Abstract
Tetraspanins are a large superfamily of glycoproteins, which are engaged in a wide range of specific molecular interactions by forming tetraspanin-enriched microdomains. Tetraspanin 9 (Tspan9) is a previously poorly studied tetraspanin gene, which was predominantly identified as an amplified gene in serous Fallopian tube carcinoma. However, the expression and role of Tspan9 in gastric cancer have yet to be fully elucidated. The aim of the present study was to evaluate the expression and clinical significance of Tspan9 in gastric cancer. In the present study, 105 gastric cancer tissue samples and corresponding adjacent normal samples were detected for Tspan9 expression using immunohistochemistry; furthermore, the association between clinical characteristics and Tspan9 expression was also analyzed. Tspan9 expression was determined to be significantly lower in cancer samples compared with those in corresponding adjacent normal samples (P<0.001). However, its increased levels of expression in cancer samples appeared to demonstrate a poorer prognostic tendency, which is associated with deeper tumor depth (P=0.025), more nodal involvement (P=0.01), more advanced tumor/lymph node/metastasis (TNM) stages (P=0.017) and a larger tumor size (P=0.026). Additionally, multivariate analysis demonstrated that high expression of Tspan9 was an independent prognostic factor for poor overall survival (P<0.01). These results suggested that Tspan9 may be used as a potential prognostic factor in gastric cancer.
Keywords: Tspan9, tetraspanins, gastric cancer, immunhistochemistry, survival analysis, prognosis
Introduction
Gastric cancer (GC) is a common cancer worldwide. Approximately 951,600 new patients were diagnosed with GC, and 723,100 deaths occurred, in 2012; over 25% of the new cases occurred in Eastern Asia (1). With the development of mass screening for GC, detecting malignant stomach lesions in the early stages has become a reality, and numerous patients with GC are now able to receive timely curative surgery or effective drug treatment (2). However, there remain a great number of patients who are diagnosed only in advanced stages, and thus have an unfavorable prognosis. Therefore, identifying and evaluating GC biomarkers is important for risk stratification, predicting survival and reducing mortality.
Tetraspanins, also called tetraspans or the transmembrane 4 superfamily (TM4SF), are a large family of evolutionarily conserved, four-transmembrane domain proteins (3). A burgeoning number of studies have discussed their involvement in tumor cell motility, invasiveness and the different aspects of the organization of tetraspanin microdomains (4–7). In 1999, seven new tetraspanins were identified from the EST database, named as new EST tetraspan NET-1 to NET-7 (8). The overexpression of NET-1 has been noted in human hepatocellular carcinoma, skin squamous cell carcinoma and cervical cancer (9–11). Upregulation of NET-6 has been demonstrated to be associated with a favorable outcome in prostate cancer (12). The previously poorly studied amplified gene, tetraspanin 9 (Tspan9; also known as NET-5) was predominantly found in serous Fallopian tube carcinoma (FTC) (13). The function of tetraspanins has not yet been fully elucidated, although their putative value as potential prognostic markers, including Tspan9, has been recognized.
To date, no studies have been published on the expression of Tspan9 in human GC tissue, and its clinical significance. In the present study, the expression of Tspan9 in GC tissues and adjacent non-cancerous tissues was detected by immunohistochemistry (IHC), and the association between the expression level of Tspan9 and various clinicopathological characteristics was also investigated.
Materials and methods
Specimen source and patient information
A total of 105 patients with GC who had undergone radical gastrectomy in the Affiliated Hospital of Medical College, Qingdao University between April 2009 and November 2010 were enrolled in the present study. The samples were fixed in 10% formaldehyde solution and embedded in paraffin. None of the patients had received any preoperative treatment.
