Abstract
Although accumulating evidence has suggested that microRNAs (miRNAs) have a serious impact on cognitive function and are associated with the etiology of several neuropsychiatric disorders, their expression in sevoflurane-induced neurotoxicity in the developing brain has not been characterized. In the present study, the miRNAs expression pattern in neonatal hippocampus samples (24 h after sevoflurane exposure) was investigated and 9 miRNAs were selected, which were associated with brain development and cognition in order to perform a bioinformatic analysis. Previous microfluidic chip assay had detected 29 upregulated and 24 downregulated miRNAs in the neonatal rat hippocampus, of which 7 selected deregulated miRNAs were identified by the quantitative polymerase chain reaction. A total of 85 targets of selected deregulated miRNAs were analyzed using bioinformatics and the main enriched metabolic pathways, mitogen-activated protein kinase and Wnt pathways may have been involved in molecular mechanisms with regard to neuronal cell body, dendrite and synapse. The observations of the present study provided a novel understanding regarding the regulatory mechanism of miRNAs underlying sevoflurane-induced neurotoxicity, therefore benefitting the improvement of the prevention and treatment strategies of volatile anesthetics related neurotoxicity.
Keywords: sevoflurane, microRNA, hippocampus, neonatal rat, bioinformatic analysis
Introduction
MicroRNAs (miRNAs) are single-stranded, non-coding RNAs recognized as endogenous regulators of post-transcriptional gene expression, as these small RNAs are capable of controlling gene expression by mediating either mRNA degradation or translation inhibition (1). These non-coding RNAs are involved in numerous biological processes, including cell cycle, cell proliferation, apoptosis, cellular response to stress and the immune response (2), and increasing evidence indicates that miRNAs are dysregulated in numerous diseases. Previous studies have demonstrated that microRNAs have an impact on cognitive function (3). Furthermore, they have been demonstrated to be involved in the etiology of numerous neuropsychiatric disorders, including schizophrenia (4), mental retardation (or intellectual disability) (5) as well as autism spectrum disorders (6).
Volatile anesthetics are widely used in the clinic with sevoflurane being the most commonly used inhalation anesthetic for pediatric anesthesia (7). Sevoflurane is suitable for infants and children because it has a low blood and gas partition coefficient, rapid onset and offset, low airway irritation and an aromatic odor (8–10). However, recent studies investigate the potential risks of such volatile anesthetics on brain development. A number of preclinical studies have demonstrated that, sevoflurane exposure in neonatal animals induces neurological deficits such as cognitive decline (11–16) and abnormal social behaviors in adulthood (17).
Evidence has also indicated that sevoflurane causes widespread apoptotic neurodegeneration and hippocampal synaptic function deficits (18,19). However, the elaborated mechanisms of sevoflurane neurotoxicity in the developing brain are largely unknown. Kalenka et al revealed that sevoflurane exposure may result in long-lasting changes in protein expression of septin 8 in the brain, using a high-throughput proteomic analysis (20). Additionally, in the genomic studies, Pan et al demonstrated that sevoflurane induced long-term expression changes of the numerous genes in the hippocampus, which may be associated with neurodevelopmental impairment and cognitive dysfunction (21). Considering the pivotal role of miRNA in gene expression regulation and cognitive dysfunction, it was hypothesized that there is an altered miRNA expression pattern in the hippocampus of neonatal rats' associated with sevoflurane neurotoxicity.
MiRNA microarray technology is an efficient method used to generate miRNA expression profiles. This approach has been previously applied to study the functional linkage between miRNAs and physiological and pathological processes (22).
The present study investigated the miRNA expression changes on neonatal rat hippocampi induced by sevoflurane using the miRNA microarray technique. Furthermore, certain altered miRNAs were selected which were associated with cognitive function and brain development in order to perform bioinformatics research. This research included target gene prediction, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis and GO term enrichment that were used in order to reveal the potential role of these miRNAs in the hippocampus of neonatal rats' in order to reveal the mechanisms of sevoflurane neurotoxicity in a developing brain.
