Abstract
More than 180 individual phages infecting hosts in the phylum Actinobacteria have been sequenced and grouped into Cluster A because of their similar overall nucleotide sequences and genome architectures. These Cluster A phages are either temperate or derivatives of temperate parents, and most have an integration cassette near the center of the genome containing an integrase gene and attP. However, about 20% of the phages lack an integration cassette, which is replaced by a 1.4 kbp segment with predicted partitioning functions, including plasmid-like parA and parB genes. Phage RedRock forms stable lysogens in Mycobacterium smegmatis in which the prophage replicates at 2.4 copies/chromosome and the partitioning system confers prophage maintenance. The parAB genes are expressed upon RedRock infection of M. smegmatis, but are down-regulated once lysogeny is established by binding of RedRock ParB to parS-L, one of two centromere-like sites flanking the parAB genes. The RedRock parS-L and parS-R sites are composed of eight directly repeated copies of an 8 bp motif that is recognized by ParB. The actinobacteriophage parABS cassettes span considerable sequence diversity and specificity, providing a suite of tools for use in mycobacterial genetics.
Graphical Abstract
Introduction
A large collection of sequenced actinobacteriophage genomes provides high-resolution insights into their genetic diversity and evolution (Pope et al., 2015). The more than 1,300 sequenced phage genomes (http://phagesdb.org), can be organized into over two-dozen clusters (Cluster A, B, C, etc.) composed of distinct nucleotide sequences, many of which can be divided into subclusters based on their overall sequence similarities. There is substantial diversity within subclusters, and their architecturally mosaic genomes reflect a long evolutionary history of genetic exchange (Hatfull et al., 2010, Pope et al., 2015). The genetic diversity is reflected in considerable biological novelty and these phages have provided numerous insights into gene expression, regulation, and function (Hatfull, 2010, Hatfull, 2012, Hatfull, 2014).
Many of the actinobacteriophages are either temperate, or are recent derivatives of temperate parents (Hatfull, 2014). A majority of the phages encode an integrase of either the tyrosine- or serine- class of site-specific recombinases that mediate prophage formation by chromosomal integration (Hatfull, 2014). Several phages (particularly those organized in Clusters P, N, and I) employ an unusual system of life cycle regulation in which the phage attachment site for integration (attP) is located within the repressor gene, such that the 3’ end of the repressor that encodes a degradation tag is recombinationally dissociated from the rest of the gene, resulting in repressor stabilization (Broussard et al., 2013). In other phage genomes such as L5 and Bxb1 (both organized in Cluster A), prophage integration is independent of repressor synthesis, although the immunity system is unusual in that the repressor binds to a large number (∼25–30) of repressor binding sites distributed across the genomes (Jain & Hatfull, 2000, Brown et al., 1997, Pope et al., 2011). Repressor binding interferes with transcriptional progression and these sites are known as ‘stoperators’, in contrast to the operator sites that regulate transcription initiation (Brown et al., 1997).
Cluster A is the largest group of actinobacteriophages, and currently can be divided into 17 subclusters (A1 – A17). However, these all share a common organization in which the virion structure and assembly genes are organized with a common synteny in the left arms, and the right arms contain DNA metabolism and regulatory genes, along with a large number of small open reading frames of unknown function [see Fig. 1A, (Hatfull & Sarkis, 1993, Hatfull, 2012)]. The left arm genes are transcribed rightwards, and the right arm genes are typically transcribed leftwards, although some of the phages (primarily within Subcluster A1) also have up to six genes at the right ends of the genomes that are also transcribed rightwards (Hatfull, 2012). Most of the Cluster A phages encode an integrase gene and a closely-linked attP site situated at the center of the genomes, between the left and right arm genes (Hatfull, 2012, Hatfull, 2014).
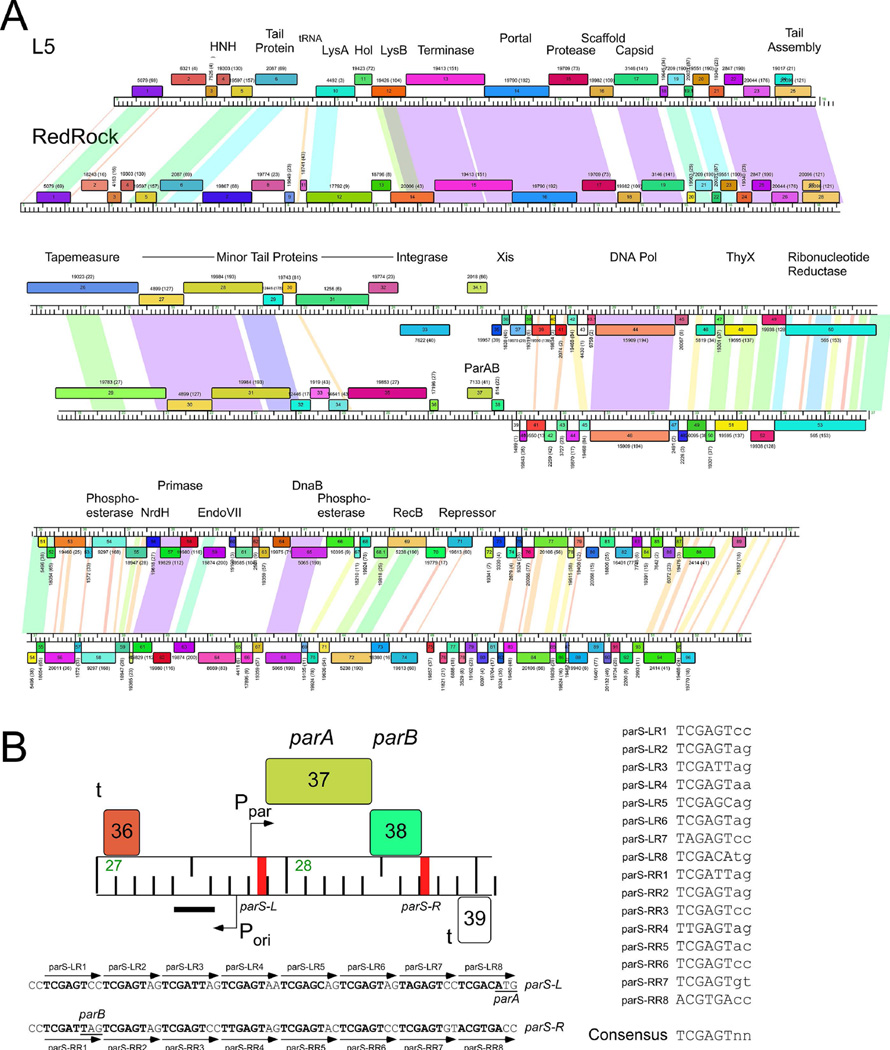
Figure 1.
The parABS system of mycobacteriophage RedRock. A. Alignment of genome maps of mycobacteriophages RedRock and L5. Both phages are grouped in Subcluster A2 and share similar genomic organizations and sequence similarity. However, at the centers of the genomes L5 contains an integration cassette (int, attP), whereas RedRock has parAB genes. Maps were generated using Phamerator (Cresawn et al., 2011). Predicted genes are shown as boxes transcribed rightwards or leftwards (shown above or below the genome ruler, respectively) with the gene number inside each box. Genes were assorted into phamilies (phams) of related sequences as described elsewhere (Cresawn et al., 2011, Pope et al., 2015), and colored according to pham membership; pham numbers are shown above or below each gene with the numbers of members in parentheses. Shading between genomes indicates nucleotide sequence similarity determined by BlastN and is spectrum colored with violet being the most similar. B. Detailed view of parABS organization. The parAB genes are transcribed rightwards and are flanked by centromere-like sites parS-L and parS-R. The location of putative promoters (P) and terminators (t) are shown, as well as a region (black bar) in which a non-coding RNA is highly expressed. Shown below are the sequences for parS-L and parS-R, with individual 8 bp repeat sequence motifs indicated, which are aligned to provide a consensus sequence on the right.
Bacterial chromosomes and many plasmids – especially those replicating at low copy number – encode a partitioning system that enables segregation of plasmid molecules into both daughter cells at division, resulting in stable maintenance of the plasmid in a population of cells (Baxter & Funnell, 2014, Ebersbach & Gerdes, 2005). The partitioning systems are highly diverse but can be organized into three main groups, Type I (subdivided into Ia and Ib), II and III, each of which typically contains a centromere binding protein (CBP), an ATPase or GTPase partitioning protein, and a centromere-like DNA site (Reyes-Lamothe et al., 2012, Wang et al., 2013, Baxter & Funnell, 2014, Ebersbach & Gerdes, 2005). The CBP binds to the centromere-like site, and the NTPase uses nucleotide hydrolysis to move DNA throughout the cell (Baxter & Funnell, 2014). Partitioning systems have also been described in temperate phages such as P1 and N15 of Escherichia coli, which replicate as extrachromosomal circular and linear prophages, respectively (Sternberg & Austin, 1981, Lobocka et al., 2004, Ravin & Lane, 1999, Ravin, 2011, Ravin et al., 2000). Although extrachromosomally-replicating prophages seem to be a relatively uncommon life style among temperate phages compared to chromosomal integration, related systems have been described in phages of diverse bacterial hosts, including Leptospira interrogans [lpc3, (Zhu et al., 2015)], Streptomyces sp. [pZL12, (Zhong et al., 2010)], Vibrio vulnificus [pVv01, (Hammerl et al., 2014)] Yersinia enterocolitica [PY54, (Hertwig et al., 2003)], Vibrio parahaemolyticus [VP58.5, (Zabala et al., 2009)], and Halomonas aquamarina [ΦHAP-1, (Mobberley et al., 2008)].
Although the vast majority of phages infecting actinobacterial hosts (i.e. actinobacteriophages) that group in Cluster A encode an integration cassette, a subset do not, and instead have a putative partitioning cassette located at a similar genomic position to the integration functions of closely related phages (Hatfull, 2014, Stella et al., 2013). These phages must presumably encode an origin of prophage replication, although no RepA-like proteins or other such replication functions have been identified. Here we characterize the partitioning systems of mycobacteriophage RedRock and related phages. We show that RedRock forms lysogens carrying extrachromosomal prophages replicating at an average copy number of 2.4 copies/chromosome, and that a Type Ib partitioning system encoding ParA and ParB promotes prophage stability. The parAB genes of four different phages are expressed in lysogens and expression is autoregulated by ParB binding to a parS site upstream of parA. A putative origin of prophage replication lies adjacent to parAB and is associated with a highly expressed non-coding RNA in lysogenic cells. Two parS sites flank the parAB genes and are composed of multiple copies of an 8 bp directly repeated sequence motif that is recognized by ParB. Phylogenetic analysis of Par proteins shows that they span considerable sequence variation, and may be under selective pressures directed by prophage incompatibility, which we demonstrate for several pairs of par-containing phages.