IHC
The expression of Tspan9 was detected by IHC analysis with the SP method, using an SP-0023 Histostain-Plus kit (Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China). Rabbit monoclonal antibody against Tspan9 (rabbit anti-human; cat. no. ab106412) was purchased from Abcam, Inc. (Cambridge, MA, USA). The paraffin-embedded tissue blocks were sectioned in 3–4 µm slides and placed on glass slides (Jiangsu Huida Medical Instruments Co., Ltd., Jiangsu, China). For IHC staining, tissue sections were dewaxed in xylene and rehydrated with graded ethanol; subsequently, the slides were rinsed in phosphate-buffered saline (PBS). Antigen retrieval treatment was performed at 121°C for 2 min in 10 nmol/l sodium citrate buffer (pH 9.0). Endogenous peroxidase was then inactivated by incubating with 0.3% hydrogen peroxide in methanol for 20 min. Non-specific binding was blocked by treating the slides with normal goat serum fluid for 20 min at room temperature. Subsequently, the specimens were incubated with the anti-Tspan9 antibody (diluted 1:100 in PBS) at 37°C in an incubator for 1 h. The sections were then washed three times with PBS (3 min each wash), and antibody binding was detected using the Histostain-Plus kit. Subsequently, the sections were washed three times (3 min each wash) with PBS, followed by the addition of diaminobenzidine as a chromogen. Positive control tissues were used to optimize the antibodies, according to the manufacturer's protocol. In negative controls, the primary antibody was replaced with PBS. According to the manufacturer's recommended criteria, the samples were independently scored for intensity of Tspan9 staining by two pathologists without knowledge of the clinical outcome at ×100 and ×200 magnification. The staining intensity was divided into three grades (using a scoring system of 0–3): No staining (0), slightly yellowish (1), brownish yellow (2) and dark-brown (3). The multiplications of the two scores were graded as follows: 0 (0 score), 1+ (1–4 score), 2+ (5–8 score) and 3+ (9–12 score). Intensity scores of 0 or 1+ were designated as low expression, whereas those of 2+ or 3+ were designated as high expression.
Statistical analysis
The χ2 statistics test or Fisher's exact test were used to assess the correlation between Tspan9 expression and the clinicopathological characteristics. Overall survival curves were estimated using the Kaplan-Meier method, and the log-rank test was used to compare survival curves in the patient groups. A Cox proportional hazards model was used to examine the prognostic value of Tspan9 expression and other clinicopathological factors. Multivariate analysis was performed to determine the prognostic factors. P<0.05 was considered to indicate a statistically significant value. The SPSS 17.0 statistical software program (SPSS, Inc., Chicago, IL, USA) was used for the analyses.
Results
Clinicopathological characteristics
The clinicopathological characteristics are shown in Table I. Of the patients, 75 were men and 30 were women, with a median follow-up period of 43 months (range 1–64 months) and a mean age of 53 years (range 27–75 years). The classification was based on the seventh edition of the tumor-lymph node-metastasis (TNM) staging system published by Biondi and coworkers (14). There were 33 cases with an extent of T1, 21 cases with T2, 8 cases with T3 and 43 cases with T4. There were 40 classified cases in stage I, 25 cases in stage II, 40 cases in stage III and 0 cases in stage IV. The histological type was poorly differentiated for 78 cases, moderately differentiated for 21 cases, and well differentiated for 6 cases.
Table I.
Relationship between TSPAN9 expression and clinicopathological factors.
| Characteristics | No. | TSPAN9-lowa (n=55) | TSPAN9-higha (n=50) | P-value |
|---|---|---|---|---|
| Gender | 0.217 | |||
| Male | 75 | 41 | 34 | |
| Female | 30 | 14 | 16 | |
| Age (year) | 0.724 | |||
| <60 | 36 | 18 | 18 | |
| ≥60 | 69 | 37 | 32 | |
| Tumor differentiation | 0.034 | |||
| Well/moderate | 27 | 16 | 11 | |
| Poor/no differentiation | 78 | 28 | 50 | |
| Tumor size | 0.026 | |||
| <5 cm | 56 | 35 | 21 | |
| ≥5 cm | 49 | 20 | 29 | |
| T factor | 0.025 | |||
| T1/T2 | 54 | 34 | 20 | |
| T3/T4 | 51 | 21 | 30 | |
| Lymph node metastasis | 0.01 | |||
| Negative | 47 | 30 | 17 | |
| Positive | 58 | 25 | 33 | |
| TNM stage | 0.017 | |||
| I/II | 65 | 40 | 25 | |
| III | 40 | 15 | 25 | |
| Lymphovascular invasion | 0.312 | |||
| Negative | 88 | 48 | 40 | |
| Positive | 17 | 7 | 10 | |
| Lauren's classification | 0.199 | |||
| Intestinal | 51 | 30 | 21 | |
| Diffusion | 54 | 25 | 29 | |
| Her-2 expression | 0.04 | |||
| Negative | 96 | 52 | 44 | |
| Positive | 10 | 2 | 8 |
TSPAN-low and TSPAN-high represent the low and high expression groups of TSPAN9, respectively. Her-2, human epidermal growth factor receptor 2; TSPAN9, tetraspanin 9; TNM stage, tumor/lymph node/metastasis stage.
