Abstract
Recently, the homeobox (HOX) gene family has been reported as a factor in tumorigenesis. In the human genome, the HOX gene family contains 4 clusters with 39 genes and multiple transcripts. Mutation or abnormal expression of genes is responsible for developmental disorders. In addition, changes in the levels and activation of certain HOX genes has been associated with the development of cancer. Long non-coding RNAs (lncRNAs) have also been identified to serve critical functions in cancer. Although a limited number of lncRNAs have been previously investigated, the list of functional lncRNA genes has recently grown. Two of the most important and well-studied lncRNAs and HOX transcript genes are HOX transcript antisense RNA (HOTAIR) and HOXA distal transcript antisense RNA (HOTTIP). The present study aimed to review not only the function of the HOTAIR and HOTTIP genes in certain forms of cancer, but also to review other HOX genes and protein functions in cancer, particularly HOX family genes associated with lncRNAs.
Keywords: homeobox genes, homeobox A, proliferation, transcript
1. Introduction
The homeobox (HOX) gene family is a large group of comparable genes that function during early embryonic development to direct the formation of various body structures (1,2). In humans, it is estimated that the HOX gene family contains 65 pseudogenes and 235 functional genes (3). HOX genes are present in every human chromosome, and are commonly organized in clusters. A number of subfamilies and classes of HOX genes have been described, although these categories are used inconsistently.
Various genes of the HOX family are involved in a broad range of crucial developmental activities (4–7). Tumor suppressing regions of HOX genes have been demonstrated to inhibit cell growth in an uncontrolled manner (4). These genes commonly perform important functions, and mutation or abnormal expression of these genes has been associated with developmental disorders and several forms of cancer (5–7).
In the human genome, only 2% of the sequence encodes proteins that function in biological progression (8), with ~90% consisting of non-coding RNAs (ncRNAs), which are described as the ‘noise’ of the genome (9–12). Over the last few years, an increasing number of long ncRNAs (lncRNAs) have been investigated by microarray and sequencing technology, and it has been proposed that they may regulate various major biological processes involved in metabolism, differentiation and tumor development (13). ncRNAs are divided into two subgroups, including small ncRNAs (sncRNAs) and lncRNAs. ncRNAs are defined as sncRNAs when the RNA length is <200 nt, which includes microRNAs (also named as miRNAs/miRs). Conversely, lncRNAs contain >200 nt of transcript. Currently, lncRNAs are becoming more popular and attracting more attention in scientific research (14–16). Two important platforms have been used for analysis of lncRNAs; advanced sequencing platforms estimated that there are ~20,000 distinct lncRNAs in humans (17,18). The other platform is the commercial lncRNAs ChIP, consisting of ~30,000 lncRNA transcripts, and is an ideal tool for making comparisons between tumorous and non-tumorous samples (19,20). It has been suggested that lncRNAs have a critical role in cancer development. However, the biological functions of a large proportion of lncRNAs remain to be elucidated. Previous studies reported that lncRNAs are involved in fundamental cellular processes, including apoptosis, cell cycle and cell proliferation (21–23), chromatin modification (24,25), genomic reprogramming (26,27), gene imprinting (28) and RNA processing (29). It was demonstrated that tumor or non-tumor tissues exhibited different profiles in various forms of human cancer, and altered lncRNA expression has been functionally linked to tumorigenesis; thus suggesting that certain lncRNAs function as tumor promoters or suppressors (17,30,31). Various studies have determined that certain lncRNAs may be detected through their altered expression in different forms of cancer (32–35). The identification of cancer-associated lncRNAs (CA-lncRNAs), and interactions between the CA-lncRNAs and target genes, are beneficial to supplement the current knowledge of signaling pathways and are conducive to a better understanding of cancer development.