Materials and methods
Sample preparation
All procedures were approved by the Institutional Animal Care and Use Committee of Wuhan University (Wuhan, China) and conformed to the guidelines for ethical treatment of animals. Efforts were made to minimize the number of animals used. Experiments required collecting RNA samples of neonatal rats' hippocampus and arterial blood that were isolated from postnatal 7 day male rats, with the day of birth considered as postnatal day 0.
The rats were randomly allocated in two groups, each group consisting of 20 male Sprague-Dawley rats (7 days old; 16–18 g; provided by the Experimental Animal Center of Wuhan University). A total of 4 rats of each group were used for the miRNA microarray assay and the remaining were used to perform arterial blood gas analysis (n=6 for each group). In the sevoflurane group, rats were allowed to breathe spontaneously and housed in an anesthesia chamber supplied with 2.4% sevoflurane (Maruishi Pharmaceutical, Osaka, Japan) and air-oxygen mixture (fraction of inspired oxygen=0.4) at the rate of 5 l/min for 4 h. In the control group, rats were only supplied with air-oxygen at the rate of 5 l/min for 4 h. In the meantime a heat pad was used to keep the rats' body temperature at ~37°C. During the experiment, the concentration of sevoflurane and carbon dioxide was continuously monitored by an anesthesia gas monitor (Draeger Medical GmbH, Lübeck, Germany) and the rectal temperature measured.
A total of 6 pups of each group underwent arterial blood sampling from the left cardiac ventricle and the samples were transferred to heparinized glass capillary tubes. The blood pH, PaCO2, PaO2 and SaO2 were analyzed immediately after blood collection using a GEM Premier 3000 analyzer (Instrumentation Laboratory, Lexington, MA, USA). Samples were obtained immediately after the rats were anesthetized. The remaining pups (n=4 for each group) were sacrificed by decapitation following anesthesia using 30–60 mg/kg pentobarbital (Venus Biotechnology, Inc., Wuhan, China) at 24 h after sevoflurane exposure for the miRNA microarray assay. The brains of the rats were quickly removed and the hippocampi were separated from the brain on an ice pad. The hippocampus samples were immediately diluted in RNAlater solution (Applied Biosystems, Foster City, CA, USA).
RNA extraction
After 24 h, the samples were rapidly stored in a −80°C refrigerator until RNA extraction. The frozen samples were quickly homogenized in 700 µl QIAZOL lysis reagent (Qiagen China Co., Ltd., Shanghai, China) using a TissueRuptor (Qiagen China Co., Ltd.) for 30 sec at 10°C. The RNA fractions were isolated with the miRNeasy mini kit (Qiagen China Co., Ltd.) following the manufacturer's instructions. Next, the concentration of RNA was determined by measuring the absorbance at 260 nm (A260) in a ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Finally, the value of A260/A280 provided an estimate of the purity of RNA. When the RNA samples complied with an A260/A280 ratio of 1.8–2.0, the RNA analysis could proceed.
