Abstract
Increasing evidence has shown that microRNA (miRNA) is extensively involved in the pathophysiology of lung cancer. Microarray data demonstrated the increasing levels of miR-19a in the peripheral blood from patients suffering from lung cancer, which is closely associated with poor prognosis of lung cancer. However, the underlying molecular mechanism of miR-19a remains to be determined. The results of the present study showed a higher expression of miR-19a compared with normal bronchial epithelial cells. Furthermore, lentivirus vectors were constructed to establish cell lines that overexpressed and knocked out miR-19a in order to study the role of miR-19a on the metastasis and proliferation of lung cancer cells. Investigation into the underlying mechanism of miR-19a, revealed that MXD1 may be the key gene targeting miR-19a, participating in the process of proliferation and metastasis of lung cancer cells.
Keywords: lung cancer, microRNA, MXD1, metastasis
Introduction
Lung cancer is a leading cause of cancer mortalities worldwide, from which approximately 80% of patients have non-small cell lung cancer (NSCLC) with a 5-year-survival rate of <15.9% (1,2). The poor prognosis of patients with lung cancer is closely associated with distant metastasis. Once diagnosed with lung cancer, the survival of patients may be <1 year (2,3). MicroRNAs (miRNAs) are a class of 22-nucleotide non-coding RNAs, which are evolutionarily conserved and function as negative regulators of gene expression (4). miRNAs are important in the process of individual development, maturation, senility and the pathological process of various types of diseases (4). Increasing evidence has shown that miRNAs play crucial roles in the pathological process of NSCLC and its occurrence, development, and metastasis (5–7). The abnormal regulation of miRNAs function as different factors in the process of NSCLC. Some of these miRNAs, including let-7, miR-34, miR-15/16, miR-143/145, miR-449a, miR-212 and miR-494, were identified as inhibitors of NSCLC in proliferation and metastasis. However, others, including miR-21, miR-221/222, miR-196a, miR-31 and miR141, may act as a promoter of NSCLC (4,7–13).
miR17-92 cluster is located on the 13th chromosome and is a genome with seven subtypes icluding 17-5p, 17-3p, 18a, 92a, 19b, and 19a. Previuos findings have shown that these subtypes have a close relationship between developments of tumor (14). It is well established that the miR-17-92 family is characterized as an oncogene, is upregulated in cancer cells and promotes cancer cell proliferation (14,15). miR-19a is an important subtype in the miR-17-92 family. miR-19a may play an important role in the regulation of the STST3/IL-6 signaling pathway and inhibit SOCS1, which was regarded as a negative factor of this signaling cascade (14,15) and a suppressor of cell apoptosis. It was also reported that miR-19a/b may upregulate the expression of Bim, and plays a crucial role in preventing apoptosis and promoting tumor cell proliferation (14).
In the present study, we discussed the role of miR-19a in the process of lung cancer and the underlying mechanisms, and demonstrated a close association between miR-19a and lung cancer.
Materials and methods
Cell culture
Normal human lung epithelial cell line BEAS-2B, and NSCLC cell lines (H1299 and H460) were purchased from the Institute of Biochemistry and Cell Biology, CAS (Shanghai, China). BEAS-2B cells were grown in Dulbecco's modified Eagle's medium (DMEM) and the NSCLC cell lines were grown in RPMI-1640 supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 100 U/ml penicillin-streptomycin. The cultured cells were maintained at 37°C in a humidified environment with 5% CO2 in air.
Establishment of stable overexpression and knockdown of miR-19a in cell lines
miR-19a overexpression and knockdown lentiviral vectors were prepared by GenePharma Co., Ltd. (Shanghai, China). H1299 and H460 were cultured in a 12-well plate for a confluence of 60%. The previous culture medium was discarded, and the cells were incubated with fresh RPMI-1640 for 2 h. After infecting the cells with lentivirus, the cells with a stable expression or knockdown of miR-19a were screened with puromycin.
Cell proliferation assay
The cell proliferation level was detected using the CCK-8 method. The cells were cultivated in a 96-well plate at a density of 2×103/well and incubated for 24, 48 and 72 h at 37°C, 5% CO2. Subsequently, the cells were incubated with CCK-8 for 1 h and then detected at 450 nm with a microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Flow cytometry assay for apoptosis
The cells were collected and washed with cold PBS, then stained with PI (BD Biosciences, San Diego, CA, USA) according to the production manuals. Cells were analyzed using flow cytometry (Coulter, Luton, UK).