Tspan9 expression in GC and adjacent non-cancerous tissues
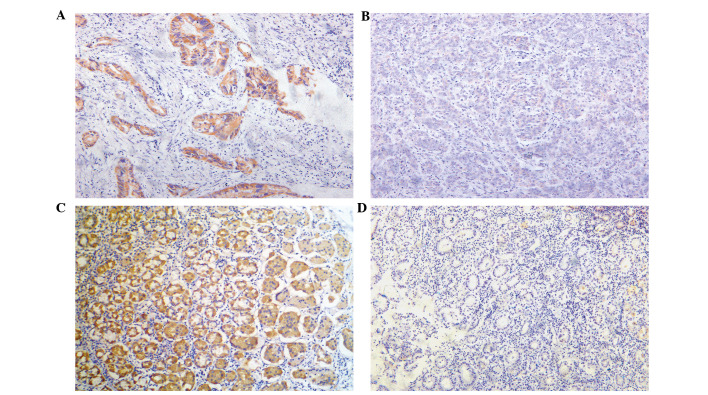
In the present study, sparse and partial Tspan9 expression was identified in GC tissue (Fig. 1A and B) and adjacent non-cancerous tissues (Fig. 1C and D). High expression levels of Tspan9 were identified in 50/105 GC tissues, which was significantly decreased compared with that in adjacent non-cancerous tissues (86/105; P<0.001), as shown in Table II.
Figure 1.
Tspan9 expression in GC and adjacent tissue identified by immunohistochemistry. Tspan9 was predominantly localized in the cytoplasm at different expression levels. (A) High expression of Tspan9 in GC tissues, (B) Low expression of Tspan9 in GC. (C) High expression of Tspan9 in adjacent normal tissue. (D) Negative expression of Tspan9 in adjacent tissue. (A-D) Magnification, ×200. GC, gastric cancer; Tspan9, tetraspanin 9..
Table II.
TSPAN9 expression in GC tissues and normal tissues.
| TSPAN9 expression | GC tissues (n=105) | Normal tissues (n=105) | P-value |
|---|---|---|---|
| Low | 55 | 19 | P<0.001 |
| High | 50 | 86 |
GC, gastric cancer; TSPAN9, tetraspanin 9.
Correlation between Tspan9 expression and clinicopathological characteristics
The association of Tspan9 expression (high expression in 50 patients; low expression in 55 patients) with clinicopathological characteristics was subsequently analyzed. As shown in Table I, the Tspan9 high-expression group had a more advanced TNM stage (P=0.017), more nodal involvement (P=0.01), larger tumor sizes (P=0.026), a greater invasion factor (P=0.025), poorer tumor differentiation (P=0.034), and more positive expression levels of human epidermal growth factor receptor 2 (Her-2; P=0.04) compared with the Tspan9-low group. However, no significant association was observed between the expression level of Tspan9 and gender (P=0.217), age (P=0.724), lymphovascular invasion (P=0.312) or Lauren's classification (P=0.199).
Survival analysis of patients with GC according to Tspan9 expression
The 4-year survival rate of the Tspan9 low-expression group was 78%, which was significantly increased compared with the Tspan9 high-expression group (38%; P<0.01).
The association between the expression level of Tspan9 and prognosis
As shown in Fig. 2, univariate analyses, which were performed to estimate the clinical significance of prognostic factors that might influence the survival of the studied population, revealed that the TNM stage (P<0.001), Tspan9 expression (P=0.008), node metastasis (P<0.001) and T factor (P=0.006) were statistically significant risk factors affecting the overall survival of patients with GC. On the basis of the multivariate analysis data shown in Table III, Tspan9 expression was an independent prognostic marker for GC (P=0.029), in addition to the TNM stage (P<0.001; Table III).
Figure 2.
Kaplan-Meier survival analysis, revealing that patients with a high expression of Tspan9, positive lymph node metastasis, a greater T factor and advanced TNM stage had significantly worse overall survival rates. TNM, tumor/lymph node/metastasis; Tspan9, tetraspanin 9; Cum., cumulative.
Table III.
Univariate and multivariate analysis of survival with clinicopathological factors.
| Multivariate analysis | |||
|---|---|---|---|
| Characteristic | Univariate P-value | HR 95% CI | P-value |
| Gender | 0.139 | ||
| Age (year) | 0.214 | ||
| Tumor differentiation | 0.251 | ||
| Tumor diameter | 0.197 | ||
| T factor | 0.006 | ||
| Lymph node metastasis | <0.001 | ||
| TNM stage | <0.001 | 4.125 | <0.001 |
| 2.025–8.405 | |||
| Lymphovascular invasion | 0.361 | 1.522 | 0.422 |
| 0.547–4.236 | |||
| Lauren's classification | 0.049 | 1.951 | 0.062 |
| 0.966–3.942 | |||
| Her-2 expression | 0.093 | 1.754 | 0.208 |
| 0.731–7.208 | |||
| Tspan9 expression | 0.008 | 2.166 | 0.029 |
| 1.082–4.334 |
HR, hazard ratio; CI, confidence interval; TNM, tumor/lymph node/metastasis; Her-2, human epidermal growth factor receptor 2; TSPAN9, tetraspanin 9.