miRNA expression screening test
After the purity of the total RNA was verified, an µParaflo microfluidic chip (LC-Bio Co., Ltd., Hangzhou, China) was used in order to screen the miRNAs. The microfluidic chip included 727 known Rattus norvegicus miRNAs corresponding to miRNA transcripts in Sanger miRBase release 20.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). The control probes used were as follows: Rattus norvegicus 5s rRNA, PUC2PM-20B and PUC2MM-20B (Venus Biotechnology, Inc.). The aforementioned probes are used for quality controls of chip production, assay conditions and sample labeling. Among the control probes, R. norvegicus 5s rRNA was used as an inner positive control; whereas PUC2PM-20B and PUC2MM-20B served as external positive controls. There were 4–8 µg total RNA from each sample that was 3′-extended and labeled with a poly (A) tail using poly (A) polymerase (Venus Biotechnology, Inc.). Next, an oligonucleotide tag (Cloning Biotechnology, Inc., Wuhan, China) was bound with the poly (A) tail for further fluorescent dye staining. According to the µParaflo microfluidic chip manual, hybridization was performed overnight on a microfluidic chip using a microcirculation pump (Atactic Technologies Inc., Houston, TX, USA). The labeled RNA samples were hybridized to the microfluidic chip that contained a chemically modified nucleotide coding segment complementary to the target microRNAs (from miRBase) or other RNA (control or customer defined sequences), and a spacer segment of polyethylene glycol to extend the coding segment at a distance from the substrate. The detection probes were created in situ using photogenerated reagent chemistry. According to the microfluidic chip manual, hybridization used 100 µl 6X SSPE buffer [0.90 mM NaCl, 60 mM Na2HPO4, 6 mM ethylenediaminetetraacetic acid (pH 6.8)] containing 25% formamide at 34°C. Following RNA hybridization, a tag-conjugating Cy3 dye (Thermo Fisher Scientific, Inc.) was circulated through the microfluidic chip for dye staining. Next, fluorescence images were scanned using a GenePix 4000B laser scanner (Molecular Devices, LLC, Sunnyvale, CA, USA) and the image analysis was undertaken using Array-Pro image analysis software (version 4.5; Media Cybernetics, Inc., Rockville, MD, USA).
The signal data was derived by subtracting the background and by normalizing and analyzing the signals employing a locally-weighted regression filter. The detected signals were transformed by log2 and the ratio of the two sets of detected signals and P-values utilizing the t-test were calculated. The sign values that could be listed as detectable were required to meet at least two conditions: A signal higher than the background value plus a three-fold standard deviation and a coefficient of variation (CV=signal standard deviation/signal intensity) <0.5. These were considered valuable and reliable. Hierarchical clustering was performed by average linkage and Euclidean distance metric. Finally, the graphic displays were visualized as heat map results of hierarchical clustering.
Quantitative polymerase chain reaction (qPCR)
To validate the accuracy of the µParaflo microfluidic chip data, single qPCR was performed for several candidate miRNAs using the ABI PRISM 7900 Real-Time PCR System (Applied Biosystems). According to a series of previously published articles, several miRNAs were selected, which are considered to be relevant to brain development and cognitive function (23–31). These candidate representative miRNAs also had the following three characteristics: P<0.05, signal value >500 and fold change >0.5.
At first, 2 µg total RNA was reverse-transcribed to cDNA using a One Step PrimeScripts miRNA cDNA Synthesis kit (D350A; Takara Bio, Inc., Otsu, Japan) and miRNA-specific stem-loop RT primers. The reaction consisted of 10 ml 2 µl miRNA reaction buffer, 2 ml 0.1% bovine serum albumin, 2 ml miRNA PrimeScripts RT enzyme mix, 2 mg total RNA and RNAe-free dH2O to a final volume of 20 ml. The reaction was incubated at 37°C for 15 min, 85°C for 5 min, then held at 4°C using an Applied Biosystems 9700 Thermocycler.
Next, qPCR was performed with SYBRs Premix Ex Taq kit (DRR081A; Takara Bio, Inc.,) using a standard SYBR Green real-time PCR master mix (QPK-201; Toyobo Co., Ltd., Osaka, Japan). In each reaction, 20 µl final reaction mixture contained 10 µl 2X SYBR Premix Ex Taq, 0.8 ml PCR forward primer (10 mm), 0.8 ml Uni-miR qPCR primer (10 mm), 0.4 µl 50X ROX reference dye, 2 µl cDNA (100 ng/µl) and dH2O. Next, the reaction mixture was incubated at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 45 sec in a 96-well optical plate. All reactions were run in triplicate and rno-U87 was used as an endogenous reference. The ∆∆Ct method (32) was used to determine the expression level differences between the sevoflurane and the control groups. P<0.05 was used to indicate a statistically significant difference.
miRNA target prediction and bioinformatics analysis
To explore the potential relevant function of all differentially-expressed miRNAs, the TargetScan (version 6.2; http://www.targetscan.org/vert/), miRanda (August 2010 release; http://www.microrna.org/microrna/home.do) and PicTar (http://pictar.mdc-berlin.de/) were used to predict their target mRNA. The final target genes were the intersection of the results of these three software packages.