Transwell assay
The cells were incubated with the culture medium containing FBS at a density of 1×105/ml, and 200 µl was added in the upper chamber. RPMI-1640 supplemented with 20% FBS was added in the lower chamber with 400 µl. After incubation for 24 h at 37°C, 5% CO2, cells on the upper surface of the filters were removed and cells adhering to the undersurface of the filter membrane were stained with 0.5% crystal violet for 30 min. The crystal violet was washed with PBS three times. The cells in the lower chamber were counted under a microscope for four fields randomly, and the mean cell numbers were recorded and analyzed.
Reverse transcription quantitative PCR
Total RNA was extracted from the cells at a density of 75% confluence using TRIzol total RNA isolation reagent (Gibco, Waltham, MA, USA) as per the manufacturers instructions. Total RNA (2 µg) was transcribed with stem-loop RT primers. The primer sequences used were: miR-19a (5-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCGGAAC-3); U6 (5-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3).
The produced cDNA was quantified by SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan). The primer sequences used were: miR-19a, 5-TGCGGTTCACAGTGGCTAAG-3; U6 forward, 5-TGCGGGTGCTCGCTTCGGCAGC-3 and reverse, 5-CCAGTGCAGGGTCCGAGGT-3. The reactions were incubated at 95°C for 5 min (95°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec) for 40 cycles and maintained at 10°C. The expression of mRNA was quantified by 2−ΔΔCT.
Western blotting
The cells were harvested (1.5×105/ml) and lysed by RIPA with PMSF. After incubation for 20 min, the cell lysate was centrifuged for 10 min at 2,580 × g and later separated on 10% SDS-PAGE (50 µg/lane) and transferred onto nitrocellulose membranes. After blocking with 5% BSA, the target proteins were probed with the primary antibodies at 4°C overnight. After three 5-min washes with TBST, the bound antibodies were detected using HRP-conjugated secondary antibodies and were visualized using the enhanced chemiluminescent reagent. The relative levels of each protein were determined by densitometric scanning (Amersham WB System, GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). The antibodies used for the study were: MXD1 antibody (rabbit polyclonal, IgG, dilution 1:1000, Proteintech, Rosemont, IL, USA; catalog no.: 17888-1-AP), GAPDH antibody (rabbit polyclonal, IgG, dilution 1:20000, Proteintech, Rosemont, IL, USA; catalog no.: 10494-1-AP). The secondary antibody was goat F(ab')2 anti-rabbit IgG (Fab')2 (HRP) preadsorbed (Abcam, Cambridge, MA, USA; catalog no.: ab6112).
Statistical analysis
Data were analyzed using SPSS 19.0 software (IBM, Armonk, NY, USA). Data were presented as means ± standard deviation. Comparisons were conducted using one-way ANOVA followed by the post-hoc (LSD) test. P<0.05 indicated a statistically significant difference.
Results
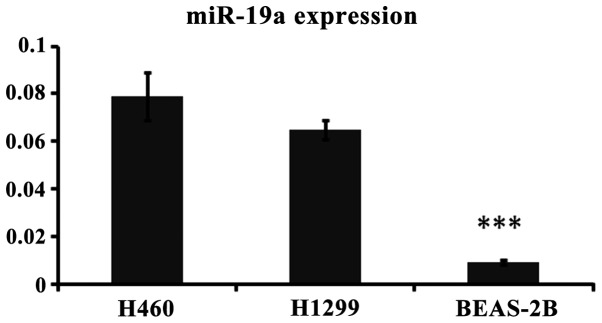
miR-19a expression is significantly higher in H460 and H1299 compared with the normal cell line BEAS-2B
RT-PCR was used to detect the expression of miR-19a in lung cancer and normal cells to determine the association between miR-19a and lung cancer. The results showed a significant difference between the normal and lung cancer cells (Fig. 1).
Figure 1.
miR-19a expression in the H460, H1299 and BEAS-2B cell lines. ***P<0.05, statistically signifiicant.