Discussion
Tetraspanins are considered to function as ‘organizers’ in forming the type of specialized membrane domain known as a tetraspanin-enriched microdomain (15). Except for tetraspanins, the microdomains contain various adhesion receptors (e.g. integrins), transmembrane enzymes, immunoglobulin superfamily proteins and various other single and multi-transmembrane proteins (3). In terms of the function of tetraspanins, they are implicated in a number of cellular processes under physiological and pathological conditions, such as virus-induced syncytium, differentiation, cell adhesion, motility, tumor-cell metastasis, signal transduction, and so forth (16–20). Tetraspanins, including cluster of differentiation 9 (CD9) and Tspan24, have been associated with the prognosis of GC, as previously reported (21,22).
Tspan9, a previously poorly studied tetraspanin molecule, drew our attention. Previously, our intention was to assay for the different levels of gene expression between drug-sensitive and drug-resistant cancer samples using gene microarray, and it was observed that the expression level of Tspan9 was increased in drug-resistant GC samples (data not shown). Therefore, at the outset of the present study our hypothesis was that Tspan9 was potentially a prognostic factor for chemoresistance, and that it may be associated with a poor prognosis of GC.
Tspan9 is one of TM4SF members with the N- and C-termini located in the cytoplasm, and it has been highly conserved during evolution (23). In 2013, a study published by Ooi et al (24) investigated host factors that were involved in alphavirus infection, and it was revealed that Tspan9 is critical for the efficient fusion of the virus in the endosome. Another previous study, which aimed to identify tetraspanin components in platelets, demonstrated that Tspan9 is a novel component of tetraspanin microdomains on the platelet surface (23). A study which aimed to investigate the detailed molecular interactions between primary serous ovarian carcinoma and FTC using a dedicated multiplex ligation-dependent probe amplification probe set identified the previously poorly studied gained/amplified gene, Tspan9, in FTC (13). To the best of our knowledge, no previous studies have demonstrated whether Tspan9 has a role in tumor progression.
The present study represents our first study on Tspan9 expression in GC. In this study, Tspan9 expression was revealed to be markedly decreased in GC tissues compared with the adjacent non-cancerous tissues. However, it was of particular note that the overexpression of Tspan9 was associated with a poor prognosis in GC. The 4-year survival rates in the Tspan9 high-expression group were clearly lower than those in the Tspan9 low-expression group, indicating that Tspan9 was a crucial factor in terms of clinical outcome in patients with GC. In the present study, the expression of Tspan9 was revealed to be an independent predictor of the overall survival rate. When the expression level of a factor in cancer tissue is lower than that in para-cancer tissue, it is more customary to associate the overexpression of the factor with a relatively good prognosis. The results obtained in the present study were therefore contrary to our expectations. In this study, the patients were divided into a Tspan9 low-expression group and a Tspan9 high-expression group, according to the results of the IHC. The association between the low or high expression of Tspan9 and clinicopathological features was then investigated. The results of the present study demonstrated that the high expression of Tspan9 was positively associated with an advanced TNM stage, lymph node metastasis, low differentiation, positive Her-2 expression and a diffuse Lauren classification, but was not associated with age, gender and lymphovascular invasion. Taken together it is possible to conclude that Tspan9 may contribute towards the development of GC.