One of the most important and well-studied lncRNA and HOX transcript gene is HOTAIR, which is located on chromosome 12 within the HOXC gene cluster and is 2,158 nt long (32,36). HOTAIR is unique in that it is overexpressed in the vast majority of cancer types and has been recognized as an oncogenic lncRNA (32). Recently, it was reported that HOTAIR induced proliferation and metastasis in a variety of tumors and functioned as a negative prognostic indicator for several forms of cancer (32,36). In addition, it was observed that the lncRNA interacted with polycomb repressive complex 2 (PRC2) and inhibited multiple genes, particularly metastasis-suppressing genes, in cancer tissues (36,37). Subsequent studies demonstrated that HOTAIR serves as a molecular scaffold for at least two distinct histone modification complexes, coordinating their functions in transcription repression (25). Several transcriptome-wide association studies have detected extensive alterations in cellular transcript levels in response to HOTAIR inhibition, indicating that HOTAIR may regulate hundreds of genes (33–35), thus providing insight into the underlying mechanisms of this lncRNA in cancer. Although transcript frequencies are informative, they do not necessarily reflect the level of cellular protein; this is due to the fact that protein activity is affected by an array of post-transcriptional regulatory mechanisms and the association between mRNA and protein levels is generally modest (34,38).
LncRNAs have been frequently investigated in recent years and certain lncRNAs associated with HOX genes have been detected in sequencing and ChIP platform research. It is expected that an increasing number of HOX gene-associated lncRNAs may be identified in the coming years, which will further clarify the association between lncRNAs and HOX in tumorigenesis. The present review discusses HOX genes and research regarding their association with lncRNAs, particularly in the context of carcinogenesis.
2. HOX genes and cancer
HOX genes were originally identified in Drosophila melanogaster (39). These genes most importantly function as transcriptional regulators during normal morphogenesis in the process of cell-to-cell communication, the modification of which may contribute to the development of cancer (40,41). The HOX gene homology domain is able to bind to specific DNA sequences and regulate gene transcription (42). However, possible mechanisms underlying the function of HOX genes in tumorigenesis have not yet been elucidated. Notably, abnormal expression of various HOX genes has been observed in a number of solid tumors and hematological malignancies (43–46), and studies have begun to distinguish the biological mechanisms associated with their expression (47–50). The human HOX gene family contains 4 HOX gene clusters, which are presented in Table I.
Table I.
Four gene clusters of the human HOX gene family.
| Cluster | Chromosome | Genes |
|---|---|---|
| HOXA | 7 | HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11, HOXA13 |
| HOXB | 17 | HOXB1, HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, HOXB13 |
| HOXC | 12 | HOXC4, HOXC5, HOXC6, HOXC8, HOXC9, HOXC10, HOXC11, HOXC12, HOXC13 |
| HOXD | 2 | HOXD1, HOXD3, HOXD4, HOXD8, HOXD9, HOXD10, HOXD11, HOXD12, HOXD13 |
HOX, homeobox.
A study by Hur et al (51) profiled 39 HOX genes involved in breast cancer, but only 25 of these genes were examined in malignant and non-malignant tissues. A total of 14/25 genes exhibited differential expression levels in malignant breast tissues compared with non-malignant breast tissues (51). Several studies have demonstrated that various HOX genes are differentially expressed (either up- or downregulated) in patients with breast cancer, indicating the involvement of these genes in carcinogenesis and breast cancer metastasis (43,52,53). In addition, HOXA5 and HOXA9 have been implicated as regulators of p53 and breast cancer 1 (BRCA1), respectively, which are important factors in cancer development (54,55). Furthermore, the function of HOXA1, -B7 and -B9 in breast tumorigenesis has been investigated; HOXB7 was identified as a key factor regulating cell proliferation, invasion and tamoxifen resistance (56–58), and the overexpression of HOXA1 and HOXB9 contributed to breast cancer tumorigenicity (59,60). Additional studies have reported an association between poor prognosis and overexpression of HOXD3 and HOXB13 (61,62).