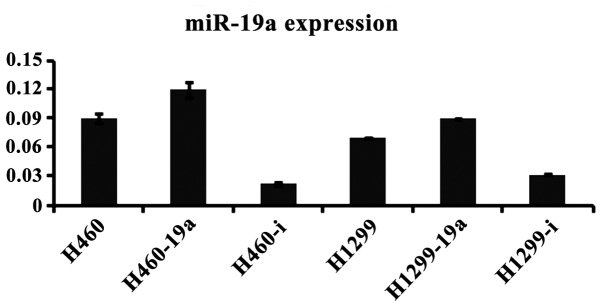
miR-19a plays a key role in lung cancer cell proliferation
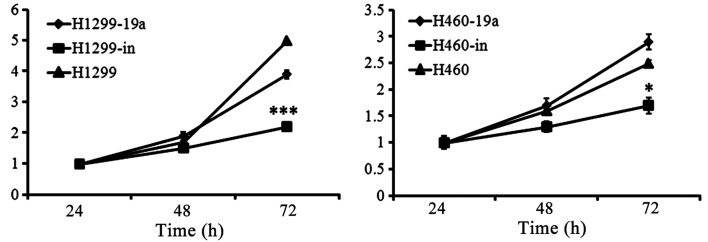
To determine the effects of miR-19a on proliferation in the different cell lines, we constructed the lentivirus vectors to establish the cells lines with overexpressed and knocked down miR-19a. The results of RT-PCR indicated that the cells lines infected by lentivirus were successfully established (Fig. 2). Subsequently, we detected cell proliferation using CCK-8 to determine the effects of miR-19a on proliferation. The cells were cultivated in a 96-well plate at a density of 2×103/well and incubated for 24, 48 and 72 h. There were no significant differences in proliferation at 24 and 48 h, whereas a significant difference was detected by 72 h, as observed by the inhibition of proliferation of the miR-19a knockdown cell lines, compared with the related control cell line and the overexpressed one (Fig. 3).
Figure 2.
Expression of miR-19a after infection by lentivirus. The suffix of ‘a’ indicates the overexpression of miR-19a while the suffix of ‘I’ indicates the knockdown of miR-19a.
Figure 3.
Lung cancer cell lines with overexpressed miR-19a show a significantly higher proliferation while the ones with miR-19a knockdown indicated a lower proliferation compared with the non-transfected control cells (***P<0.001; *P<0.05).
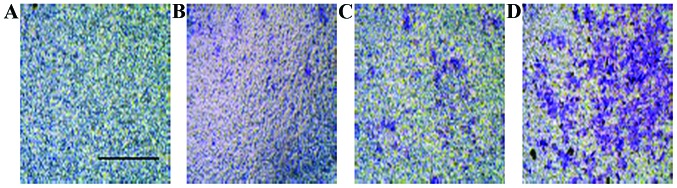
miR-19a stimulates the lung cancer cell migration
In addition to the proliferative function of miR-19a, the migratory influence of miR-19a was examined using an invasion assay. As shown in Fig. 4, miR-19a overexpressed cells had a significant increasing number of cells in the lower chamber compared with the non-transfected control cells. The invasion data indicated that miR-19a likely played an important role in promoting lung cancer metastasis.
Figure 4.
Crystal violet staining for migrated cells of non-transfected controls. (A) H460, (C) H1299 and the overexpressed cell lines, (B) H460 and (D) H1299.
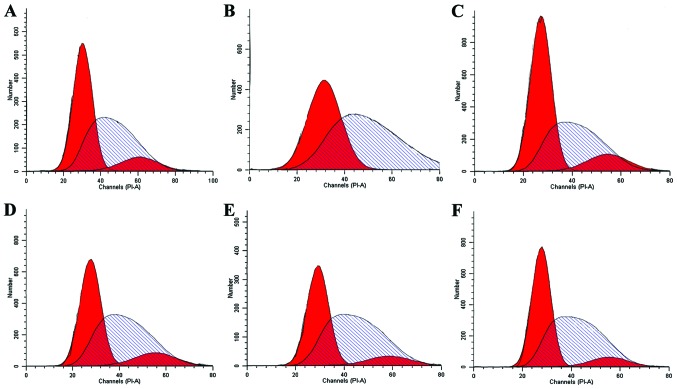
Effect of miR-19a on the cell cycle in lung cancer cells
The aforementioned results identified the upregulation of miR-19a in lung cancer cells and a higher expression of miR-19a may function as a promoter in cell proliferation and migration. Thus, the effect of miR-19a on the cell cycle was assessed. Using flow cytometry and PI staining, G2/M arrest in cell lines with miR-19a knockdown was identified, compared with the non-transfected control. However, the cell lines with miR-19a overexpression did not show any significant difference between the non-transfected control (Fig. 5).