Following this, the miRNA function was explored further using the Gene Ontology (GO) Database (http://www.geneontology.org/), via GO enrichment analysis. It was used as an analysis tool for target genes of miRNAs that were predicted. The ontology covers three domains: Cellular component, molecular function and biological process. The basic unit of GO is the GO term, each of which belongs to one type of ontology. It may provide a GO term significantly enriched in the predicted target genes involving the corresponding biological function. Therefore, the result may reveal the functions that are associated with the predicted target candidates of miRNAs. GO enrichment analysis of functional significance applies an ultra-geometric test to map all of the differentially-expressed genes to terms in the GO database. Furthermore, it looks for significantly enriched GO terms in genes and compares to the genome background. The formula used for calculating is the following:
Where N is the number of all the genes with GO annotation; n is the number of target predicted in N; M is the number of significant genes that are annotated to the certain GO terms and m is the number of significant genes that are found in M. All the GO terms with P≤0.01 were regarded as significantly enriched in the target genes (33).
A KEGG functional annotation analysis could also be performed using the DAVID (version 6.3; https://david.ncifcrf.gov/) bioinformatics resources in order to further analyze the pathways of the miRNA targets. As with the GO enrichment, the KEGG enrichment analysis of functional significance applies an ultra-geometric test to map all the differentially-expressed genes to terms in the DAVID bioinformatics resources, looking for significantly enriched KEGG terms in genes and comparing to the genome background. It is capable of identifying a significantly enriched metabolic or signal transduction pathway in target candidates. The calculating formula is the same as that of the GO enrichment analysis. However, N is the number of all the genes with KEGG annotation; n is the number of target predicted in N; M is the number of significant genes that are annotated to the certain KEGG terms and m is the number of significant genes that are found in M. KEGG analysis could reveal the main pathway that the target gene candidates are involved in.
Statistical analysis
SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA) was used to perform statistical analyses. Values were expressed as the mean ± standard error of the mean. Unpaired Student's tests were used to compare the arterial blood gas analyses and body temperature data between the control and sevoflurane groups. P<0.05 was used to indicate a statistically significant difference. Tukey test was applied to identify genes and miRNAs that demonstrated a significant differential expression upon exposure to sevoflurane.
Results
Arterial blood gas analyses and body temperature data
The color of the pups appeared pink throughout the 4-h sevoflurane exposure, indicating no change in respiration. The rats recovered rapidly from anesthesia and did not present any neurological symptoms or signs that indicate discomfort. Furthermore, no rats died during or after anesthesia. In order to examine the ventilation and oxygenation efficiency, blood gas analyses were performed at the end of anesthesia on six rats. Rats exposed to an air-oxygen mixture alone during the same period served as controls. The results of the blood gas analyses did not indicate signs of respiratory impairment. Furthermore, an analysis of several parameters, including pH, arterial oxygen, arterial CO2 tension, saturation of arterial blood oxygen and body temperature were not significantly different to the values observed for the control (t-test, all P-values >0.05; Table I).
Table I.
Arterial blood gas analyses and body temperature data for the control and sevoflurane anesthesia groups.
| Measurement | Control (n=6) | Sevoflurane (n=6) |
|---|---|---|
| pH | 7. 42±0.05 | 7.45±0.05 |
| PaCO2 (mmHg) | 35.33±6.02 | 38.67±7.31 |
| PaO2 (mmHg) | 146.50±22.53 | 132.83±21.59 |
| SaO2 (%) | 99.38±0.19 | 99.00±0.50 |
| Body temperature (°C) | 37.29±0.32 | 37.46±0.26 |
No between-group differences were statistically significant. Data represent the mean ± standard deviation for six animals in each group.
miRNA expression screening analysis
The results of the µParaflo miRNA microarray assay was presented in Table II. From the 727 known preload R. norvegicus miRNAs microarray, 291 miRNAs were detected. There were 141 out of 291 (48%) detected miRNAs that were differentially expressed with P<0.01 in this microarray. Among these differentially-expressed miRNAs, 53 miRNAs expressed signal values >500. Following sevoflurane exposure, 29 out of 53 miRNAs were upregulated in the sevoflurane group and 24 were downregulated compared with the control (Table II).