Accumulating evidence indicates that various tumors exhibit low levels of HOXA11 expression, which has been described as a tumor suppressor (63). A study by Bai et al (64) observed that the methylation frequencies of HOXA11 in gastric cancer tissues and adjacent cancer tissues were higher than those in normal gastric mucosa (P<0.05). Furthermore, HOXA11 expression was downregulated following hypermethylation of the promoter region (64); this suggests that HOXA11 may function as an important repressor in gastric cancer tumorigenesis, and aberrant promoter methylation may be the primary cause behind the loss or downregulation of HOXA11 expression, subsequently resulting in gastric cancer carcinogenesis. The study also observed that hypermethylation of HOXA11 was significantly associated with lymph node metastasis and Tumor-Node-Metastasis stage in gastric cancer, and in vitro experiments demonstrated that HOXA11 may control cell growth with its defect enhancing cell proliferation (64).
HOXC11 expression has been observed to be significantly higher in renal cell carcinoma (RCC) compared with normal kidney tissues (64). It was identified that different expression levels of HOXC11 represented different functions, with overexpression promoting proliferation and downregulation inhibiting proliferation (64). In addition, the study reported that high immunohistochemical expression of HOXC11 was associated with T stage, N stage and Ki67 level in RCC (65). The function of HOX genes remains unknown in oral tumorigenesis (66). However, HOXB2, HOXB7 and HOXB13 have been reported to be important in oral tissues, and dysregulation of HOXB7 results in increased tumor cell proliferation in oral squamous cell carcinoma tissues (67). Previous studies have demonstrated that HOXA1 serves an important role in tumorigenesis. It has been reported that the disorder expression of HOXA1 in several cancer types (68) is involved in regulating multiple cellular processes, including proliferation, apoptosis, etc. (69), and is therefore closely associated with the shorter overall survival of patients after resection. In addition, HOXA1 expression was correlated with shorter overall survival in patients with small cell lung cancer (70).
HOXA1 overexpression is associated with cell growth and tumor formation in mice (60). A previous study reported that elevated HOXA1 expression levels inhibited differentiation, and resulted in transformation and the development of acute myeloblastic leukemia in lethally irradiated mice (71). Increased levels of HOXA1 in the mammary carcinoma MCF-7 cell line have also been demonstrated to dramatically increase proliferation via the signal transducers and activators of transcription pathway (72) and transcriptional upregulation of B-cell lymphoma 2 (Bcl-2) (60). HOXA1 controls the transcriptional upregulation of Bcl-2, cyclin D1 and c-Myc (73). However, various studies have reported that HOX1A overexpression does not affect adhesion, invasion or apoptosis (60,69,71,74). The gene is dependent on the Meis, Prep or homeothorax protein cofactors to activate and/or repress transcription (75). Increased HOXA1 expression results in promoting the proliferation and decreasing the apoptosis of mammary carcinoma cells (69), thus contributing to the acceleration of cancer growth.
Previous studies have demonstrated that the upregulated expression of HOXA1 promotes proliferation, whereas downregulated HOXA1 expression decreases cell proliferation (71–73). In addition, HOXA1-positive cells correlate with N stage, T stage, proliferation and differentiation, and poor survival (76).
3. HOX genes and lncRNAs
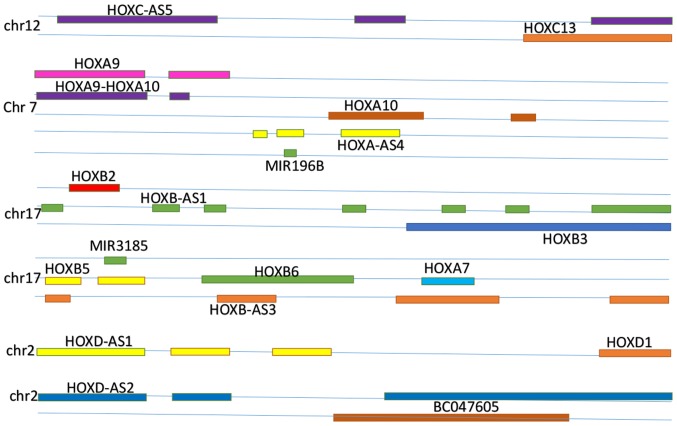
A number of non-coding RNAs are located in and associated with HOX gene clusters, including HOTAIRM1, HOXC5, HOXA-AS3, HOXA10, HOXB-AS3, HOXA10-HOXA9, HOXB-AS5, HOXD-AS2, HOXD-AS1, HOXA-AS3, HOXA-AS4, pre-B-cell leukemia HOX4, HOXA distal transcript antisense RNA (HOTTIP) and HOTAIR. The associations between downstream and upstream HOX gene clusters with lncRNAs are presented in Fig. 1.