Results
Cluster A actinobacteriophages encode diverse partitioning cassettes
Over 1,300 completely sequenced phages of actinobacterial hosts are deposited in the phagesdb database (http://phagesdb.org), including 706 entries that are fully annotated and available from GenBank or http://phagesdb.org. We therefore focused our analyses on these 706 phages, 183 of which are grouped in Cluster A, and which can be further divided into 17 subclusters (A1-A17) based on their overall genomic similarities. Most of these infect Mycobacterium smegmatis although one (the sole member of Subcluster A13) infects Mycobacterium phlei and three (all within Subcluster A15) infect Gordonia terrae (http://phagesdb.org). Nine of the subclusters (A2, A6, A9, A11, A13, A14, A15, A16, and A17) contain phages encoding putative homologues of previously described parA and parB genes; in each instance parA and parB are closely linked and are genomically located where the integration cassette – containing an integrase gene and attP attachment site – is typically located (Fig. 1A). For example, the temperate mycobacteriophages L5 and RedRock (both in Subcluster A2) share substantial nucleotide sequence similarity and similar overall genomic organization. However, whereas the integration cassette is located between the rightwards-transcribed virion structural and assembly genes and the leftwards-transcribed right arm genes of L5 (Hatfull & Sarkis, 1993), in RedRock this position is occupied by the parA and parB genes (Fig. 1A), as it is also in the previously described phages 20ES, 40AC, and First (Stella et al., 2013). The shared synteny is consistent with the integration and partitioning cassettes conferring the same biological function of prophage stability, although unlike the integrating phages, par-containing prophages presumably replicate autonomously and extrachromosomally.
In RedRock, genes 37 and 38 encode the parA and parB functions respectively, and are organized into an apparent operon (Fig. 1B). The genes are flanked to their immediate left and right by ∼70 bp sites that include multiple copies of an 8 bp repeated motif (5’-TCGAGTnn). This organization is reminiscent of the centromere-like parS sites of some Type Ib plasmid partitioning systems – e.g. pSM19035 (Dmowski et al., 2006) and TP228 (Zampini et al., 2009) – and we designate these as parS-L and parS-R for the left and right sites respectively (Fig. 1B). We note that there are other circular permutations of the 8 bp repeat including those on the opposite strand (because of its partial palindromic nature), but six of the positions are highly conserved (present in at least 13 of the 16 repeat units at positions 1–6, in 5’-TCGAGTnn) and two (positions 7 and 8) are more varied (Fig. 1B). Similar characteristics of parS loci have been described in pSM19035 and TP228. Presumably one protomer of RedRock ParB recognizes each of the 8 bp motifs.
We identified 42 genomes among the sequenced actinobacteriophages containing partitioning cassettes; these include 37 M. smegmatis phages, 1 M. phlei phage, 3 G. terrae phages, and a previously described extrachromosomally replicating phage of Streptomyces, pZL12 (Fig. 2; details of phages used in the analysis are shown in Table S1) (Stella et al., 2013, Zhong et al., 2010). We examined the predicted ParA and ParB proteins of each of these for helix-turn-helix DNA-binding motifs (Dodd & Egan, 1990), conserved domains, and structural motifs using HHpred (Soding, 2005), and compared them to 41 previously identified partitioning systems representing the three major types (Fig. 2, S2). Several lines of evidence suggest that all of the Cluster A cassettes belong to Type Ib (Fig. 2). First, the predicted ParA proteins contain a particular variant of the Walker A ATPase motif common to Type I ParA proteins, but do not contain a predicted DNA binding motif common to Type Ia. Second, a structural motif analysis of the ParB proteins predicts C-terminal ribbon-helix-helix (RHH) motifs. The strongest hits are to pSM19035 omega (Murayama et al., 2001) and TP228 ParG (Golovanov et al., 2003), structurally defined members of Type Ib systems, at probabilities greater than 98% (except for Echild and 40AC). These motifs are found in Type Ib but not Type Ia systems, and are located at the C-terminal ends of the proteins (Baxter & Funnell, 2014). More broadly, the structural motif analysis shows closest similarity of the ParA and ParB proteins to domains that are specific to Type Ib ParA and ParB proteins and lack domains specific to Type Ia, II, or III proteins (see Materials and Methods). Lastly, the ParA and ParB proteins range in size from 159–228 amino acids and 84–104 amino acids, respectively, both of which are within the common size ranges for Type Ib proteins, but smaller than common Type Ia proteins.
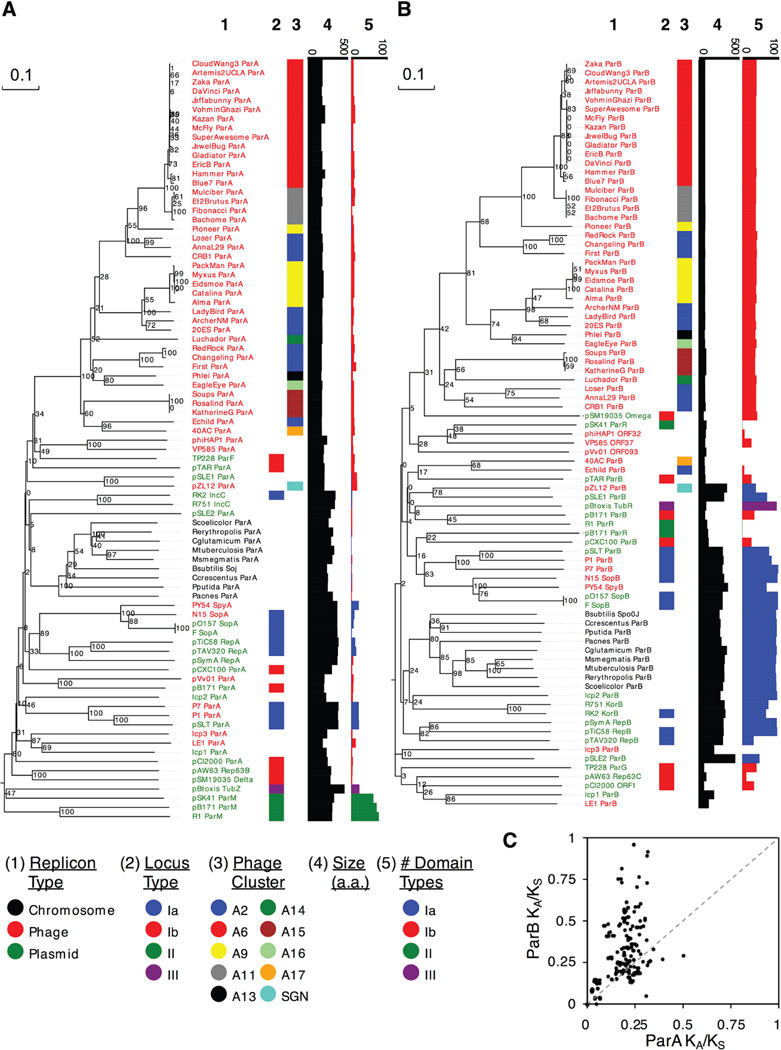
Figure 2.
Phylogenetic comparison of ParA and ParB proteins. Phylogenetic comparison was performed on NTPase ParA-related proteins (A) and ParB-like centromere binding proteins (B) from 83 characterized and predicted partitioning systems representing different types of partitioning system (Ia, Ib, II, and III) and derived from various types of replicons (chromosomal, plasmid, and phage). Bootstrap values are indicated. For each sequence, the subcluster designation in the Actinobacteriophage_706 database is represented by color (if applicable), the amino acid size is displayed in a horizontal bar graph, and the frequency of categorized structural domains identified by HHpred analysis is displayed in a colored horizontal bar graph. We note that Leptospira lcp1, lcp2, and lcp3 have been predicted to have partitioning cassettes (Zhu et al., 2015), but their roles in segregation have not yet been reported. Although lcp3 ParB has no par type domains and no close relatives, lcp1 ParB groups with LE1 ParB, which has been shown to play a role in plasmid stability (Bourhy et al., 2005), and is positioned in a clade of Type Ib ParBs from TP228, pAW63, and pCI2000 replicons. In contrast, lcp2 is positioned elsewhere in the tree and has many Type Ia domains; likewise, the predicted ParB of the Streptomyces plasmids pZL12 (Zhong et al., 2010), pSLE1, and pSLE2 (Gomez-Escribano et al., 2015), also have many Type Ia domains. The cassettes from ΦHAP-1, pVv01 and Vp58.5 (Mobberley et al., 2008, Hammerl et al., 2014, Zabala et al., 2009), exhibit Type Ib similarities, including ParB-like genes adjacent to parA that were not previously predicted. C. Rate of evolution of ParA and ParB genes. For 27 actinobacteriophages with non-redundant ParA and ParB DNA sequences, a KA/KS analysis was performed to compare rates of evolution between the two genes. For all pairwise comparisons of partitioning cassettes, the resulting ParA and ParB KA/KS ratios were graphed as a scatter plot. The dotted line reflects how the data would be distributed if both genes experienced similar selective forces.
In most of the actinobacteriophage systems we identified, putative parS sites are located to the left and right of parAB, as in RedRock (Figs. 1B, S2A), although some lack either parS-L or parS-R. Motif searches identified putative repeated sequences related to the 8 bp identified in the RedRock parS sites (Fig. S2B).
The number and diversity of the partitioning systems identified here provides an opportunity to examine their evolutionary patterns. We compared the ratios of non-synonymous substitutions (KA) to synonymous substitutions (KS) for parA and parB between the par-containing actinobacteriophages (Fig. 2C) and observed that although the KA/KS ratios of parA genes are rarely above 0.3 – suggesting they are under strong purifying selection – the KA/KS ratios of parB are highly varied, and in some cases approach 1.0; furthermore, there is no apparent correlation of the parA and parB KA/KS ratios. The parB ratios are outside of the range typically regarded as indications of strong diversifying selection (KA/KS > 1.0), although diversifying selection may act only on parts of the sequences, thus moderating the overall signal. Nevertheless, there appears to be different evolutionary pressures exerted on ParA and ParB. This is consistent with the hypothesis that the CBP proteins of these Type Ib partitioning systems evolve rapidly to develop new parS specificities, presumably because phages with similar partitioning cassettes will exhibit incompatibility and prophage loss (Ebersbach & Gerdes, 2005, Hyland et al., 2014, Radnedge et al., 1996, Sergueev et al., 2005). Prior analyses suggested that ParA and ParB phylogenies tend to mirror each other (Petersen et al., 2009, Stella et al., 2013), but quantitatively comparing their evolutionary rates is complicated because ParB homologs tend to be much more diverse and difficult to predict than their ParA counterparts (Gerdes et al., 2000, Fothergill et al., 2005).
The RedRock prophage replicates extrachromosomally
All of the actinobacteriophages with partitioning cassettes are predicted to be temperate, and encode repressors related to those of other Cluster A phages such as L5 and Bxb1 (Donnelly-Wu et al., 1993, Jain & Hatfull, 2000); the exceptions are phages Jeffabunny, Jewelbug, and Phlei in which the repressor gene appears to have been deleted. We successfully generated stable lysogens for ten par-containing phages (Alma, ArcherNM, DaVinci, EagleEye, Et2Brutus, Gladiator, LadyBird, Mulciber, Pioneer, and RedRock) representing nearly all major clades of the ParB phylogeny (Fig. 2B), and showed that all of them are temperate, and not only form turbid plaques on M. smegmatis, but also form stable lysogens (data not shown); plaque turbidity varies considerably (the least turbid being Alma and Pioneer, the most turbid being EagleEye) likely reflecting variations in lysogeny frequency. Phage Echild forms turbid plaques but we were unable to propagate a stable lysogen. RedRock and Echild also infect and form turbid plaques on M. tuberculosis, Alma, Ladybird, and Pioneer infect M. tuberculosis at a reduced efficiency of plating (ranging from 10−4-10−6), and EagleEye, Gladiator, and Mulciber do not infect M. tuberculosis.
