 2010;38:689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moskwa P., Buffa F.M., Pan Y., Panchakshari R., Gottipati P., Muschel R.J., Beech J., Kulshrestha R., Abdelmohsen K., Weinstock D.M., Gorospe M., Harris A.L., Helleday T., Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell. 2011;41(2):210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi Y.E., Pan Y., Park E., Konstantinopoulos P., De S., D'Andrea A., Chowdhury D. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. eLife. 2014;3:e02445. doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun C., Li N., Yang Z., Zhou B., He Y., Weng D., Fang Y., Wu P., Chen P., Yang X., Ma D., Zhou J., Chen G. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J. Natl. Cancer Inst. 2013;105(22):1750–1758. doi: 10.1093/jnci/djt302. [DOI] [PubMed] [Google Scholar]
- 37.Tan X., Peng J., Fu Y., An S., Rezaei K., Tabbara S., Teal C.B., Man Y.G., Brem R.F., Fu S.W. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014;16(5):435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia A.I., Buisson M., Bertrand P., Rimokh R., Rouleau E., Lopez B.S., Lidereau R., Mikaélian I., Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 2011;3(5):279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L., Dai T., Xie Y., Wang C., Lin C., Wu Z., Ying Z., Wu J., Li M., Li J. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J. Mol. Cell Biol. 2012;4(2):108–117. doi: 10.1093/jmcb/mjr046. [DOI] [PubMed] [Google Scholar]
- 40.Girardi C., De Pittà C., Casara S., Sales G., Lanfranchi G., Celotti L., Mognato M. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS One. 2012;7(2):e31293. doi: 10.1371/journal.pone.0031293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao L., Gu H., Chang J., Wu J., Wang D., Chen S., Yang X., Qian B. MicroRNA-383 Regulates the Apoptosis of Tumor Cells through Targeting Gadd45g. PLoS One. 2014;9(11):e110472. doi: 10.1371/journal.pone.0110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahman M., Lovat F., Romano G., Calore F., Acunzo M., Bell E.H., Nana-Sinkam P. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J. Biol. Chem. 2014;289(38):26406–26416. doi: 10.1074/jbc.M114.573592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Wan G., Mlotshwa S., Vance V., Berger F.G., Chen H., Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70(18):7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasparini P., Lovat F., Fassan M., Casadei L., Cascione L., Jacob N.K., Carasi S., Palmieri D., Costinean S., Shapiro C.L., Huebner K., Croce C.M. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA. 2014;111(12):4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vergoulis T., Vlachos I.S., Alexiou P., Georgakilas G., Maragkakis M., Reczko M., Gerangelos S., Koziris N., Dalamagas T., Hatzigeorgiou A.G. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40(Database issue):D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Huang J.W., Calses P., Kemp C.J., Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012;72(16):4037–4046. doi: 10.1158/0008-5472.CAN-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J.W., Wang Y., Dhillon K.K., Calses P., Villegas E., Mitchell P.S., Tewari M., Kemp C.J., Taniguchi T. Systematic Screen Identifies miRNAs that Target RAD51 and RAD51D to Enhance Chemosensitivity. Mol. Cancer Res. 2013;11(12):1564–1573. doi: 10.1158/1541-7786.MCR-13-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu K., Chen Z., Qin C., Song X. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014;7:325–332. doi: 10.2147/OTT.S59364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le M.T., The C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H.F., Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu W., Chan C.S., Wu R., Zhang C., Sun Y., Song J.S., Tang L.H., Levine A.J., Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 2010;38(5):689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar M., Lu Z., Takwi A.A., Chen W., Callander N.S., Ramos K.S., Young K.H., Li Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30(7):843–853. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Y., Yin S., Hao Y., Yang J., Zhang H., Sun C., Ma M., Chang Q., Xi J.J. miR-19b promotes tumor growth and metastasis via targeting TP53. RNA. 2014;20(6):765–772. doi: 10.1261/rna.043026.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N., Wei X., Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587(15):2346–2351. doi: 10.1016/j.febslet.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 54.Ren Z.J., Nong X.Y., Lv Y.R., Sun H.H., An P.P., Wang F., Li X., Liu M., Tang H. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis. 2014;5:e1387. doi: 10.1038/cddis.2014.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fortunato O., Boeri M., Moro M., Verri C., Mensah M., Conte D., Caleca L., Roz L., Pastorino U., Sozzi G. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. 2014. [DOI] [PMC free article] [PubMed]
- 56.Gu H., Yang T., Fu S., Chen X., Guo L., Ni Y. MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochem. Biophys. Res. Commun. 2014;444(1):104–108. doi: 10.1016/j.bbrc.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Du B., Wang Z., Zhang X., Feng S., Wang G., He J., Zhang B. MicroRNA-545 Suppresses Cell Proliferation by Targeting Cyclin D1 and CDK4 in Lung Cancer Cells. PLoS One. 2014;9(2):e88022. doi: 10.1371/journal.pone.0088022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W., Zhao L.J., Tan Y.X., Ren H., Qi Z.T. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33(5):1113–1120. doi: 10.1093/carcin/bgs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao J.X., Lu Y., Qi J.J., An G.S., Mao Z.B., Jia H.T., Li S.Y., Ni J.H. MiR-630 inhibits proliferation by targeting CDC7 kinase, but maintains the apoptotic balance by targeting multiple modulators in human lung cancer A549 cells. Cell Death Dis. 2014;5:e1426. doi: 10.1038/cddis.2014.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon S., Han E., Choi Y.C., Kee H., Jeong Y., Yoon J., Baek K. Inhibition of cell proliferation and migration by miR-509-3p that targets CDK2, Rac1, and PIK3C2A. Mol. Cells. 2014;37(4):314–321. doi: 10.14348/molcells.2014.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H., Zhu L.J., Yang Y.C., Wang Z.X., Wang R. MiR-224 promotes the chemoresistance of human lung adenocarcinoma cells to cisplatin via regulating G1/S transition and apoptosis by targeting p21WAF1/CIP1. Br. J. Cancer. 2014;111(12):2383. doi: 10.1038/bjc.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen D., Huang J., Zhang K., Pan B., Chen J., De W., Wang R., Chen L. MicroRNA-451 induces epithelial-mesenchymal transition in docetaxel-resistant lung adenocarcinoma cells by targeting proto-oncogene c-Myc. Eur. J. Cancer. 2014;50(17):3050–3067. doi: 10.1016/j.ejca.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Zhu W., Xu H., Zhu D., Zhi H., Wang T., Wang J., Jiang B., Shu Y., Liu P. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother. Pharmacol. 2012;69:723–731. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 64.Zhu W., Shan X., Wang T., Shu Y., Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int. J. Cancer. 2010;127(11):2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 65.Ryan J., Tivnan A., Fay J., Bryan K., Meehan M., Creevey L., Lynch J., Bray I.M., O'Meara A., Tracey L., Davidoff A.M., Stallings R.L. MicroRNA-204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br. J. Cancer. 2012;107(6):967–976. doi: 10.1038/bjc.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srivastava N., Manvati S., Srivastava A., Pal R., Kalaiarasan P., Chattopadhyay S., Gochhait S., Dua R., Bamezai R.N. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 2011;13(2):R39. doi: 10.1186/bcr2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Hui L., Xu W. miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44(3):269–277. doi: 10.1093/abbs/gmr128. [DOI] [PubMed] [Google Scholar]