Figure 1.
Associations between HOX gene clusters and long non-coding RNA. HOX, homeobox; Chr, chromosome; MIR. microRNA.
3.1. HOTAIR
DNA-binding proteins target ncRNAs, such as HOTAIR, to specific sites in chromatin (77). HOTAIR is a 2.2-kb lncRNA implicated in the suppression of the HOX loci, which subsequently promotes breast cancer metastasis (33,78). Whether lncRNAs directly or indirectly regulate the expression of HOX gene family members has gained increasing attention in the context of development and tumorigenesis (24,78,79). Furthermore, altered HOX gene expression has been observed in certain forms of cancer (80). In a recent study, Xu et al used a high-throughput microarray to analyze the lncRNAs and mRNA expression profiles in lung adenocarcinoma (81). A total of 564 dysregulated lncRNAs and 83 HOX subtypes (including 35 HOX genes and 48 HOX ncRNAs) were screened out, which indicated that there is a close relationship between HOX genes and lncRNAs in lung adenocarcinoma (81). Taurine upregulated 1 (TUG1) was able to modulate HOXB7 expression, which may be partially responsible for TUG1-mediated proliferation regulation, thus affecting the proliferation of non-small cell lung cancer (NSCLC) in vitro and in vivo (82). The signals of p53/TUG1/PRC2/HOXB7 may serve as targets for NSCLC diagnosis and therapy (82). A previous study observed that when HOXB7 expression was inhibited, proliferation was also subsequently inhibited (83). HOXB7 primarily regulates cell growth through the phosphoinositide 3-kinase/Akt and mitogen-activated protein kinase pathways (83). HOXB7 is able to promote cell proliferation in different types of tumors through its high levels of expression (83–85). In addition, HOXB7 has been demonstrated to be an oncogene (57,83,86,87), and its involvement is essential in various forms of cancer.
A number of studies have reported that the HOX family genes serve as classic modification targets of the polycomb complex during development, with all four gene clusters being highly enriched in histone 3 lysine 27 trimethylation (88,89). HOTAIR mediates epigenetic silencing by guiding PRC2 to the HOX loci (78). Various ncRNAs are located within the HOX gene clusters, with ~231 ncRNAs estimated to be present in humans (24). HOTAIR, a HOX ncRNA, is transcribed from the antisense of canonical HOXC genes, which interact with PRC2 and silence the subsequent transcription of the HOXD locus (78).
3.2. HOTTIP
HOTTIP is a ncRNA that targets genes through chromosomal looping (77); it is crucial for metastasis of various forms of human cancer, and its overexpression results in PRC2 silencing HOXD9, a locus involved in developmental pattern (78). A previous study observed that HOTAIR expression is increased in human breast cancer, which targets PRC2 in the genome and results in changes in gene expression (33). Overexpression of HOTAIR has also been documented in several other types of human cancer, including hepatocellular (90) and pancreatic (35) cancer, suggesting that this lncRNA has an oncogenic function. However, its potential involvement in gastric cancer remains to be elucidated. Further evidence supporting the involvement of HOTAIR in epithelial-mesenchymal transition (EMT) is the interaction between Twist and miR-10b. miR-10b is an miRNA transcribed from the 3′ HOXD locus and may be induced by Twist (36).