Figure 5.
The cell cycle is detected by flow cytometry. (A) H1299, (B) H1299 with miR-19a overexpression, (C) H1299 with miR-19a knockdown, (D) H460, (E) H460 with miR-19a overexpression and (F) H460 with miR-19a knockdown.
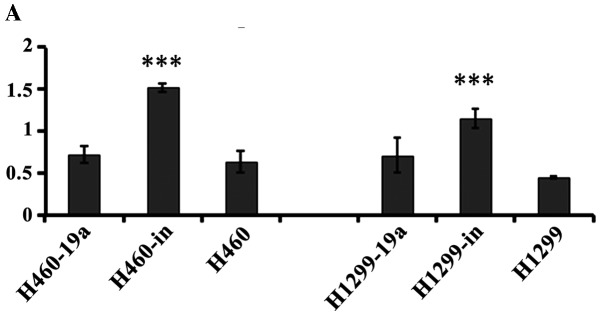
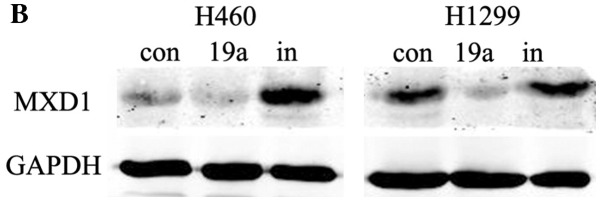
miR-19a inhibits MXD1 expression at the gene and protein level
MXD1 was capable of binding with MAX competitively, which inhibited the formation of the c-Myc/MAX heterodimers, and the transcriptional activity of c-Myc was downregulated (16). Our results found that following the overexpression of miR-19a in cancer cells, MXD1 expression was significantly downregulated, which indicated that miR-19a may regulate MXD1 expression negatively, and thus promote the formation of c-myc/max heterodimers indirectly, thereby enhancing cell proliferation. (Fig. 6).
Figure 6.
Expression of MXD1 at the gene and protein level. Suffix of ‘a’ indicates an overexpression of miR-19a while suffix of ‘I’ indicating a knockdown of miR-19a (***P<0.001).
Discussion
In the present study, we found that the expression of miR-19a was significantly higher in the lung cancer cell lines compared to the normal human lung epithelial cell line. In addition, we demonstrated that the high expression of miR-19a may act as a promoter in lung cancer cell proliferation and migration. To examine the underlying mechanisms of miR-19a in lung cancer cell lines, we identified the expression of MXD1, which was regaredd as an obligate partner to prevent the c-Myc/Max was negatively associated with the expression of miR-19a. Additionally, in miR-19a knockdown cell lines, we demonstrated the arrest of cell cycle at G2/M phase, indicating the miR-19a may target protein associated with the cell cycle.
miRNA are evolutionarily conserved and consist of 20–25 nucleotides, which bind to the UTR of mRNA, blocking or degrading the mRNA, indicating that it plays crucial roles in the process of cell-life activities, which have been demonstrated in various diseases, including cancer (17). miR-19a is an important member of the miR-17-92 RNA family, is considered an oncogene and is controlled by c-Myc (18,19). These characteristics of the miR-17-92 RNA family have been identified in various types of cancer, including lung cancer. Thus, miR-19a had common characteristics in promoting the proliferation and migration of cancer cells (14). Higher levels of miR-19a in the serum were also detected from the patients suffering from lung cancer (15).
There are several genes that miR-19a targets, including atrogin-1, MuRF-1, TNF-α, SOCS1, PTEN and CUL5 (16,20–25). In addition, a close association between miR-19a and MXD1 has been demonstrated in gastric cancer (16). It has been established that MXD1 is an obligate partner involved in the prevention of c-Myc/Max, playing a key role in preventing the process of cancers (26,27). In the present study, we indicated that miR-19a may function as a promoter in cell proliferation and migration underlying the mechanisms of downregulating the expression of MXD1 in lung cancer cell lines. Apart from the present study, in vivo and clinical research of miR-19a and other cancer cell lines should be conducted in future to clarify the important role that miR-19a plays on physiological and pathological processes of cancer cells.