To address specifically whether a RedRock prophage in a lysogenic strain replicates extrachromosomally we isolated total DNA from a lysogen and performed whole genome sequencing. We reasoned that the sequencing reads mapping to the RedRock genome should have characteristics that are distinct from those of an L5 lysogen, in which the prophage is chromosomally integrated (Hatfull & Sarkis, 1993). First, if lytic growth is tightly down-regulated then both samples should contain few if any reads corresponding to the precise ends of the viral genomes, which are otherwise readily recognized in sequence reads of viral DNA alone (Table 1). As expected, for both L5 and RedRock few reads mapped to the precise viral genome ends (Table 1). In addition, fewer than 2% of reads across the attachment sites map to a L5 viral attP site, with the rest mapping to the integrated attL and attR sites, consistent with the sample containing only minor amounts of viral DNA (Table 1). Second, the L5 prophage has average sequence coverage that is the same as that for the rest of the M. smegmatis genome, consistent with it being chromosomally integrated (Table 1), whereas in the RedRock lysogen the prophage coverage is 2.4 fold higher than the bacterial chromosome (Fig. S1). We conclude that the RedRock prophage replicates as an extrachromosomal circle with an average copy number of 2.4 per host chromosome, and note that other extrachromosomally-replicating prophages such as P1 also replicate at low copy number [P1 has a copy number of 1.6/chromosome in standard growth conditions, (Lobocka et al., 2004, Prentki et al., 1977)].
Table 1.
Whole genome sequence coverage of L5 and RedRock lysogens
| Phage reads1 |
viral end reads (#)2 |
attL/attR reads (#) |
attP reads (#) |
Coverage: Phage |
Coverage: M.smegmatis |
Phage:Host coverage |
|
|---|---|---|---|---|---|---|---|
| mc2155(L5) | 41,053 | 23 | 141 | 2 | 58.9 | 62.5 | 0.94 |
| mc2155(RedRock) | 105,978 | 0 | N/A | N/A | 149 | 61.8 | 2.41 |
Reads mapping to phage genome out of a total of 5.8 million per sample
Reads beginning at precisely the terminus of viral genomic DNA
The RedRock genome must presumably contain two origins of DNA replication, one for lytic growth and one for extrachromosomal prophage replication, whereas L5 and other integrating phages require only a lytic replication origin. Comparison of the L5 and RedRock genomes (Fig. 1A) shows close relationships in both the left-arm and right-arm sets of genes, such that the likely location of a prophage origin is nearby the parAB cassette. There are no additional reading frames in this region (or elsewhere in the genome) encoding putative plasmid replication-like functions [similar to the RepA protein of P1, (Chattoraj, 2000)], and the only gene for consideration of this role is gene 36, which is downstream of a transcriptional terminator at the end of gene 35 (Fig. 1B). However, the 62-residue gp36 protein has no database matches and has no known function; it is conserved in phages such as Rebeuca, Larenn, and Serenity, that encode integration rather than partitioning systems, and is thus unlikely to be associated with partitioning or replication functions. It is plausible that the replication origin is located in the non-coding gap between genes 36 and 37 (parA) and that only RNA products are required for initiation and regulation of replication. We also note that RedRock lacks additional functions associated with P1 prophage plasmid maintenance, such as a site-specific recombination system to resolve plasmid dimers [as in loxP/Cre of P1, (Austin et al., 1981)], or a toxin-antitoxin addiction module [as in P1 doc/phd (Lehnherr et al., 1993)]. Finally, the G-C and A-T skew patterns of RedRock and L5 are very similar, suggesting the prophage replication origin does not contribute substantially to overall biases in nucleotide composition (Fig. S3). This is consistent with the hypothesis that the integration and ori/partitioning cassettes are actively exchanging among this group of genomes.
RedRock parAB confers plasmid stability
To show that the RedRock parABS cassette is required for prophage stability we attempted to construct mutant phage derivatives in which parA, parB, or both parA and parB are deleted using BRED mutagenesis (Marinelli et al., 2008). For all mutant constructions we were able to identify the presence of mutant alleles in screening of primary plaques, but were unable to purify the mutants to homogeneity. The parA and parB constructions were repeated using a complementation system, but we again failed to purify the mutants to homogeneity (Fig. S4). Complementation of mycobacteriophage mutants has previously been found to be somewhat fickle (Dedrick et al., 2013), but the general profile of mutagenesis is consistent with the parABS cassette being required for RedRock lytic growth; it could also be accounted for toxicity associated with loss of function.
Although we were unable to construct mutant phage derivatives, we determined whether the RedRock parABS system is able to confer stability to mycobacterial plasmids carrying an origin of replication (oriM) derived from plasmid pAL5000 (Rauzier et al., 1988). These plasmids replicate at moderate copy numbers in M. smegmatis of about 22 copies/cell (Huff et al., 2010) but are somewhat unstable, with about 35% plasmid loss following 30 generations of unselected growth (Lee et al., 1991). We used an oriM plasmid (pLO87) containing an mCherry gene driven by the strong hsp60 promoter (Oldfield & Hatfull, 2014), which confers a visible red color to M. smegmatis colonies and liquid cultures. This plasmid confers a notable growth disadvantage to M. smegmatis and in the absence of selection is lost quickly, and less than 1% of colonies retain the plasmid after 52 generations of unselected growth (Table 2); the liquid culture also lost all visible fluorescence (data not shown). In contrast, plasmid pMO01 – a derivative of pLO87 carrying the parABS cassette (RedRock coordinates 27,720 – 28,898) – maintained fluorescence in unselected liquid growth, and plasmid loss was constrained to about 20% after 52 generations of unselected growth (Table 2). In similar experiments in which the plasmids lack the hsp60-mCherry cassette and are better tolerated (e.g. pMO20), we observed only 13% loss after 52 generations of unselected growth in the absence of parABS, and full stabilization with inclusion of parABS (pMO21; Table 2).
Table 2.
par-mediated plasmid stabilization
| Plasmid | Reporter | par locus | Plasmid retention (%)1 |
|---|---|---|---|
| pLO87 | hsp60-mCherry | None | <1 |
| pMO01 | hsp60-mCherry | parSL-parA-parB-parSR | 822 |
| pMO02 | hsp60-mCherry | parSL- -parB-parSR | 7 |
| pMO05 | hsp60-mCherry | parSL-parA-parB | 712 |
| pMO20 | None | None | 873 |
| pMO21 | None | parSL-parA-parB-parSR | 1003 |
Plasmid retention determined by percentages of red (pLO87, pMO01, pMO02, pMO05) or kanamycin-resistant (pMO20, pMO21) colonies after 52 generations of unselected growth.
Difference in pMO01 and pMO05 retention levels is not significant (p-value 0.25).
Difference in pMO20 and pMO21 retention is statistically significant (p-value 0.002).
Interruption of the parA gene in plasmid pMO01 destroys the plasmid stabilization effect confirming that ParA is required for stability (Table 2). We were unable to test the requirement for parB in this assay, as a plasmid derivative in which parB was interrupted fails to transform M. smegmatis. Removal of parS-L confers the same phenotype, and we hypothesize that ParB plays a regulatory role in binding to parS-L, and interruption of this regulation confers the non-transformable phenotype, perhaps due to toxicity of unregulated parA overexpression from an upstream promoter. Deletion of parS-R shows that it is not required for stabilization, exhibiting no significant change in plasmid retention from pMO01 (Table 2).
Expression and regulation of RedRock parAB
Because RedRock parABS is required for prophage stability we predict that the parAB genes are expressed during lysogenic growth. RNAseq analysis of a RedRock lysogen shows that the parAB genes are indeed expressed, although at a level approximately 5-fold lower than the phage repressor, gene 74 (Fig. 3A); the phage lytic genes are tightly down-regulated as expected. A precise transcription start site cannot be readily identified – perhaps due to RNA processing or degradation – although transcription likely begins in the 36-37 intergenic region. We also note that there is low level of expression of gene 36 (Fig. 3B).
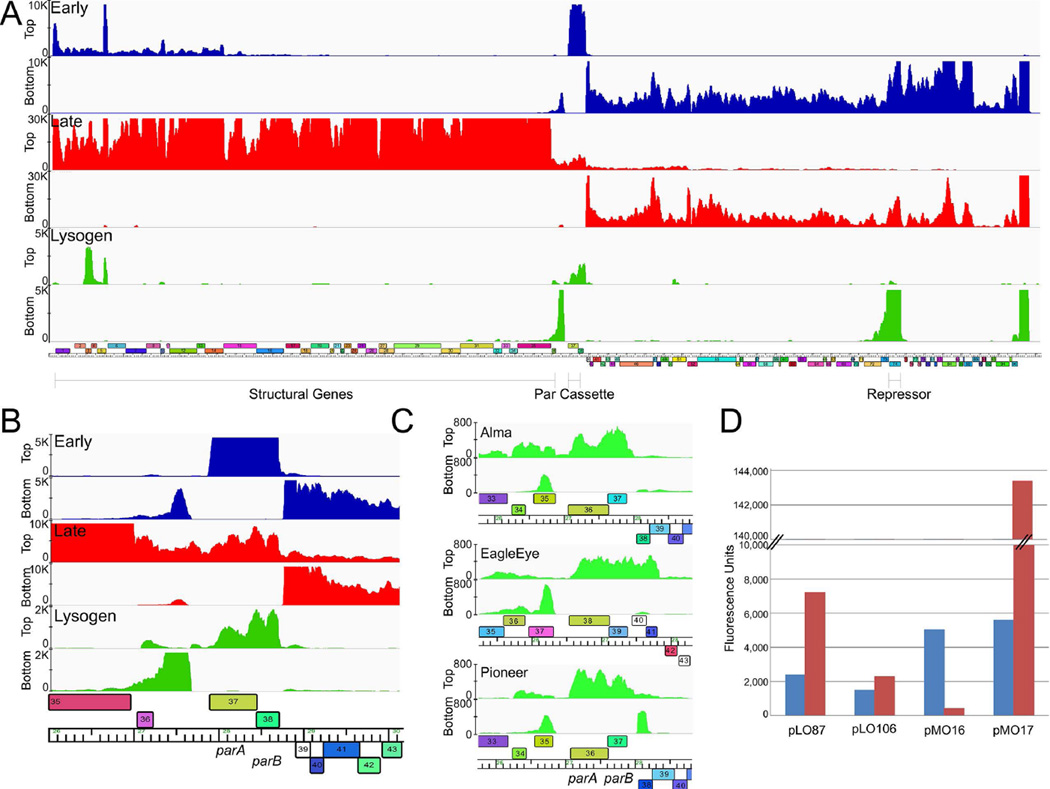
Figure 3.