HOTTIP resides at the 5′ end of the HOXA locus; it has been has been identified as one of the 231 ncRNAs associated with the human HOX loci (24) and has been associated with the activation of multiple 5′ HOXA genes in vivo (34). HOTTIP was previously considered as a negative prognostic factor in patients with liver cancer, and its increased expression was associated with enhanced liver cancer metastasis (38). It was observed that inhibition of HOTTIP potentiated the antitumor effects of gemcitabine in vitro and in vivo. Furthermore, HOXA13-knockdown by RNA interference (siHOXA13) demonstrated that HOTTIP promoted pancreatic ductal adenocarcinoma (PDAC) cell proliferation, invasion and chemoresistance, at least partially through regulating HOXA13 (91). In addition, immunohistochemical analysis revealed that high HOXA13 expression correlated with poor histological differentiation, lymph node metastasis and decreased overall survival in patients with PDAC (91).
A large number of HOX genes function as transcriptional regulators, and govern cell proliferation and differentiation (87). Furthermore, HOX genes have a distinctive gene network organization (four chromosomal loci: A, B, C and D) and represent the most repeat-poor regions within the human genome. The HOXA locus consists of a cluster of 11 HOX genes with a graded expression pattern along the body's appendages from proximal (close to the main body) to distal (appendage tip) (78,88). Among the HOXA genes, HOXA13, which is a marker of gut primordial posteriorization during development (78), serves an essential role in tumorigenesis of the liver and bladder, and in esophageal cancer (33,78,90). To examine the association between HOTTIP dysregulation and HOXA cluster expression, HOTTIP-knockdown studies were performed, and the expression level of the HOXA cluster (HOXA7, A9, A10, A11, and A13) was further evaluated by polymerase chain reaction (92). Although HOXA7, A9, A10, A11 and A13 gene expression was lower in the HOTTIP-knockdown group compared with the control, HOXA13 was the most significantly inhibited gene within the HOXA locus following depletion of HOTTIP in pancreatic cancer cells (92). Furthermore, it was demonstrated that the expression levels of HOTTIP and HOXA13 were significantly positively correlated with 90 PDAC tissues and paired adjacent non-neoplastic tissues (92). In addition, small interfering RNA-mediated HOXA13-knockdown inhibited the proliferation, invasion and EMT of PDAC cells, which was consistent with the functional changes that occurred after silencing the expression of HOTTIP in PDAC cells (92). Targeted inhibition of HOXA13 in two pancreatic cancer cell lines led to a decrease in the HOXA13 mRNA level and lower HOTTIP expression, which was consistent with the results of another study (93).
4. HOX gene clusters encoding miRNAs
Human HOX gene clusters exhibit a low density of interspersed repeats, and cis-regulatory elements effect HOX gene expression (94). The enhancer of the clusters is able to regulate HOXD temporal co-linearity (95). Previous studies have demonstrated that ncRNAs are involved in the regulation of HOX gene expression (96), and certain microRNAs participate in this regulation (97), including miR-196 (98) and miR-10 (99). Due to the technical limitations, earlier lncRNA studies have been performed in only two species (mouse and human), such as the previous studies of HOTAIR (33,78) and HOTAIRM1 (100). However, one study performed an integrated analyses of HOX gene clusters between the kangaroo family and eutherians (101). Well-studied miRNAs, including miR-196a/b, miR-10a/b and lncRNAs (such as HOTAIR, HOTAIRM1 and HOXA11-AS), were observed to dysregulate gene expression and were associated with tumor development (24,32,36). miR-464, miR-10 and miR-414 are located in the tammar HOX clusters and were identified to have an effect on these, for example inhibiting lymphangiogenesis (miR-414) and promoting active tumor cell invasion (miR-10) (102,103). In addition, certain novel miRNAs are transcribed from elsewhere on the tammar genome and regulate the expressions of HOXB and HOXD clusters by specifically interacting with the mRNAs transcribed from them (104).