- 68.Zhang R., Li Y., Dong X., Peng L., Nie X. MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1 in breast cancer. Med. Oncol. 2014;31(12):347. doi: 10.1007/s12032-014-0347-3. [DOI] [PubMed] [Google Scholar]
- 69.Ke G., Liang L., Yang J.M., Huang X., Han D., Huang S., Zhao Y., Zha R., He X., Wu X. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2013;32(25):3019–3027. doi: 10.1038/onc.2012.323. [DOI] [PubMed] [Google Scholar]
- 70.Ye X., Bai W., Zhu H., Zhang X., Chen Y., Wang L., Yang A., Zhao J., Jia L. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47(5):268–273. doi: 10.5483/BMBRep.2014.47.5.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang F., Zhao W., Zhou L., Liu Z., Li W., Yu D. MiR-222 targeted PUMA to improve sensitization of UM1 cells to cisplatin. Int. J. Mol. Sci. 2014;15(12):22128–22141. doi: 10.3390/ijms151222128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie Y., Tobin L.A., Camps J., Wangsa D., Yang J., Rao M., Witasp E., Awad K.S., Yoo N., Ried T., Kwong K.F. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2013;32(19):2442–2451. doi: 10.1038/onc.2012.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarasin A., Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat. Res. 2008;659(1-2):49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 74.Brown E.T., Holt J.T. Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol. Carcinog. 2009;48(2):105–109. doi: 10.1002/mc.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schild D., Wiese C. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 2010;38(4):1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiao G.B., Wu Y.L., Yang X.N., Zhong W.Z., Xie D., Guan X.Y., Fischer D., Kolberg H.C., Kruger S., Stuerzbecher H.W. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br. J. Cancer. 2005;93(1):137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitra A., Jameson C., Barbachano Y., Sanchez L. Kote- Jarai, Z.; Peock, S.; Sodha, N.; Bancroft, E.; Fletcher, A.; Cooper, C.; Easton, D.; Eeles, R.; Foster, C.S. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009;55(6):696–704. doi: 10.1111/j.1365-2559.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbano R., Copetti M., Perrone G., Pazienza V., Muscarella L.A., Balsamo T., Storlazzi C.T., Ripoli M., Rinaldi M., Valori V.M., Latiano T.P., Maiello E., Stanziale P., Carella M., Mangia A., Pellegrini F., Bisceglia M., Muda A.O., Altomare V., Murgo R., Fazio V.M., Parrella P. High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient's outcome. Int. J. Cancer. 2011;129(3):536–545. doi: 10.1002/ijc.25736. [DOI] [PubMed] [Google Scholar]
- 79.Wiegmans A.P., Al-Ejeh F., Chee N., Yap P.Y., Gorski J.J., Da Silva L., Bolderson E., Chenevix-Trench G., Anderson R., Simpson P.T., Lakhani S.R., Khanna K.K. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5(10):3261–3272. doi: 10.18632/oncotarget.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borst P., Rottenberg S., Jonkers J. How do real tumors become resistant to cisplatin? Cell Cycle. 2008;7(10):1353–1359. doi: 10.4161/cc.7.10.5930. [DOI] [PubMed] [Google Scholar]
- 81.Xing J., Wu X., Vaporciyan A., Spitz M.J. Prognostic significance of ataxia-telangiectasia mutated, DNA-dependent protein kinase catalytic subunit, and Ku heterodimeric regulatory complex 86-kD subunit expression in patients with nonsmall cell lung cancer. Cancer. 2008;112:2756–2764. doi: 10.1002/cncr.23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willmore E., Elliott S., Mainou-Fowler T., Summerfield G., Jackson G., O’Neill F., Lowe C., Carter A., Harris R., Pettitt A., Cano-Soumillac C., Griffin R.J., Cowell I.G., Austin C.A., Durkacz B.W. DNA-dependent protein kinase is a therapeutic target and an indicator of poor prognosis in B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 2008;14:3984–3992. doi: 10.1158/1078-0432.CCR-07-5158. [DOI] [PubMed] [Google Scholar]