Expression and regulation of the RedRock parABS cassette. A. Strand-specific RNAseq analysis of a RedRock lysogen (green) and at early (30 mins; blue) and late (150 mins; red) times post-infection. Lanes corresponding to top and bottom strand sequence reads and scale maxima are shown at the left. A map of the RedRock genome is shown below. B. Details of parAB expression. Profiles are colored as in panel A. C. Detailed view of the RNAseq profiles of the parAB genes from Alma, EagleEye, and Pioneer lysogens. The full genome profiles are shown in Fig. S5. D. Reporter gene expression of par-promoters. The fluorescence of M. smegmatis strains containing mCherry reporter plasmids, as indicated, is shown. Strains also lacked (blue bars) or contained (red bars) integrating plasmid pMO15 in which RedRock ParB is expressed from the hsp60 promoter. Reporter plasmids contain the following promoters: hsp60 (pLO87), phage BPs p6 (pLO106), Ppar (pMO16), and Pori (pMO17) (see Fig. 1).
Interestingly, there is a region of strong leftwards transcription just to the left of the parAB genes (Fig. 3A, B). The peak of strongest signal is approximately 200 bp long and corresponds to about 70% of the level of repressor RNA, although weaker signals extend leftwards for about another 900 bp. There are no predicted leftwards open reading frames in this region and we propose that the RNA product is either a functional or a regulatory component of the prophage origin of replication. RNAseq analysis of the lysogens of three other par-containing phages, Alma, EagleEye, and Pioneer, show that all make a similar leftwards-transcribed RNA adjacent to the parAB genes (Figs. 3C, S5), in addition to parAB expression. For Alma, EagleEye, and Pioneer this coincides with a predicted, rightwards-transcribed gene (35, 37, and 35 respectively), but all three have very weak coding potential and are likely mis-annotations. Insertion of a 1.68 kbp fragment from RedRock (coordinates 27,232 – 28,911) that spans the parABS locus and intergenic transcript into a non-replicating vector did not support autonomous replication in M. smegmatis, so other prophage-encoded functions are likely required (data not shown).
Two other regions of RedRock prophage expression are observed. One includes genes 3, 4 and 5, with transcription starting within gene 2 and ending in the gene 5-6 intergenic gap. Gene 4 is predicted to encode an HNH endonuclease, gene 5 encodes a putative virion tail protein, and gene 3 is of unknown function. It is unclear whether any of the genes could be associated with prophage replication. The second region is within a non-coding region at the right end of the genome, and its role is unclear although similar expression has been observed in related but non-parABS phages, and is therefore also unrelated to prophage maintenance [our unpublished information; (Halleran et al., 2015)]. Similar profiles are observed for the Alma, EagleEye, and Pioneer lysogens, although the expression at the genome right end is not seen in EagleEye (Fig. S5).
We also examined expression in RedRock-infected cells, both early (30 mins) and late in infection (2.5 hours). At the early time, expression is predominantly of the right arm genes, although some expression of genes at the beginning of the left arm is also observed (Fig. 3A). At the late time, expression of the right genes remains, although the left arm genes encoding the virion structure and assembly genes are expressed strongly. Interestingly, the parAB genes are among the most highly expressed genes early post-infection, although less so at the later time (Fig. 3A). A plausible explanation is that during liquid infection (using a multiplicity of infection of 3) a proportion of infected cells are in the process of establishing lysogeny, and that parAB expression is vigorous until it is subsequently down-regulated once lysogeny is established.
To further explore the regulation of parAB expression we constructed reporter fusion plasmids and determined promoter activity in the presence and absence of ParB (Fig. 3D). Initially a 178 bp fragment containing parS-L and the upstream sequences (RedRock coordinates 27,720 – 27,897) was inserted into vector pLO106 (Villanueva et al., 2015) in either orientation relative to the mCherry reporter gene. In the forward orientation (pMO16) substantial promoter activity was observed at a level greater than that of the strong hsp60 promoter (Oldfield & Hatfull, 2014), indicating that a highly active promoter for parAB expression is in this region. However, this activity is strongly down-regulated in a strain expressing RedRock ParB (Fig. 3D), consistent with transcriptional repression by binding of ParB to parS-L. Interestingly, when the same DNA segment in pMO16 is inserted in the opposite orientation (pMO17), a similarly active promoter is observed, and it appears to be up-regulated by ParB (Fig. 3D). There are no clear bioinformatic signals as to the precise promoter locations, but the promoter is presumably located upstream of parS-L (Fig. 3B). We designate these promoters as Ppar and Pori respectively (See Fig. 1B).
Binding of RedRock ParB to parS DNA
To investigate RedRock ParB binding to the repeated sequences designated as parS-L and parS-R, we overexpressed the ParB protein and purified it to near homogeneity. Using a DNA substrate spanning the entire 666 bp 36-37 intergenic region (coordinates 27,232 – 27,897), ParB binds and forms complexes separable by native gel electrophoresis (Fig. 4A). ParB binding affinity varies somewhat between experiments (Kd = 100 nM – 300 nM), and forms discernible complexes at lower protein concentrations, but at high protein concentrations forms indistinct complexes with much slower mobilities. A similar pattern is seen using a 194 bp fragment that includes parS-R although the complexes are less distinct (Fig. 4A). A simple explanation is that RedRock ParB binds specifically to parS sequences, but due to the complexity of the locus – similar to what has been observed with the Type Ib partitioning cassettes of pSM19035 (Dmowski et al., 2006) and TP228 (Zampini et al., 2009, Carmelo et al., 2005) – ParB may recognize individual sites with different affinities and with cooperativity at higher concentrations, forming a variety of complexes.
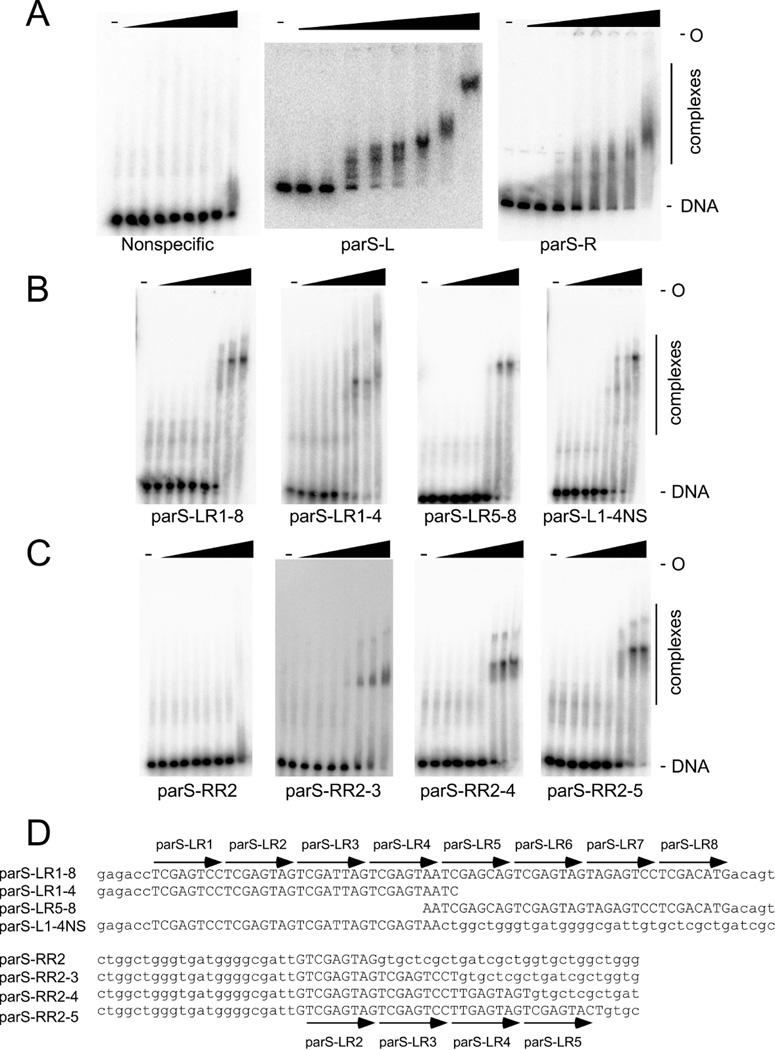
Figure 4.
Binding of RedRock ParB to parSA. Binding of RedRock ParB to parS-L and parS-R. ParB was incubated at different concentrations to a 45 bp nonspecific DNA fragment (left), a 666 bp fragment containing parS-L (middle) and a 194 bp fragment containing parS-R (right), and the complexes separated from unbound DNA by native acrylamide gel electrophoresis. Protein concentrations from left to right are : 0, 8.4, 25, 76, 228, 684, 2054, 6163, and 18490 nM. B. ParB binding to DNA fragments (from left to right) containing all 8 repeats of parS-L (60 bp), the four leftmost repeats (38 bp), the four rightmost repeats (38 bp), and the four leftmost repeats with flanking nonspecific DNA (60 bp). Protein concentrations are as in panel A. C. ParB binding to DNA fragments containing varying numbers of 8 bp repeats. From left to right, increasing the parS-L repeat from 1 to 4. Protein concentrations are as in panel A. D. The substrates used in panels B and C with the parS-L repeats identified.
RedRock ParB binds to a 75 bp substrate containing the eight 8bp repeat motifs of parS-L to form a single complex (Fig. 4B). ParB binds substrates containing the four leftmost or four rightmost repeats similarly, such that these are equivalent for recognition. ParB also binds to a 75 bp substrate containing just the four leftmost repeats and nonspecific DNA, and forms a complex with similar mobility to the 75 bp 8-motif DNA (Fig. 4B).