5. Future prospects
A large number of studies have investigated lncRNAs in recent years, but only a few lncRNAs associated with HOX genes have been detected with sequencing and ChIP microarray. It is expected that an increasing number of HOX genes and related lncRNAs may be identified in the coming years, with further analysis required to determine their specific functions. Further studying the associations between lncRNAs and HOX will lead to a deeper understanding to the mechanism of cancer development.
Acknowledgements
This study was supported by The Key Clinical Specialty Discipline Construction Program of Fujian, The Specialized Science and Technology Key Project of Fujian Province (grant no. 2013YZ0002-3), The Science and Technology Infrastructure Construction Program of Fujian Province (grant no. 2014Y2005), The Natural Science Foundation of Fujian Province (grant no. 2015J05174), The Scientific Research Project of Health and Family Planning Commission of Fujian Province (grant no. 2015-1-94), The Scientific Foundation of Fuzhou Health Department (grant no's. 2014-S-139-3, 2014-S-w19 and 2013-S-wp1) and the Postdoctoral Science Foundation of Fujian Medical University (grant no. 149318).
References
- 1.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence PA, Morata G. Homeobox genes: Their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 3.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duboule D. Vertebrate Hox genes and proliferation: An alternative pathway to homeosis? Curr Opin Genet Dev. 1995;5:525–528. doi: 10.1016/0959-437X(95)90058-O. [DOI] [PubMed] [Google Scholar]
- 5.Peverali FA, D'Esposito M, Acampora D, Bunone G, Negri M, Faiella A, Stornaiuolo A, Pannese M, Migliaccio E, Simeone A, et al. Expression of HOX homeogenes in human neuroblastoma cell culture lines. Differentiation. 1990;45:61–69. doi: 10.1111/j.1432-0436.1990.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 6.Cillo C, Barba P, Freschi G, Bucciarelli G, Magli MC, Boncinelli E. HOX gene expression in normal and neoplastic human kidney. Int J Cancer. 1992;51:892–897. doi: 10.1002/ijc.2910510610. [DOI] [PubMed] [Google Scholar]
- 7.Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 9.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 10.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kugel JF, Goodrich JA. Non-coding RNAs: Key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, Gao W, Zheng S, Zhao X, Chen T, Chen R. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137. doi: 10.1186/s12967-015-0490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18:111–123. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, et al. Human cancer long non-coding RNA transcriptomes. PloS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan Y, Chang HY. HOTAIR: Flight of noncoding RNAs in cancer metastasis. Cell Cycle. 2010;9:3391–3392. doi: 10.4161/cc.9.17.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorge S, Ha N, Polychronidou M, Friedrich J, Bezdan D, Kaspar P, Schaefer MH, Ossowski S, Henz SR, Mundorf J, et al. The cis-regulatory code of Hox function in Drosophila. EMBO J. 2012;31:3323–3333. doi: 10.1038/emboj.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahanger SH, Srinivasan A, Vasanthi D, Shouche YS, Mishra RK. Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae. Nucleic Acids Res. 2013;41:804–816. doi: 10.1093/nar/gks1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javed S, Langley SE. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. 2014;113:535–540. doi: 10.1111/bju.12269. [DOI] [PubMed] [Google Scholar]
- 42.McGrath SE, Michael A, Pandha H, Morgan R. Engrailed homeobox transcription factors as potential markers and targets in cancer. FEBS Lett. 2013;587:549–554. doi: 10.1016/j.febslet.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 43.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto K, et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673–679. [PubMed] [Google Scholar]
- 44.Kanai M, Hamada J, Takada M, Asano T, Murakawa K, Takahashi Y, Murai T, Tada M, Miyamoto M, Kondo S, Moriuchi T. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
- 45.Bodey B, Bodey B, Jr, Gröger AM, Siegel SE, Kaiser HE. Immunocytochemical detection of homeobox B3, B4, and C6 gene product expression in lung carcinomas. Anticancer Res. 2000;20:2711–2716. [PubMed] [Google Scholar]