- 83.Beskow C., Skikuniene J., Holgersson A., Nilsson B., Lewensohn R., Kanter L., Viktorsson K. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br. J. Cancer. 2009;101:816–821. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng Z., Ng W.L., Zhang X., Olson J.J., Hao C., Curran W.J., Wang Y. RNAi-mediated targeting of noncoding and coding sequences in DNA repair gene messages efficiently radiosensitizes human tumor cells. Cancer Res. 2012;72(5):1221–1228. doi: 10.1158/0008-5472.CAN-11-2785. [DOI] [PubMed] [Google Scholar]
- 85.Valeri N., Gasparini P., Fabbri M., Braconi C., Veronese A., Lovat F., et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R., French A.J., Kabat B., Foster N.R., Torri V., Ribic C., Grothey A., Moore M., Zaniboni A., Seitz J.F., Sinicrope F., Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28(20):3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aebi S., Fink D., Gordon R., Kim H.K., Zheng H., Fink J.L., Howell S.B. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clin. Cancer Res. 1997;3(10):1763–1767. [PubMed] [Google Scholar]
- 88.Fedier A., Schwarz V.A., Walt H., Carpini R.D., Haller U., Fink D. Resistance to topoisomerase poisons due to loss of DNA mismatch repair. Int. J. Cancer. 2001;93(4):571–576. doi: 10.1002/ijc.1356. [DOI] [PubMed] [Google Scholar]
- 89.Papouli E., Cejka P., Jiricny J. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 2004;64(10):3391–3394. doi: 10.1158/0008-5472.CAN-04-0513. [DOI] [PubMed] [Google Scholar]
- 90.Ferry K.V., Hamilton T.C., Johnson S.W. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem. Pharmacol. 2000;60(9):1305–1313. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 91.Olaussen K.A., Dunant A., Fouret P., Brambilla E., André F., Haddad V., Taranchon E., Filipits M., Pirker R., Popper H.H., et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 92.Kim M.K., Cho K-J., Kwon G.Y., Park S-I., Kim Y.H., Kim J.H., Song H-Y., Shin J.H., Jung H.Y., Lee G.H., et al. ERCC1 predicting chemoradiation resistance and poor outcome in oesophageal cancer. Eur. J. Cancer. 2008;44:54–60. doi: 10.1016/j.ejca.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 93.Barckhausen C., Roos W.P., Naumann S.C., Kaina B. Malignant melanoma cells acquire resistance to DNA interstrand cross-linking chemotherapeutics by p53-triggered upregulation of DDB2/XPC-mediated DNA repair. Oncogene. 2014;33(15):1964–1974. doi: 10.1038/onc.2013.141. [DOI] [PubMed] [Google Scholar]
- 94.Hartwell L.H., Kastan M.B. Cell cycle control and cancer. Science. 1994;266(5192):1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 95.Burandt E., Grünert M., Lebeau A., Choschzick M., Quaas A., Jänicke F., Müller V., Scholz U., Bokemeyer C., Petersen C., Geist S., Paluchowski P., Wilke C., Heilenkötter U., Simon R., Sauter G., Wilczak W. Cyclin D1 gene amplification is highly homogeneous in breast cancer. Breast Cancer. 2014 doi: 10.1007/s12282-014-0538-y. [DOI] [PubMed] [Google Scholar]
- 96.Sewify E.M., Afifi O.A., Mosad E., Zaki A.H., El Gammal S.A. Cyclin D1 amplification in multiple myeloma is associated with multidrug resistance expression. Clin. Lymphoma Myeloma Leuk. 2014;14(3):215–222. doi: 10.1016/j.clml.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 97.Hochhauser D., Schnieders B., Ercikan-Abali E., Gorlick R., Muise-Helmericks R., Li W.W., Fan J., Banerjee D., Bertino J.R. Effect of cyclin D1 overexpression on drug sensitivity in a human fibrosarcoma cell line. J. Natl. Cancer Inst. 1996;88(18):1269–1275. doi: 10.1093/jnci/88.18.1269. [DOI] [PubMed] [Google Scholar]