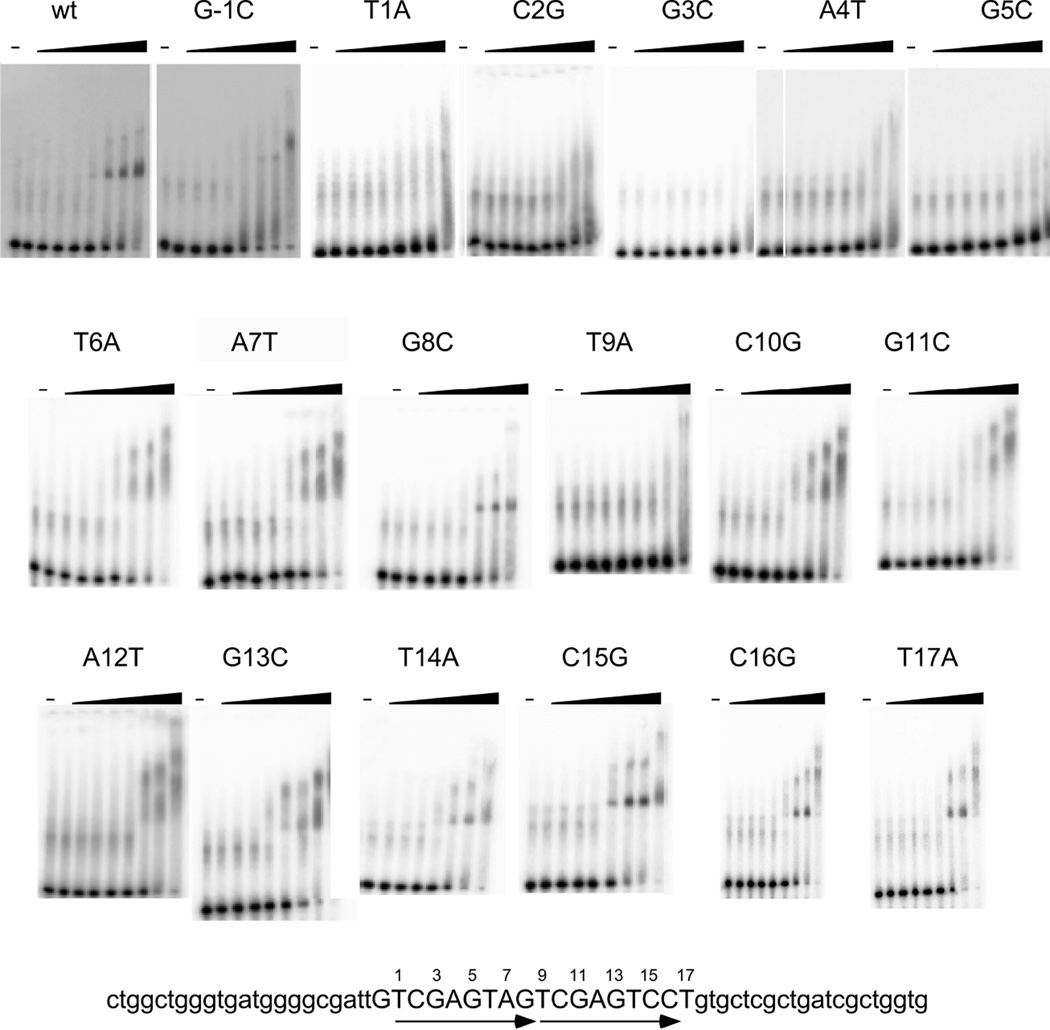
ParB binds poorly, if at all, to a DNA substrate containing only a single repeat unit 5’-TCGAGTAG, and binding is enhanced when two or more repeats are present (Fig. 4C). We note, however, that there are alternative circularly permuted versions of the 8 bp repeat motif, and that 5’-TCGAGTAG is only one particular configuration. It is likely that ParB binds primarily as a dimer based on RHH structures, and as reported for other Type Ib systems (Huang et al., 2011, Schreiter & Drennan, 2007); it could also recognize sites in inverted repeat orientation as described for ParG of TP228 (Zampini et al., 2009). To investigate ParB-parS interactions, we constructed several substrates based on the parent substrate with two repeat units (Fig. 5), each with single base substitutions. Substitutions at the G in the −1 position and at the T in the +17 position in Figure 5 both show little or no impact on ParB binding, suggesting that these are either not important for recognition, or are not elements of intact binding sites in the substrates. In contrast, the T’s at positions 1 and 9 both show reduced binding, such that this T position likely corresponds to the first and not the last position in the 8 bp repeat unit. Interestingly, substitutions at positions 2 – 5 and 10 – 13 have notably different impacts depending on whether they are in the first or the second 8 bp unit of the substrate. A plausible explanation is that ParB binds more weakly to the second than it does to the first of the repeats, such that binding is nearly undetectable in the 2 – 5 mutants, but is only modestly impacted by mutations at positions 10 – 13. We also note that ParB binding to the parent substrate as well as to the mutations at positions 8, 15, 16, and 17 forms complexes with discrete mobilities, whereas many of the other mutant substrates form complexes with indiscrete mobilities. In addition, although the T at position 6 is well conserved, substituting with an A at either position 9 or 14 has little impact on binding (Fig. 5). While it is not clear what causes these patterns, it has been shown that TP228’s ParG can bind to nonspecific sequence adjacent to a half-site (Carmelo et al., 2005) and RedRock’s ParB may be exhibiting a similar phenomenon, resulting in nucleoprotein complexes that are unstable during electrophoresis. Nonetheless, these observations are consistent with the repeated unit that is recognized by a ParB protomer being defined as 5’-TCGAGnnn.
Figure 5.
Mutational analysis of parS. The substrate for all EMSAs shown here is at the bottom of the figure. It contains the 18 bp predicted parS site and is flanked by nonspecific DNA. This sequence contains two copies of a tandem repeated sequence (shown by arrows). ParB binds to this substrate, but when single base mutations are made and binding is assayed, it is clear that the first repeat sequence is more important for binding. ParB protein concentrations are the same as in Figure 4.
Specificities of ParB-parS binding
The 42 partitioning systems identified in actinobacteriophages span considerable sequence diversity (Fig. 2). In general, the ParA and ParB proteins of each system appear to be co-evolving, consistent with a model in which the two proteins interact directly, as described for other partitioning systems (Baxter & Funnell, 2014). We predict that the parS sites similarly co-evolve and the entire systems are likely to be under selection for diversity to avoid incompatibility, as described for plasmid systems (Baxter & Funnell, 2014). We note for example that mycobacteriophages Pioneer and Alma are generally closely related (both are grouped in Subcluster A9; Fig. S6) but the partitioning cassettes are more highly diverged than the rest of the genomes, and the ParA and ParB proteins share only 60% and 44% identity, respectively.
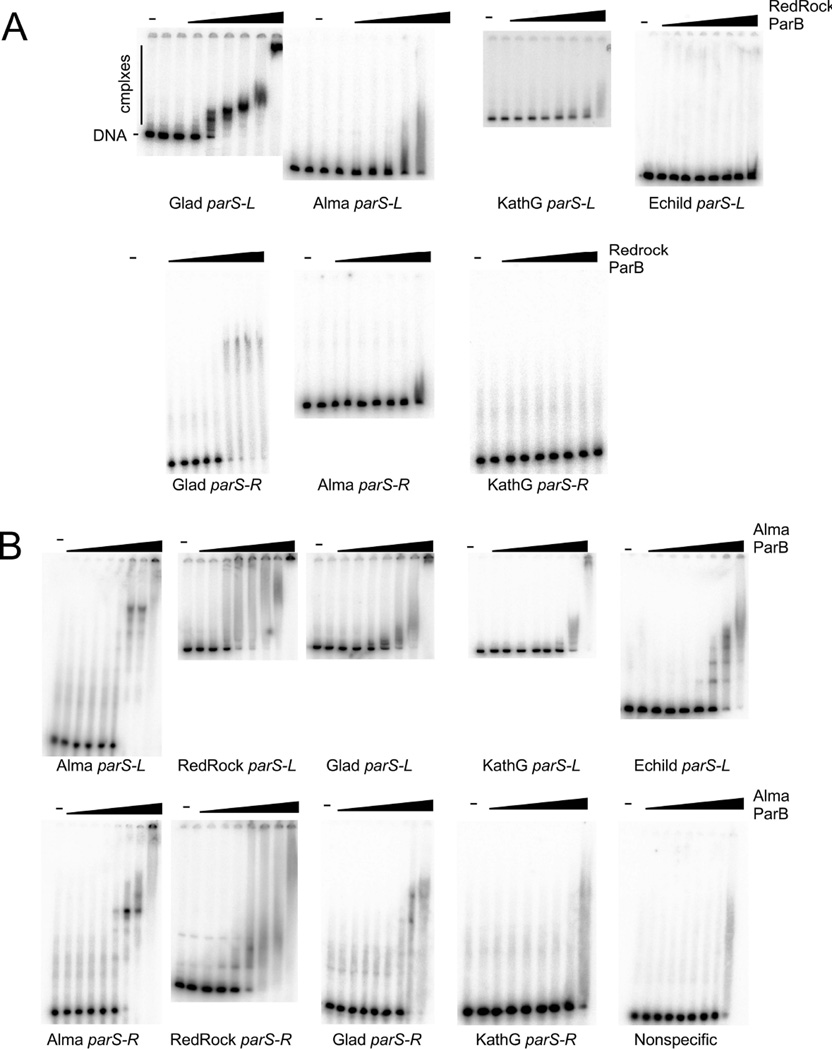
To further explore the specificity of ParB-parS interactions, we examined the binding of RedRock ParB to the parS sites of a variety of other phages. First we asked whether RedRock ParB binds to Gladiator parS sites, because although the ParB proteins share only 51% identity, the consensus repeat motifs are similar (Figs. 1, S7). RedRock ParB binds to both parS-L and parS-R with only modest reduction in affinity relative to its own sites, and forms slow moving indistinct complexes as seen for RedRock parS (Figs. 4, 6A). We also tested RedRock ParB binding to the parS sites of mycobacteriophages Alma and Echild, as well as Gordonia phage KatherineG. The ParB proteins of these phages are more distant relatives of RedRock ParB (45%, 17%, and 37% amino acid sequence identity; Fig. 2) and the parS sites have related but different 8 bp repeat consensus sequences (Fig. S7). We observed little or no RedRock ParB binding to any of these sites (Fig. 6A).
Figure 6.
Specificity of ParB binding to parSA. Binding of RedRock ParB to parS sites of mycobacteriophages Alma, Gladiator (Glad), and Echild, as well as Gordonia phage KatherineG (KathG). Gladiator, Alma, and KatherineG, all have predicted parS-L and parS-R sites flanking the parAB genes; a parS-L site is predicted upstream of Echild parA, but the sequences downstream of Echild parB are not obviously related to parS-R. Protein concentrations are as in Figure 4, panel A. B. Binding of Alma ParB to parS sites of mycobacteriophages Alma, RedRock, Gladiator (Glad), and Echild, as well as Gordonia phage KatherineG (KathG). Protein concentrations of Alma ParB from left to right are: 0, 7.8, 23.4, 70.2, 210, 632, 1896, 5690, and 17070 nM.
We also overexpressed and purified the ParB protein of phage Alma. Alma ParB binds to both Alma parS-L and parS-R sites and forms slowly migrating complexes. Somewhat surprisingly, Alma ParB is more promiscuous than RedRock ParB and is able to bind to both RedRock parS-L and parS-R sites (Figs. 6B, S7). Alma ParB also binds albeit with somewhat lower affinity to Gladiator parS sites (Fig. 6B, Fig. S7), and also to Echild parS-L despite the divergence of the motif consensus sequences, but not to KatherineG parS-R (Fig. S7). Similar to the analysis of RedRock parS individual repeat positions, these observations show that specificity of the partitioning systems is complex, and that although there is substantial sequence variation, ParB proteins can range broadly in their specificities.