Table II.
miRNAs differentially-expressed in the hippocampus between the sevoflurane and control group as analyzed by a microarray at a signal value >500 and P<0.01.
| miRNA | Control group | Sevoflurane group | Fold change [Log2(sev/con)] | P-value |
|---|---|---|---|---|
| Expressed upregulated miRNAs in the sevoflurane group | ||||
| rno-miR-466b-1-3p | 37 | 603 | 4.02 | 3.61×10−7 |
| rno-miR-196c-3p | 51 | 715 | 3.80 | 1.75×10−4 |
| rno-miR-204-5p | 6,460 | 30,380 | 2.23 | 9.28×10−5 |
| rno-miR-214-3p | 2,240 | 5,221 | 1.22 | 7.21×10−5 |
| rno-miR-6216 | 5,290 | 11,869 | 1.17 | 5.46×10−4 |
| rno-miR-1224 | 354 | 742 | 1.07 | 8.87×10−4 |
| rno-miR-328a-5p | 311 | 607 | 0.96 | 1.74×10−5 |
| rno-miR-494-3p | 689 | 1,223 | 0.83 | 7.23×10−4 |
| rno-miR-146a-5p | 3,360 | 5,830 | 0.80 | 4.61×10−4 |
| rno-miR-539-5p | 982 | 1,695 | 0.79 | 1.48×10−6 |
| rno-miR-379-3p | 287 | 451 | 0.65 | 8.84×10−4 |
| rno-miR-379-5p | 919 | 1,419 | 0.63 | 3.63×10−4 |
| rno-miR-185-5p | 415 | 630 | 0.60 | 3.80×10−3 |
| rno-miR-195-5p | 2,802 | 4,189 | 0.58 | 2.30×10−5 |
| rno-miR-411-3p | 1,148 | 1,694 | 0.56 | 3.42×10−5 |
| rno-miR-377-3p | 331 | 484 | 0.55 | 2.71×10−3 |
| rno-miR-15b-5p | 1,437 | 2,082 | 0.53 | 3.45×10−4 |
| rno-miR-384-5p | 2,693 | 3,873 | 0.52 | 3.31×10−5 |
| rno-miR-25-3p | 696 | 990 | 0.51 | 5.96×10−4 |
| rno-miR-872-5p | 451 | 633 | 0.49 | 5.72×10−3 |
| rno-miR-98-5p | 1,630 | 2,278 | 0.48 | 2.35×10−3 |
| rno-miR-382-3p | 589 | 794 | 0.43 | 9.27×10−4 |
| rno-miR-151-5p | 1,378 | 1,852 | 0.43 | 1.21×10−3 |
| rno-miR-487b-3p | 2,778 | 3,713 | 0.42 | 6.72×10−3 |
| rno-miR-411-5p | 737 | 907 | 0.30 | 5.82×10−4 |
| rno-miR-335 | 4,610 | 5,664 | 0.30 | 6.28×10−3 |