- 98.Zhou X., Zhang Z., Yang X., Chen W., Zhang P. Inhibition of cyclin D1 expression by cyclin D1 shRNAs in human oral squamous cell carcinoma cells is associated with increased cisplatin chemosensitivity. Int. J. Cancer. 2009;124(2):483–489. doi: 10.1002/ijc.23964. [DOI] [PubMed] [Google Scholar]
- 99.Jardin F., Jais J.P., Molina T.J., Parmentier F., Picquenot J.M., Ruminy P., et al. Diffuse large B cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood. 2010;116(7):1092–1104. doi: 10.1182/blood-2009-10-247122. [DOI] [PubMed] [Google Scholar]
- 100.Nesbit C.E., Tersak J.M., Prochownik E.V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 101.Campos L., Rouault J.P., Sabido O., Oriol P., Roubi N., Vasselon C., Archimbaud E., Magaud J.P., Guyotat D. High expression of bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Blood. 1993;81(11):3091–3096. [Google Scholar]
- 102.An J., Chervin A.S., Nie A., Ducoff H.S., Huang Z. Overcoming the radioresistance of prostate cancer cells with a novel Bcl-2 inhibitor. Oncogene. 2007;26(5):652–661. doi: 10.1038/sj.onc.1209830. [DOI] [PubMed] [Google Scholar]
- 103.Hussein R.M., Haemel A.K., Wood G.S. Apoptosis and melanoma: molecular mechanisms. J. Pathol. 2003;1299:275–288. doi: 10.1002/path.1300. [DOI] [PubMed] [Google Scholar]
- 104.Kashkar H. X-linked inhibitor of apoptosis: a chemoresistance factor or a hollow promise. Clin. Cancer Res. 2010;16:4496–4502. doi: 10.1158/1078-0432.CCR-10-1664. [DOI] [PubMed] [Google Scholar]
- 105.Henry J.C., Azevedo-Pouly A.C., Schmittgen T.D. Microrna replacement therapy for cancer. Pharm. Res. 2011;28:3030–3042. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 106.Chorn G., Klein-McDowell M., Zhao L., Saunders M.A., Flanagan W.M., Willingham A.T., Lim L.P. Single-stranded microrna mimics. RNA. 2012;18:1796–1804. doi: 10.1261/rna.031278.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 108.Judge A., MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19(2):111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 109.Mansour W.Y., Bogdanova N.V., Kasten-Pisula U., Rieckmann T., Köcher S., Borgmann K., Baumann M., Krause M., Petersen C., Hu H., Gatti R.A., Dikomey E., Dörk T., Dahm-Daphi J. Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiother. Oncol. 2013;106(1):147–154. doi: 10.1016/j.radonc.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 110.Lal A., Pan Y., Navarro F., Dykxhoorn D.M., Moreau L., Meire E., et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang Y., Huang J.W., Li M., Cavenee W.K., Mitchell P.S., Zhou X., et al. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol. Cancer Res. 2011;9:1100–1111. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Husain A., He G., Venkatraman E.S., Spriggs D.R. BRCA1 up-regulation is associated with repair-mediated resistance to cisdiamminedichloroplatinum(II). Cancer Res. 1998;58:1120–1123. [PubMed] [Google Scholar]
- 113.He X., Xiao X., Dong L., Wan N., Zhou Z., Deng H., Zhang X. MiR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1. Tumour Biol. 2015;36(3):2065–2075. doi: 10.1007/s13277-014-2814-z. [DOI] [PubMed] [Google Scholar]
- 114.Valeri N., Gasparini P., Fabbri M., Braconi C., Veronese A., Lovat F., et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pouliot L.M., Chen Y.C., Bai J., Guha R., Martin S.E., Gottesman M.M., Hall M.D. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72(22):5945–5955. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu Y., Xia F., Ma L., Shan J., Shen J., Yang Z., Liu J., Cui Y., Bian X., Bie P., Qian C. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310(2):160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 117.Dong Y., Zhao J., Wu C.W., Zhang L., Liu X., Kang W., Leung W.W., Zhang N., Chan F.K., Sung J.J., Ng S.S., Yu J. Tumor suppressor functions of miR-133a in colorectal cancer. Mol. Cancer Res. 2013;11(9):1051–1060. doi: 10.1158/1541-7786.MCR-13-0061. [DOI] [PubMed] [Google Scholar]