References
- 1.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, DAmico TA, Demmy TL, Ganti AKP, Govindan R, et al. NCCN (National Comprehensive Cancer Network): Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10:1236–1271. doi: 10.6004/jnccn.2012.0130. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 3.Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD, Jacobsen J, Pirl WF, et al. Early palliative care for patients with metastatic non-small-cell lung cancer. N Engl J Med. 2010;363:733–742. doi: 10.1056/NEJMoa1000678. [DOI] [PubMed] [Google Scholar]
- 4.Qi J, Mu D. MicroRNAs and lung cancers: from pathogenesis to clinical implications. Front Med. 2012;6:134–155. doi: 10.1007/s11684-012-0188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang X, Liu L, Li H, Yu J, Wang C, Ren X. Clinical implication of microrna for lung cancer. Cancer Biother Radiopharm. 2013;28:261–267. doi: 10.1089/cbr.2012.1401. [DOI] [PubMed] [Google Scholar]
- 6.Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31. doi: 10.1016/j.mrfmmm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Zhang D, Chen C, Ruan Z, Li Y, Huang Y. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell. 2012;23:1423–1434. doi: 10.1091/mbc.E11-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W, Huang B, Li Z, Li H, Sun L, Zhang Q, Qiu X, Wang E. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One. 2013;8:e64759. doi: 10.1371/journal.pone.0064759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei Z, He Y, Feng J, Shi J, Du Y, Qian L, Huang Q, Jie Z. MicroRNA-141 promotes the proliferation of non-small cell lung cancer cells by regulating expression of PHLPP1 and PHLPP2. FEBS Lett. 2014;588:3055–3061. doi: 10.1016/j.febslet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Ohdaira H, Sekiguchi M, Miyata K, Yoshida K. MicroRNA-494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 2012;45:32–38. doi: 10.1111/j.1365-2184.2011.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonauer A, Dimmeler S. The microRNA-17-92 cluster: still a miRacle? Cell Cycle. 2009;8:3866–3873. doi: 10.4161/cc.8.23.9994. [DOI] [PubMed] [Google Scholar]
- 15.Lin Q, Chen T, Lin Q, Lin G, Lin J, Chen G, Guo L. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2013;107:767–771. doi: 10.1002/jso.23312. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha TY. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune Netw. 2011;11:135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, DAndrea A, Sander C, Ventura A. Genetic dissection of the miR-17-92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ODonnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 20.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 21.Song DW, Ryu JY, Kim JO, Kwon EJ, Kim H. The miR-19a/b family positively regulates cardiomyocyte hypertrophy by targeting atrogin-1 and MuRF-1. Biochem J. 2014;457:151–162. doi: 10.1042/BJ20130833. [DOI] [PubMed] [Google Scholar]
- 22.Chen B, She S, Li D, Liu Z, Yang X, Zeng Z, Liu F. Role of miR-19a targeting TNF-α in mediating ulcerative colitis. Scand J Gastroenterol. 2013;48:815–824. doi: 10.3109/00365521.2013.800991. [DOI] [PubMed] [Google Scholar]
- 23.Qin S, Ai F, Ji WF, Rao W, Zhang HC, Yao WJ. miR-19a promotes cell growth and tumorigenesis through targeting SOCS1 in gastric cancer. Asian Pac J Cancer Prev. 2013;14:835–840. doi: 10.7314/APJCP.2013.14.2.835. [DOI] [PubMed] [Google Scholar]
- 24.Pezzolesi MG, Platzer P, Waite KA, Eng C. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in cowden syndrome. Am J Hum Genet. 2008;82:1141–1149. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu XM, Wang XB, Chen MM, Liu T, Li YX, Jia WH, Liu M, Li X, Tang H. MicroRNA-19a and −19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett. 2012;322:148–158. doi: 10.1016/j.canlet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 26.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta Y, Hamada Y, Saitoh N, Katsuoka K. Effect of the transcriptional repressor Mad1 on proliferation of human melanoma cells. Exp Dermatol. 2002;11:439–447. doi: 10.1034/j.1600-0625.2002.110507.x. [DOI] [PubMed] [Google Scholar]