Partitioning-mediated prophage incompatibility
To test whether par-containing prophages exhibit incompatibility, we identified pairs of mycobacteriophages containing par cassettes in which the immunity repressors and their operator/stoperator binding sites are sufficiently different that they are heteroimmune. As a control, we used phages Bxb1 and RedRock, which are heteroimmune, but unlike RedRock, Bxb1 has a canonical integration cassette (Kim et al., 2003). Starting with a RedRock lysogen, we attempted to construct double lysogens by superinfection with Bxb1, Alma, or Pioneer, followed by propagation through several rounds of purification and growth in liquid culture, and testing for superinfection immunity to each of the phages throughout the experiment (see Materials and Methods and Fig. S8 for details). For the RedRock/Bxb1 infections, we successfully generated double lysogens, confirming they are fully compatible. In contrast, for the RedRock/Alma and RedRock/Pioneer pairs, we identified double lysogens after the initial superinfection, but were unable to propagate the strains through purification. Nearly all colonies tested resulted in a single lysogen of either Alma or Pioneer in which the RedRock prophage had been displaced. These pairs of par-containing phages thus clearly exhibit incompatibility. This is consistent with the non-reciprocal parS recognition of the Alma and RedRock ParB proteins (Fig. 6), and we predict that Pioneer ParB may behave similarly, although we cannot discount that incompatibility results from the prophage replication systems, rather than partitioning per se. In similar experiments where either RedRock or Alma superinfected a Gladiator lysogen, Alma failed to displace Gladiator, RedRock displaced Gladiator 40% of the time to generate single RedRock lysogens, and no double lysogens were obtained. Alma and L5 (an integrating phage) co-infections of wild type M. smegmatis successfully generated double lysogens, as seen with RedRock and Bxb1.
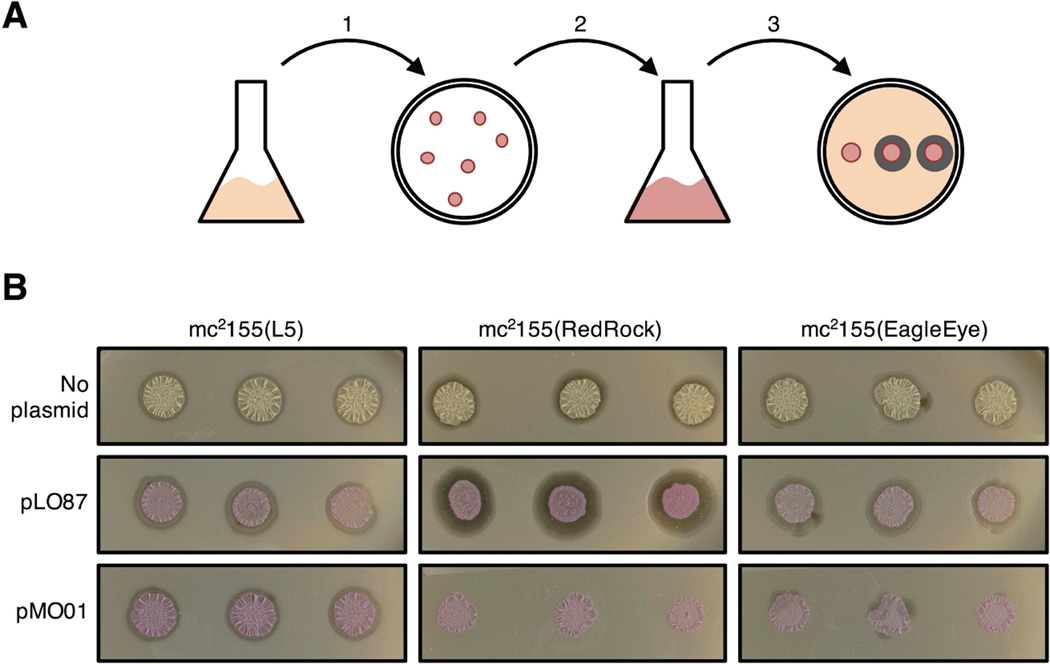
To determine whether the par systems specifically contribute to incompatibility, we tested the compatibility of plasmid pMO01 – which contains the parABS cassette but not the putative RNA-encoding replication functions to its left (see above) – with a RedRock lysogen. Plasmid pMO01 was introduced into RedRock lysogenic cells by electroporation, and kanamycin resistant transformants were selected (Fig. 7A). Three independent transformants were propagated and tested for lysogeny by a standard spontaneous phage release assay. All transformants were shown to have lost the RedRock prophage (Fig. 7B). In contrast, control transformants carrying pLO87 DNA (which lacks parABS) all maintained RedRock lysogeny, and lysogeny was maintained in L5 lysogens that carry an integrated prophage. Plasmid pMO01 thus displays incompatibility specifically with the RedRock prophage. Phage release was also observed by pMO01 transformants of the EagleEye lysogen suggesting partial incompatibility between the RedRock and EagleEye par systems. However, we note that this may be exacerbated by the relatively high plasmid copy number [∼20 copies/chromosome (Huff et al., 2010)] relative to the EagleEye prophage, which we assume has a similarly copy number of ∼2.4 copies/chromosome as RedRock.
Figure 7.
Phage RedRock par-mediated incompatibility. A. Scheme to test for parABS-mediated incompatibility. Plasmids pMO01 (carrying RedRock parABS and mCherry), pLO87 (lacking parABS) were electroporated into M. smegmatis lysogens of L5, RedRock, or EagleEye. and kanamycin-resistant transformants selected (step 1). Three individual red transformants (or un-transformed control colonies) were propagated in liquid selective media (step 2), and spotted onto a lawn of wild type M. smegmatis to test for phage release (seen as rings of infection surrounding spotted cultures) indicating lysogeny (step 3). B. Plasmid MO01 displaces the RedRock prophage. Three independent transformants carrying pMO01 show no spontaneous phage release indicating prophage loss, which is dependent on parABS (pLO87 transformants of RedRock lysogen all show phage release). All transformants and (untransformed cells) of an L5 lysogen show prophage maintenance and compatibility. Spontaneous phage release is reduced in pMO01 transformants of the EagleEye lysogen suggesting partial incompatibility between the RedRock parABS in pMO01 and the EagleEye prophage.
Discussion
Phage-encoded partitioning systems are not uncommon among temperate phages of the actinobacteria although all are found within a group of related phages defined as Cluster A [the exception is pZL12 which was identified as a plasmid in Streptomyces and is unrelated to the other phages (Zhong et al., 2010)]. Approximately 20% of the Cluster A phages have partitioning cassettes and the remainder have integration cassettes, distributed between tyrosine- and serine-integrase systems. These par systems considerably expand the number of previously described phage-encoded partitioning systems. All of the Cluster A phage par components belong to the Type Ib system, which have not been previously identified in phage genomes.
Phage RedRock forms stable lysogens in which the prophage replicates extrachromosomally at an average copy number of 2.4 copies/chromosome, with the parABS system promoting prophage maintenance. RedRock ParB binds to two parS sites flanking the parAB genes and plays a regulatory role in addition to its presumed role in prophage segregation. Upon infection, RNAseq analysis shows strong unregulated expression of parAB from a promoter located between gene 36 and parA, which is then down regulated in lysogeny through the binding of ParB to parS-L. ParB could also play a role in terminating transcription of parAB by its binding to parS-R. The inability to construct a deletion derivative of parB can be explained by the toxic consequences of parA overexpression, although the inability to construct a lytically-proficient deletion of parA is more puzzling. It is possible that parB expression is inhibitory to lytic growth unless parA is also expressed.
In general, the actinobacteriophage parS sites are composed of 5–10 copies of tandemly repeated 8 bp motifs. We propose that one ParB protomer binds to each of these motifs, but that occupancy may be stimulated by several factors, similar to other Type Ib systems, such as those described in pSM19035 (Dmowski et al., 2006; Schreiter & Drennan, 2007), TP228 (Carmelo et al., 2005; Zampini et al., 2009), and pCXC100 (Huang et al., 2011). For instance, repeats that vary in sequence can be bound with varying affinities, the repeats can occur in multiple orientations that can impact affinity, and binding of tandem sites can be cooperative. The N-terminal tail of the CBP, which tends to be unstructured, has been shown to enhance binding stability through transient interactions with the folded C-terminal region. Taken together, while the current study investigates the general binding behaviors of several actinobacteriophage partitioning systems, further investigations are needed to elucidate the binding interactions in greater detail. Additionally, we note that the phage-encoded parS sites are distinctly different from the palindromic sites that form the host parS site in Mycobacterium tuberculosis and M. smegmatis (5’-GTTTCACGTGAAAC 3’) or parS in Bacillus subtilis (5’-TGTTCCACGTGAAACT 3’) (Lin & Grossman, 1998, Jakimowicz et al., 2007).
The partitioning cassettes span considerable diversity, and we note that the partitioning cassettes of phages Echild and 40AC are noticeably different than other Cluster A phages (Fig. 2). Their parA genes can be readily identified (Fig. 2A), but their parB genes are more divergent (Fig. 2B). Structural domain analysis shows that Echild’s ParB has noticeably fewer Type Ib domain hits than other Cluster A phages, and 40AC’s ParB has no predicted domains of any partitioning type. Additionally, the tandem repeat program, etandem (Rice et al., 2000), predicts parS-L sites for Echild and 40AC, but no parS-R for 40AC, and an Echild parS-R that is distinct from its parS-L and thus may not represent a ParB recognition sequence (Fig. S2). A prior study failed to isolate 40AC stable lysogens (Stella et al., 2013), suggesting the predicted ParB may not be functional. We have successfully isolated an Echild lysogen that exhibits superinfection immunity (data not shown), although lysogeny is not stably maintained and non-lysogenic derivatives accumulate at high frequency (data not shown). The KA/KS ratios indicate that parA and parB are under selection, and is thus plausible that Echild and 40AC have a bona fide partitioning cassette, but that does not function efficiently in M. smegmatis.
The par actinobacteriophages are broadly distributed within Cluster A, and are found in over half of the component subclusters. The ParA and ParB sequences themselves span considerable sequence diversity, illustrated by the most distantly related ParB proteins (e.g. Echild and RedRock) sharing only 17% amino sequence identity. Because lysogeny could not be established by two different extrachromosomal phages in the same cell, it is likely that there is selective pressure to diversify the partitioning systems in order to avoid incompatibility, as seen in plasmid systems (Radnedge et al., 1996, Sergueev et al., 2005, Hyland et al., 2014). This could occur at two levels: by diversification of the ParB and the parS sites that they recognize, or through variation in the interactions between the interacting components of ParA and ParB. It seems likely that both play important roles, and we show here that the ParB proteins can distinguish between parS sites of distantly related phages. The pairs of phages that could be readily tested for compatibility (RedRock and Alma, RedRock and Pioneer) do show marked incompatibility, and we demonstrated par-mediated incompatibility using a RedRock par-containing recombinant plasmid and a RedRock prophage.
The par-containing actinobacteriophages replicate extrachromosomally as low copy number prophages, and thus must have an origin of replication that is absent from the integrating phages. The most obvious location for the origin is adjacent to the parABS cassette with which it can co-evolve, and this is consistent with genome comparisons of pairs of closely-related phages encoding integration and partitioning cassettes. However, none of the extrachromosomal phages encode RepA or other plasmid-like replication proteins, but RedRock as well as three other lysogens express an RNA implicated in prophage replication. The nucleotide sequences of the ∼300 bp regions to the left of parABS are highly varied, and although folded RNA structures can be predicted, there is little in common to these phages that reveals the functional components. Efforts to clone RedRock DNA fragments that promote autonomous replication have thus far been unsuccessful.