In summary, Tspan9 is downregulated in GC, and it may be a useful prognostic factor for the survival of patients with GC. Tspan9 may exert important roles in the progression of GC. Further studies are required to investigate the mechanism of action, and the predictive value, of Tspan9 in GC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China for mechanisms of tetraspanin Tspan9 in promoting 5-FU drug-resistance of gastric cancer through regulating autophagy in acidic tumors (no. 81472338) and a Project of the Shandong Province Higher Educational Science and Technology Program (no. J15LL58).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ishigami S, Arigami T, Uchikado Y, Setoyama T, Kita Y, Sasaki K, Okumura H, Kurahara H, Kijima Y, Harada A, et al. IL-32 expression is an independent prognostic marker for gastric cancer. Med Oncol. 2013;30:472. doi: 10.1007/s12032-013-0472-4. [DOI] [PubMed] [Google Scholar]
- 3.Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol. 2011;223:4–14. doi: 10.1002/path.2779. [DOI] [PubMed] [Google Scholar]
- 4.Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med. 2001;2001:1–17. doi: 10.1017/S1462399401002381. [DOI] [PubMed] [Google Scholar]
- 5.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 6.Lazo PA. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 2007;98:1666–1677. doi: 10.1111/j.1349-7006.2007.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zöller M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 8.Serru V, Dessen P, Boucheix C, Rubinstein E. Sequence and expression of seven new tetraspans. Biochim Biophys Acta. 2000;1478:159–163. doi: 10.1016/S0167-4838(00)00022-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Wang Z, Zhan X, Li DC, Zhu YY, Zhu J. Association of NET-1 gene expression with human hepatocellular carcinoma. Int J Surg Pathol. 2007;15:346–353. doi: 10.1177/1066896907306083. [DOI] [PubMed] [Google Scholar]
- 10.Hölters S, Anacker J, Jansen L, Beer-Grondke K, Dürst M, Rubio I. Tetraspanin 1 promotes invasiveness of cervical cancer cells. Int J Oncol. 2013;43:503–512. doi: 10.3892/ijo.2013.1980. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Wang J, Chen L, Wang G, Qin J, Xu Y, Li X. Expression and function of NET-1 in human skin squamous cell carcinoma. Arch Dermatol Res. 2014;306:385–397. doi: 10.1007/s00403-013-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arencibia JM, Martín S, Pérez-Rodríguez FJ, Bonnin A. Gene expression profiling reveals overexpression of TSPAN13 in prostate cancer. Int J Oncol. 2009;34:457–463. [PubMed] [Google Scholar]
- 13.Nowee ME, Snijders AM, Rockx DA, de Wit RM, Kosma VM, Hämäläinen K, Schouten JP, Verheijen RH, van Diest PJ, Albertson DG, Dorsman JC. DNA profiling of primary serous ovarian and fallopian tube carcinomas with array comparative genomic hybridization and multiplex ligation-dependent probe amplification. J Pathol. 2007;213:46–55. doi: 10.1002/path.2217. [DOI] [PubMed] [Google Scholar]
- 14.Biondi A, Hyung WJ. Seventh edition of TNM classification for gastric cancer. J Clin Oncol. 2011;29:4338–4339. doi: 10.1200/JCO.2011.36.9900. [DOI] [PubMed] [Google Scholar]
- 15.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berditchevski F, Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: Role of tetraspanins in integrin signaling. J Cell Biol. 1999;146:477–492. doi: 10.1083/jcb.146.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boismenu R, Rhein M, Fischer WH, Havran WL. A role for CD81 in early T cell development. Science. 1996;271:198–200. doi: 10.1126/science.271.5246.198. [DOI] [PubMed] [Google Scholar]
- 18.Chambrion C, Le Naour F. The tetraspanins CD9 and CD81 regulate CD9P1-induced effects on cell migration. PloS One. 2010;5:e11219. doi: 10.1371/journal.pone.0011219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charrin S, Latil M, Soave S, Polesskaya A, Chrétien F, Boucheix C, Rubinstein E. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat Commun. 2013;4:1674. doi: 10.1038/ncomms2675. [DOI] [PubMed] [Google Scholar]
- 20.Detchokul S, Williams ED, Parker MW, Frauman AG. Tetraspanins as regulators of the tumour microenvironment: Implications for metastasis and therapeutic strategies. Br J Pharmacol. 2014;171:5462–5490. doi: 10.1111/bph.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Shen C, Zhang B, Chen H, Chen Z, Chen J. Expression and clinicopathological significance of CD9 in gastrointestinal stromal tumor. J Korean Med Sci. 2013;28:1443–1448. doi: 10.3346/jkms.2013.28.10.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YM, Zhang ZW, Liu QM, Sun YF, Yu JR, Xu WX. Overexpression of CD151 predicts prognosis in patients with resected gastric cancer. PloS One. 2013;8:e58990. doi: 10.1371/journal.pone.0058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Protty MB, Watkins NA, Colombo D, Thomas SG, Heath VL, Herbert JM, Bicknell R, Senis YA, Ashman LK, Berditchevski F, et al. Identification of Tspan9 as a novel platelet tetraspanin and the collagen receptor GPVI as a component of tetraspanin microdomains. Biochem J. 2009;417:391–400. doi: 10.1042/BJ20081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog. 2013;9:e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]