- 46.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 47.Yamatoji M, Kasamatsu A, Yamano Y, Sakuma K, Ogoshi K, Iyoda M, Shinozuka K, Ogawara K, Takiguchi Y, Shiiba M, et al. State of homeobox A10 expression as a putative prognostic marker for oral squamous cell carcinoma. Oncol Rep. 2010;23:61–67. [PubMed] [Google Scholar]
- 48.Ota T, Klausen C, Salamanca MC, Woo HL, Leung PC, Auersperg N. Expression and function of HOXA genes in normal and neoplastic ovarian epithelial cells. Differentiation. 2009;77:162–171. doi: 10.1016/j.diff.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 49.Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 51.Hur H, Lee JY, Yun HJ, Park BW, Kim MH. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56:64–71. doi: 10.1007/s12033-013-9682-4. [DOI] [PubMed] [Google Scholar]
- 52.Svingen T, Tonissen KF. Altered HOX gene expression in human skin and breast cancer cells. Cancer Biol Ther. 2003;2:518–523. doi: 10.4161/cbt.2.5.441. [DOI] [PubMed] [Google Scholar]
- 53.Cantile M, Pettinato G, Procino A, Feliciello I, Cindolo L, Cillo C. In vivo expression of the whole HOX gene network in human breast cancer. Eur J Cancer. 2003;39:257–264. doi: 10.1016/S0959-8049(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 54.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 55.Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, Benezra M, Licht JD, Boudreau NJ, Tsai KK, et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535–1550. doi: 10.1172/JCI39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caré A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 57.Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, Ma XJ, Huang Y, Kallioniemi A, Landberg G, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci USA. 2012;109:2736–2741. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Care A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, Peschle C, Colombo MP. HOXB7: A key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61:6532–6559. [PubMed] [Google Scholar]
- 59.Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco Md, Wijendran V, Shioda T, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci USA. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Zhu T, Chen Y, Mertani HC, Lee KO, Lobie PE. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J Biol Chem. 2003;278:7580–7590. doi: 10.1074/jbc.M212050200. [DOI] [PubMed] [Google Scholar]
- 61.Shaoqiang C, Yue Z, Yang L, Hong Z, Lina Z, Da P, Qingyuan Z. Expression of HOXD3 correlates with shorter survival in patients with invasive breast cancer. Clin Exp Metastasis. 2013;30:155–163. doi: 10.1007/s10585-012-9524-y. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez BA, Cheng AS, Yan PS, Potter D, Agosto-Perez FJ, Shapiro CL, Huang TH. Epigenetic repression of the estrogen-regulated Homeobox B13 gene in breast cancer. Carcinogenesis. 2008;29:1459–1465. doi: 10.1093/carcin/bgn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiegl H, Windbichler G, Mueller-Holzner E, Goebel G, Lechner M, Jacobs IJ, Widschwendter M. HOXA11 DNA methylation-a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008;123:725–729. doi: 10.1002/ijc.23563. [DOI] [PubMed] [Google Scholar]
- 64.Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, Zhang H, Suo Z, Ji S. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79. doi: 10.1186/s12935-014-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu YJ, Zhu Y, Yuan HX, Zhang JP, Guo JM, Lin ZM. Overexpression of HOXC11 homeobox gene in clear cell renal cell carcinoma induces cellular proliferation and is associated with poor prognosis. Tumour Biol. 2015;36:2821–2829. doi: 10.1007/s13277-014-2909-6. [DOI] [PubMed] [Google Scholar]
- 66.Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, Zhang X, Kashiwazaki H, Yamazaki Y, Inoue N, Moriuchi T. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. 2006;16:217–224. doi: 10.3727/000000006783981080. [DOI] [PubMed] [Google Scholar]
- 67.De Souza Setubal Destro MF, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, Coletta RD. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36:141–149. [PubMed] [Google Scholar]
- 68.Abate-Shen C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, Gluckman PD, Lee KO, Lobie PE. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J Biol Chem. 2006;281:6471–6481. doi: 10.1074/jbc.M512666200. [DOI] [PubMed] [Google Scholar]