The actinobacteriophage partitioning cassettes have three potential utilities for bacterial genetics. First, the pAL5000-derived plasmid vectors commonly used in mycobacterial genetics are often poorly maintained in the absence of selection, and the parABS cassettes of RedRock and related phages can be used to confer plasmid maintenance (Table 2). There are a variety of particular applications where this may be useful, but notably where recombinants are tested in animal model systems or as live vaccine candidates, and where expression levels derived from the use of multicopy plasmids is desirable. Second, although the origin of replication has yet to be precisely determined, the ori-par functions have the potential to provide a new series of low copy number plasmid vectors that are fully compatible with extant vector systems. Lastly, the ability of ParB to bind to parS in multiple copies offers the possibility that ParB-GFP fusion proteins could be used to geographically identify chromosomal segments in the cell by introduction of parS sites into the host genome. This approach has worked well in E. coli using P1 ParB (Erdmann et al., 1999), and the variety of actinobacteriophage partitioning cassettes provide multiple systems that could be used in combination and are anticipated to be well-expressed. The phylogenetic and structural domain analyses of the partitioning systems may enhance the development of similar tools in other bacterial hosts including Leptospira [exploiting lcp1, lcp2, and lcp3, (Zhu et al., 2015)], Streptomyces [plasmids pZL12, pSLE1, and pSLE2 (Gomez-Escribano et al., 2015, Zhong et al., 2010)], and in Vibrio [using ΦHAP-1, pVv01, and Vp58.5 (Mobberley et al., 2008, Hammerl et al., 2014, Zabala et al., 2009)].
Materials and Methods
Bacteria and plasmids
Liquid cultures of M. smegmatis mc2155 and the lysogenic derivatives were grown in Middlebrook 7H9 at 37°C with shaking. Phage infection assays on solid media were performed using exponentially growing cultures plated with a soft top agar layer on Middlebrook 7H10 plates. Plasmids used in this study are listed in Table S2. Plasmid pMO01 is a derivative of pLO87 (Oldfield & Hatfull, 2014) an extrachromosomal shuttle vector with Phsp60 driving mCherry expression, with the RedRock par cassette (coordinates 27,720 – 28,898) cloned downstream of mCherry. Several derivatives of pMO01 were constructed, including pMO02 and pMO03, which contain translational termination codons early in the parA and parB open reading frames, respectively. Plasmids pMO04 and pMO05 are derivatives of pMO01 with deletions of parS-L and parS-R, respectively. Plasmid pMO20 and pMO21 are derivatives of pLO87 and pMO01, respectively, in which the hsp60-mCherry cassette has been removed. Plasmid pMO16 and pMO17 are derivatives of the extrachromosomal shuttle vector pLO106 (Villanueva et al., 2015), which contains phage BPs p6 driving mCherry expression. A 178 bp fragment of RedRock (coordinates 27,720 – 27,897) was cloned in the forward orientation (pMO16) and reverse orientation (pMO17) downstream of p6. Plasmid pMO15 is a derivative of the integration-proficient vector pJV39 carrying Phsp60 fused to RedRock parB. Plasmids pJC04 and pJC05 are derivatives of vector pLAM12 (Marinelli et al., 2008) containing RedRock parA and parB, respectively.
Plasmid Maintenance Assays
M. smegmatis transformants carrying various plasmids were grown in liquid culture with antibiotic selection for plasmid maintenance for approximately 24 hours or until saturated. Cultures were then diluted 1:10,000 into antibiotic-free media and re-grown to saturation (approximately 13 generations), and subsequent rounds of dilution were used to increase the number of rounds of unselected growth. Cultures were then plated onto solid media, and colonies were scored for plasmid maintenance (red) or for plasmid loss (white). Statistical significance of changes in plasmid retention was computed using two-sample two-tailed t-test of the retention level from three independent replicates.
Promoter Strength Assays
Individual colonies of M. smegmatis transformants carrying various plasmids were grown in liquid media with shaking, at 37°C, for 24–48 hrs. Readings of mCherry fluorescence were taken as described previously (Oldfield & Hatfull, 2014), except that the measurement of optical density was taken at 600 nm in a Beckman Coulter DU530. Fluorescence units are reported as the amount of fluorescence per area per OD600.
DNA sequencing and RNAseq
DNA from a 2 ml sample of the RedRock or L5 lysogen was extracted from late logarithmically growing cells (OD600 approximately 1.0) using the Wizard kit (Promega) according to the manufacturers’ instructions. DNA was quantified using Qubit and libraries were prepped using the TruSeq Library Kit (Illumina) according to the manufacturers’ instructions. The completed libraries were run on an Illumina MiSeq and data was evaluated using CLC Genomics software. For RNAseq, total RNA was isolated from M. smegmatis cultures in exponential growth, as well as 30 min and 2.5 hrs after infection with RedRock at a multiplicity of infection of three. DNA was removed using the DNA-free kit (Ambion) and rRNA was depleted using the Ribo-Zero kit (Illumina). Libraries were prepared using a TruSeq Stranded RNAseq kit (Illumina) and run on an Illumina MiSeq: one lane for each RedRock sample, and one multiplexed lane for wild type M. smegmatis, Alma, Pioneer, and EagleEye lysogens. The fastq reads were analyzed for overall quality using FastQC (Andrews, S. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), trimmed at the 5’ and 3’ ends with cutadapt (Martin, 2011) using a quality score threshold of 30, and then mapped simultaneously to the M. smegmatis and RedRock genomes with Bowtie2 (Langmead & Salzberg, 2012). SAMtools (Li et al., 2009) and BEDtools (Quinlan & Hall, 2010) were used to process reads that aligned to exactly one locus (as computed by Bowtie2) and calculate strand-specific genome coverage. Integrative Genomics Viewer (Thorvaldsdottir et al., 2013) was used to visualize the data. The RNAseq data set is deposited in the Gene Expression Omnibus (GEO) with accession number GSE79010.
Purification of ParB
The parB gene was PCR amplified from RedRock or Alma and inserted into plasmid pET28a (Novagen) such as to include a His6 tag at the C-termini of each protein. After verification by sequencing, the resulting plasmids (pJC02 and pWN01, respectively) were transformed into BL21* (DE3)pLysS cells and grown to an OD600 of 0.5 at 37°C in LB. His-ParB expression was induced by the addition of 1 mM IPTG at 37°C for 3 hrs. Cells were pelleted, resuspended in 5 ml/g of lysis buffer (50 mM Tris-HCL (pH8), 300 mM NaCl and 5% glycerol), and sonicated. The sonicated cells were centrifuged and the cleared lysate was applied to a nickel column using Ni-NTA column (Qiagen). The column was washed with lysis buffer, 10 mM, and 50 mM imidazole and the proteins eluted with 150 mM imidazole. Fractions were collected and dialyzed in lysis buffer containing 30–50% glycerol overnight at 4°C.
Electrophoretic Mobility Shift Assays
DNA substrates were prepared using either gel-extracted PCR substrates or annealed synthetic oligonucleotides (IDT & Invitrogen) (Table S3). Double stranded DNA substrates were 5’-end radiolabeled using ATP, [γ-32P] with T4 polynucleotide kinase (Roche) at 37°C for 30 min and cleaned up using G-50 sephadex columns. 5–10 ng of radiolabeled substrates were incubated at room temperature for 30 min with indicated concentrations of ParB in a buffer containing 20 mM Tris pH 7.5, 10 mM EDTA, 25 mM NaCl, 10 mM spermadine, 1 mM DTT and 1 µg calf thymus DNA in a total volume of 10 µl. The DNA-protein samples were then resolved on a 5% native polyacrylamide gel run at 4°C. The gel was dried and exposed to a phosphorimaging plate, then scanned using a Fuji 5,000 Phosphorimager. Dissociation constants (Kd) were calculated as the protein concentration at which 50% of the input DNA was protein-bound.
Phylogenetic analysis of partitioning cassettes
A database (Actinobacteriophage_706) was constructed using the program Phamerator as described previously (Cresawn et al., 2011, Pope et al., 2015). The database contains 706 genomes of phages infecting Actinobacterial hosts coding for 70,341 genes that are grouped according to their sequence similarity into 9,523 phamilies (phams). This database contains 42 genomes with predicted partitioning cassettes. The protein sequences for 41 other putative and characterized NTPase and CBP genes from partitioning cassettes were identified from the literature and retrieved from NCBI (see Table S1). This non-exhaustive list of par cassettes represents cassettes from each partitioning type [Ia, Ib, II, III, or unknown, depending on how cassettes have been previously categorized in (Gerdes et al., 2000, Ebersbach & Gerdes, 2005, Schumacher, 2012)], from various replicon types (chromosomal, plasmid, or phage), and from various bacterial host genera. In some replicons where there was a previously predicted NTPase but no accompanying CBP, there nevertheless tended to be an ORF immediately downstream in an apparent operon with parA, and these sequences were used as a potential CBP in the phylogeny. Protein sequences were aligned in Seaview (Gouy et al., 2010) using ClustalO and a phylogeny was created using the BioNJ algorithm with observed distances. A bootstrap analysis was performed with 100 replicates. Trees generated using other methods were comparable. Phylogenies were visualized and appended with genomic data using Evolview (Zhang et al., 2012).
Prediction of partitioning types
HHpred (Soding, 2005) was used to predict the types of partitioning system for the cassettes used in this study. First, each partitioning protein was analyzed using HHpred with the pdb70_15Feb16 database and with default settings as of February 22, 2016. The top 100 domain hits per gene that exceeded a homologous relationship probability (as computed by the program) cutoff of 90% were retained. All structural domain hits returned for the group of partitioning cassettes that have been previously categorized as Type Ia, Ib, II, or III (see Table S1) were assigned a partitioning type category as follows. If the domain was found in one or more genes from only one partitioning cassette type category, the domain was assigned the same partitioning type category, reflecting that in this analysis the domain is only found in genes of that particular partitioning type. If the domain was present in genes from more than one partitioning type, it was categorized as “nonspecific.” Next, the frequency of each domain category was calculated for partitioning genes in the entire set of 83 partitioning cassettes. Finally, stacked bar graphs of these frequencies were generated for each gene to provide a qualitative measure of how similar each partitioning gene is to previously characterized partitioning genes. The analysis was done separately for parA and parB genes.
ParA and ParB coevolution analysis
The rate of evolution of the actinobacteriophage parA and parB genes were analyzed as follows. Of the 42 phages in the Actinobacteriophage_706 database, Echild, 40AC, and pZL12 were not used; their ParA and/or ParB genes did not group with the rest of the actinobacteriophages in the protein sequence phylogenies, suggesting they were too distantly related for meaningful comparison. Of the remaining 39 phages, redundant DNA sequences of each partitioning gene were removed, reducing the list to a total of 27 phages that contain unique parA and parB sequences available for analysis (see Table S1); parA and parB genes were processed separately. DNA sequences were aligned at the codon level using webPRANK (Loytynoja & Goldman, 2010) and processed using the kaks tool in the ‘seqinr’ R package to compute the pairwise KA, KS, and KA/KS values. The KA/KS ratios for all pairwise comparisons that had a KS < 2.0 were retained, and a scatter plot of the matching parA and parB ratios was generated.