- 118.Zhao X., Yang L., Hu J. Down-regulation of miR-27a might inhibit proliferation and drug resistance of gastric cancer cells. J. Exp. Clin. Cancer Res. 2011;30:55. doi: 10.1186/1756-9966-30-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bian H.B., Pan X., Yang J.S., Wang Z.X., De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549). J. Exp. Clin. Cancer Res. 2011;30:20. doi: 10.1186/1756-9966-30-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karaayvaz M., Zhai H., Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4:e659. doi: 10.1038/cddis.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun Z., Song X., Li X., Su T., Qi S., Qiao R., Wang F., Huan Y., Yang W., Wang J., Nie Y., Wu K., Gao M., Cao F. In vivo multimodality imaging of miRNA-16 iron nanoparticle reversing drug resistance to chemotherapy in a mouse gastric cancer model. Nanoscale. 2014;6(23):14343–14353. doi: 10.1039/c4nr03003f. [DOI] [PubMed] [Google Scholar]
- 122.Cortez M.A., Valdecanas D., Zhang X., Zhan Y., Bhardwaj V., Calin G.A., Komaki R., Giri D.K., Quini C.C., Wolfe T., Peltier H.J., Bader A.G., Heymach J.V., Meyn R.E., Welsh J.W. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol. Ther. 2014;22(8):1494–1503. doi: 10.1038/mt.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kasinski A.L., Kelnar K., Stahlhut C., Orellana E., Zhao J., Shimer E., Dysart S., Chen X., Bader A.G., Slack F.J. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34(27):3547–3555. doi: 10.1038/onc.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang P., Wang L., Rodriguez-Aguayo C., Yuan Y., Debeb B.G., Chen D., Sun Y., You M.J., Liu Y., Dean D.C., Woodward W.A., Liang H., Yang X., Lopez-Berestein G., Sood A.K., Hu Y., Ang K.K., Chen J., Ma L. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat. Commun. 2014;5:5671. doi: 10.1038/ncomms6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mittal A., Chitkara D., Behrman S.W., Mahato R.I. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35(25):7077–7087. doi: 10.1016/j.biomaterials.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 126.Xue W., Dahlman J.E., Tammela T., Khan O.F., Sood S., Dave A., Cai W., Chirino L.M., Yang G.R., Bronson R., Crowley D.G., Sahay G., Schroeder A., Langer R., Anderson D.G., Jacks T. Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. USA. 2014;111(34):E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shi S., Han L., Deng L., Zhang Y., Shen H., Gong T., Zhang Z., Sun X. Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J. Control. Release. 2014;194:228–237. doi: 10.1016/j.jconrel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 128.Deng X., Cao M., Zhang J., Hu K., Yin Z., Zhou Z., Xiao X., Yang Y., Sheng W., Wu Y., Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35(14):4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 129.miRNA Therapeutics. http://www.mirnarx.com . 2012.
- 130.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Soriano A., Jubierre L., Almazán-Moga A., Molist C., Roma J., de Toledo J.S., Gallego S., Segura M.F. microRNAs as pharmacological targets in cancer. Pharmacol. Res. 2013;75:3–14. doi: 10.1016/j.phrs.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 132.Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 133.Pecot C.V., Calin G.A., Coleman R.L., Lopez-Berestein G., Sood A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Broderick J.A., Zamore P.D. MicroRNA therapeutics. Gene Ther. 2011;18(12):1104–1110. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gary D.J., Puri N., Won Y.Y. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J. Control. Release. 2007;121(1-2):64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 136.Junttila M.R., de Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]