- 70.Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang J, Yang J, Liao H, Guo L. Downregulation of HOXA1 gene affects small cell lungcancer cell survival and chemoresistance under the regulation of miR-100. Eur J Cancer. 2014;50:1541–1554. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 71.Bach C, Buhl S, Mueller D, García-Cuéllar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–2918. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 72.Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, Gluckman PD, Sukumar S, Emerald BS, Lobie PE. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007;26:3998–4008. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- 73.Zhu T, Starling-Emerald B, Zhang X, Lee KO, Gluckman PD, Mertani HC, Lobie PE. Oncogenic transformation of human mammary epithelial cells by autocrine human growth hormone. Cancer Res. 2005;65:317–324. [PubMed] [Google Scholar]
- 74.Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity (Edinb) 2006;97:88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 75.Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/S1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 76.Bitu CC, Destro MF, Carrera M, da Silva SD, Graner E, Kowalski LP, Soares FA, nd, Coletta RD. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC Cancer. 2012;12:146. doi: 10.1186/1471-2407-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanhere A, Jenner RG. Noncoding RNA localisation mechanisms in chromatin regulation. Silence. 2012;3:2. doi: 10.1186/1758-907X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 81.Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Chen J, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PloS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao WT, Jiang D, Yuan J, Cui YM, Shi XW, Chen CM, Bian XW, Deng YJ, Ding YQ. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Bian XW, Deng YJ, Ding YQ. 2011;17:3569–3578. doi: 10.1158/1078-0432.CCR-10-2533. [DOI] [PubMed] [Google Scholar]
- 84.Storti P, Donofrio G, Colla S, Airoldi I, Bolzoni M, Agnelli L, Abeltino M, Todoerti K, Lazzaretti M, Mancini C, et al. HOXB7 expression by myeloma cells regulates their pro-angiogenic properties in multiple myeloma patients. Leukemia. 2011;25:527–537. doi: 10.1038/leu.2010.270. [DOI] [PubMed] [Google Scholar]
- 85.Yuan W, Zhang X, Xu Y, Li S, Hu Y, Wu S. Role of HOXB7 in regulation of progression and metastasis of human lung adenocarcinoma. Mol Carcinog. 2014;53:49–57. doi: 10.1002/mc.21947. [DOI] [PubMed] [Google Scholar]
- 86.Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, Argani P, Sukumar S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 87.di Pietro M, Lao-Sirieix P, Boyle S, Cassidy A, Castillo D, Saadi A, Eskeland R, Fitzgerald RC. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proc Natl Acad Sci USA. 2012;109:9077–9082. doi: 10.1073/pnas.1116933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 89.Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 91.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–10852. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 95.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 96.Petruk S, Sedkov Y, Brock HW, Mazo A. A model for initiation of mosaic HOX gene expression patterns by non-coding RNAs in early embryos. RNA Biol. 2007;4:1–6. doi: 10.4161/rna.4.1.4300. [DOI] [PubMed] [Google Scholar]
- 97.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: An apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 99.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PloS One. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Sha G, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seo M, Choi JS, Rho CR, Joo CK, Lee SK. MicroRNA miR-466 inhibits Lymphangiogenesis by targeting prospero-related homeobox 1 in the alkali burn corneal injury model. J Biomed Sci. 2015;22:3. doi: 10.1186/s12929-014-0104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40:1553–1560. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 104.Yu H, Lindsay J, Feng ZP, Frankenberg S, Hu Y, Carone D, Shaw G, Pask AJ, O'Neill R, Papenfuss AT, Renfree MB. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics. 2012;13:251. doi: 10.1186/1471-2164-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]