Supplementary Material
Acknowledgments
We thank Carlos Guerrero for excellent technical assistance, Valerie Villanueva and Lauren Oldfield for experimental assistance and student supervision, and Charles Bowman for DNA sequencing assistance. This was work was supported by grants from the National Institutes of Health (GM116884) and Howard Hughes Medical Institute (54308198) to GFH, and National Science Foundation pre-doctoral fellowship to TNM (1247842). No authors have conflicts of interest. Author contributions: RMD, TNM, WLN, JCCRR, MRO, RER, DJS, DAR, designed, performed and interpreted experiments; GFH designed and interpreted experiments; RMD, TNM, and GFH wrote the manuscript.
References
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Baxter JC, Funnell BE. Plasmid Partition Mechanisms. Microbiol Spectr. 2014:2. doi: 10.1128/microbiolspec.PLAS-0023-2014. [DOI] [PubMed] [Google Scholar]
- Bourhy P, Frangeul L, Couve E, Glaser P, Saint Girons I, Picardeau M. Complete nucleotide sequence of the LE1 prophage from the spirochete Leptospira biflexa and characterization of its replication and partition functions. J Bacteriol. 2005;187:3931–3940. doi: 10.1128/JB.187.12.3931-3940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard GW, Oldfield LM, Villanueva VM, Lunt BL, Shine EE, Hatfull GF. Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches. Mol Cell. 2013;49:237–248. doi: 10.1016/j.molcel.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KL, Sarkis GJ, Wadsworth C, Hatfull GF. Transcriptional silencing by the mycobacteriophage L5 repressor. Embo J. 1997;16:5914–5921. doi: 10.1093/emboj/16.19.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmelo E, Barilla D, Golovanov AP, Lian LY, Derome A, Hayes F. The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J Biol Chem. 2005;280:28683–28691. doi: 10.1074/jbc.M501173200. [DOI] [PubMed] [Google Scholar]
- Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol Microbiol. 2013;88:577–589. doi: 10.1111/mmi.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmowski M, Sitkiewicz I, Ceglowski P. Characterization of a novel partition system encoded by the delta and omega genes from the streptococcal plasmid pSM19035. J Bacteriol. 2006;188:4362–4372. doi: 10.1128/JB.01922-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Egan JB. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly-Wu MK, Jacobs WR, Jr, Hatfull GF. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- Erdmann N, Petroff T, Funnell BE. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc Natl Acad Sci U S A. 1999;96:14905–14910. doi: 10.1073/pnas.96.26.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill TJ, Barilla D, Hayes F. Protein diversity confers specificity in plasmid segregation. J Bacteriol. 2005;187:2651–2661. doi: 10.1128/JB.187.8.2651-2661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Castro JF, Razmilic V, Chandra G, Andrews B, Asenjo JA, Bibb MJ. The Streptomyces leeuwenhoekii genome: de novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genomics. 2015;16:485. doi: 10.1186/s12864-015-1652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Halleran A, Clamons S, Saha M. Transcriptomic Characterization of an Infection of Mycobacterium smegmatis by the Cluster A4 Mycobacteriophage Kampy. PLoS One. 2015;10:e0141100. doi: 10.1371/journal.pone.0141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl JA, Klevanskaa K, Strauch E, Hertwig S. Complete Nucleotide Sequence of pVv01, a P1-Like Plasmid Prophage of Vibrio vulnificus. Genome announcements. 2014:2. doi: 10.1128/genomeA.00135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF. Mycobacteriophages: genes and genomes. Annu Rev Microbiol. 2010;64:331–356. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. The secret lives of mycobacteriophages. Adv Virus Res. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Molecular Genetics of Mycobacteriophages. Microbiology Spectrum. 2014;2:1–36. [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O’Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW. Comparative Genomic Analysis of 60 Mycobacteriophage Genomes: Genome Clustering, Gene Acquisition, and Gene Size. J Mol Biol. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- Hertwig S, Klein I, Schmidt V, Beck S, Hammerl JA, Appel B. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J Mol Biol. 2003;331:605–622. doi: 10.1016/s0022-2836(03)00763-0. [DOI] [PubMed] [Google Scholar]
- Huang L, Yin P, Zhu X, Zhang Y, Ye K. Crystal structure and centromere binding of the plasmid segregation protein ParB from pCXC100. Nucleic Acids Res. 2011;39:2954–2968. doi: 10.1093/nar/gkq915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J, Czyz A, Landick R, Niederweis M. Taking phage integration to the next level as a genetic tool for mycobacteria. Gene. 2010;468:8–19. doi: 10.1016/j.gene.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Wallace EW, Murray AW. A model for the evolution of biological specificity: a cross-reacting DNA-binding protein causes plasmid incompatibility. J Bacteriol. 2014;196:3002–3011. doi: 10.1128/JB.01811-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Hatfull GF. Transcriptional regulation and immunity in mycobacteriophage Bxb1. Mol Microbiol. 2000;38:971–985. doi: 10.1046/j.1365-2958.2000.02184.x. [DOI] [PubMed] [Google Scholar]
- Jakimowicz D, Brzostek A, Rumijowska-Galewicz A, Zydek P, Dolzblasz A, Smulczyk-Krawczyszyn A, Zimniak T, Wojtasz L, Zawilak-Pawlik A, Kois A, Dziadek J, Zakrzewska-Czerwinska J. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology. 2007;153:4050–4060. doi: 10.1099/mic.0.2007/011619-0. [DOI] [PubMed] [Google Scholar]
- Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol. 2003;50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R S. Genome Project Data Processing. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lobocka MB, Rose DJ, Plunkett G, 3rd, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. Genome of bacteriophage P1. J Bacteriol. 2004;186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobberley JM, Authement RN, Segall AM, Paul JH. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol. 2008;82:6618–6630. doi: 10.1128/JVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution. J Mol Biol. 2001;314:789–796. doi: 10.1006/jmbi.2001.5157. [DOI] [PubMed] [Google Scholar]
- Oldfield LM, Hatfull GF. Mutational analysis of the mycobacteriophage BPs promoter PR reveals context-dependent sequences for mycobacterial gene expression. J Bacteriol. 2014;196:3589–3597. doi: 10.1128/JB.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Pradella S. Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ Microbiol. 2009;11:2627–2638. doi: 10.1111/j.1462-2920.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF G. Science Education Alliance Phage Hunters Advancing, S. Evolutionary, R. Phage Hunters Integrating, Education, & C. Mycobacterial Genetics. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife. 2015;4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD, Anderson AG, Ang AAS, Ares M, Jr, Barber AJ, Barker LP, Barrett JM, Barshop WD, Bauerle CM, Bayles IM, Belfield KL, Best AA, Borjon A, Jr, Bowman CA, Boyer CA, Bradley KW, Bradley VA, Broadway LN, Budwal K, Busby KN, Campbell IW, Campbell AM, Carey A, Caruso SM, Chew RD, Cockburn CL, Cohen LB, Corajod JM, Cresawn SG, Davis KR, Deng L, Denver DR, Dixon BR, Ekram S, Elgin SCR, Engelsen AE, English BEV, Erb ML, Estrada C, Filliger LZ, Findley AM, Forbes L, Forsyth MH, Fox TM, Fritz MJ, Garcia R, George ZD, Georges AE, Gissendanner CR, Goff S, Goldstein R, Gordon KC, Green RD, Guerra SL, Guiney-Olsen KR, Guiza BG, Haghighat L, Hagopian GV, Harmon CJ, Harmson JS, Hartzog GA, Harvey SE, He S, He KJ, Healy KE, Higinbotham ER, Hildebrandt EN, Ho JH, Hogan GM, Hohenstein VG, Holz NA, Huang VJ, Hufford EL, Hynes PM, Jackson AS, Jansen EC, Jarvik J, Jasinto PG, Jordan TC, Kasza T, Katelyn MA, Kelsey JS, Kerrigan LA, Khaw D, Kim J, Knutter JZ, Ko C-C, Larkin GV, Laroche JR, Latif A, et al. Expanding the Diversity of Mycobacteriophages: Insights into Genome Architecture and Evolution. PLoS ONE. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P, Chandler M, Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977;152:71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radnedge L, Davis MA, Austin SJ. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 1996;15:1155–1162. [PMC free article] [PubMed] [Google Scholar]
- Rauzier J, Moniz-Pereira J, Gicquel-Sanzey B. Complete nucleotide sequence of pAL5000, a plasmid from Mycobacterium fortuitum. Gene. 1988;71:315–321. doi: 10.1016/0378-1119(88)90048-0. [DOI] [PubMed] [Google Scholar]
- Ravin N, Lane D. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J Bacteriol. 1999;181:6898–6906. doi: 10.1128/jb.181.22.6898-6906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV. N15: the linear phage-plasmid. Plasmid. 2011;65:102–109. doi: 10.1016/j.plasmid.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol. 2000;299:53–73. doi: 10.1006/jmbi.2000.3731. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Nicolas E, Sherratt DJ. Chromosome replication and segregation in bacteria. Annu Rev Genet. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- Schumacher MA. Bacterial plasmid partition machinery: a minimalist approach to survival. Curr Opin Struct Biol. 2012;22:72–79. doi: 10.1016/j.sbi.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergueev K, Dabrazhynetskaya A, Austin S. Plasmid partition system of the P1par family from the pWR100 virulence plasmid of Shigella flexneri. J Bacteriol. 2005;187:3369–3373. doi: 10.1128/JB.187.10.3369-3373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Stella EJ, Franceschelli JJ, Tasselli SE, Morbidoni HR. Analysis of novel mycobacteriophages indicates the existence of different strategies for phage inheritance in mycobacteria. PLoS One. 2013;8:e56384. doi: 10.1371/journal.pone.0056384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Austin S. The maintenance of the P1 plasmid prophage. Plasmid. 1981;5:20–31. doi: 10.1016/0147-619x(81)90075-5. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva VM, Oldfield LM, Hatfull GF. An Unusual Phage Repressor Encoded by Mycobacteriophage BPs. PLoS One. 2015;10:e0137187. doi: 10.1371/journal.pone.0137187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala B, Hammerl JA, Espejo RT, Hertwig S. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J Virol. 2009;83:9313–9320. doi: 10.1128/JVI.00672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampini M, Derome A, Bailey SE, Barilla D, Hayes F. Recruitment of the ParG segregation protein to different affinity DNA sites. J Bacteriol. 2009;191:3832–3841. doi: 10.1128/JB.01630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Cheng Q, Tian X, Zhao L, Qin Z. Characterization of the replication, transfer, and plasmid/lytic phage cycle of the Streptomyces plasmid-phage pZL12. J Bacteriol. 2010;192:3747–3754. doi: 10.1128/JB.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wang J, Zhu Y, Tang B, Zhang Y, He P, Zhang Y, Liu B, Guo X, Zhao G, Qin J. Identification of three extra-chromosomal replicons in Leptospira pathogenic strain and development of new shuttle vectors. BMC Genomics. 2015;16:90. doi: 10.1186/s12864-015-1321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.