| rno-miR-323-3p | 971 | 1,173 | 0.27 | 7.48×10−3 |
| rno-miR-342-3p | 1,781 | 2,149 | 0.27 | 4.04×10−3 |
| rno-miR-26a-5p | 10,297 | 12,154 | 0.24 | 3.61×10−3 |
| Expressed downregulated miRNAs in the sevoflurane group | ||||
| rno-miR-125b-5p | 21,793 | 17,586 | −0.31 | 3.94×10−4 |
| rno-miR-103-3p | 3,418 | 2,750 | −0.31 | 3.79×10−3 |
| rno-miR-708-5p | 515 | 408 | −0.34 | 4.61×10−3 |
| rno-miR-124-3p | 7,939 | 5,998 | −0.40 | 7.19×10−4 |
| rno-miR-128-3p | 12,516 | 9,354 | −0.42 | 1.57×10−7 |
| rno-miR-29b-3p | 543 | 390 | −0.48 | 1.49×10−3 |
| rno-miR-341 | 597 | 427 | −0.48 | 8.53×10−3 |
| rno-miR-153-3p | 986 | 643 | −0.62 | 1.50×10−3 |
| rno-miR-376a-3p | 654 | 425 | −0.62 | 1.56×10−3 |
| rno-miR-331-3p | 689 | 440 | −0.65 | 6.47×10−4 |
| rno-miR-127-3p | 2,956 | 1,875 | −0.66 | 8.38×10−4 |
| rno-miR-9a-5p | 20,004 | 12,387 | −0.69 | 4.73×10−5 |
| rno-miR-101a-3p | 494 | 304 | −0.70 | 1.06×10−3 |
| rno-miR-218a-5p | 2,393 | 1,410 | −0.76 | 2.58×10−3 |
| rno-miR-17-5p | 7,110 | 4,180 | −0.77 | 6.03×10−6 |
| rno-miR-21-5p | 590 | 332 | −0.83 | 5.65×10−4 |
| rno-miR-20a-5p | 679 | 376 | −0.85 | 8.96×10−6 |
| rno-miR-106b-5p | 527 | 284 | −0.89 | 5.15×10−5 |
| rno-miR-29a-3p | 621 | 317 | −0.97 | 3.52×10−5 |
| rno-miR-135b-5p | 457 | 232 | −0.98 | 1.89×10−4 |
| rno-miR-19b-3p | 788 | 364 | −1.11 | 2.25×10−5 |
| rno-miR-138-5p | 1,515 | 693 | −1.13 | 2.24×10−3 |
| rno-miR-137-3p | 1,791 | 656 | −1.45 | 1.81×10−3 |
| rno-miR-551b-3p | 531 | 148 | −1.85 | 2.62×10−5 |
miRNA, microRNA.
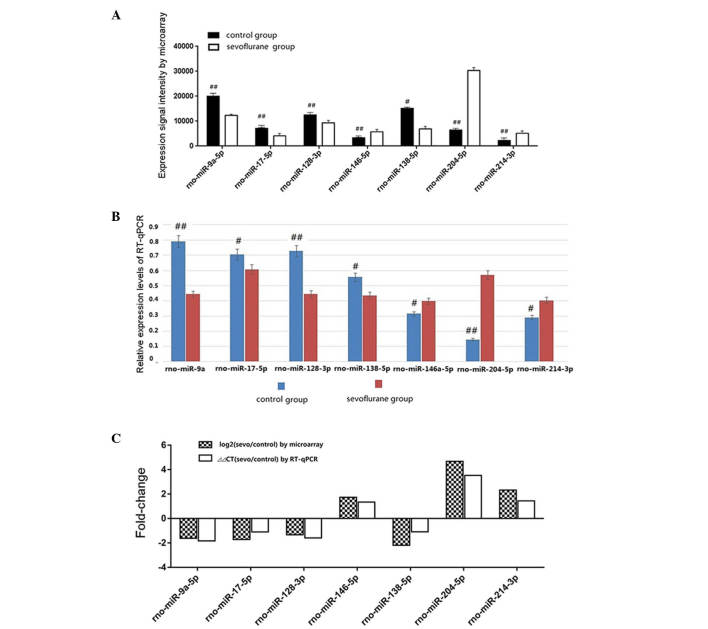
A heat map is a graphical representation of data in which individual values contained in a matrix are represented by colors. Differentially-expressed miRNAs were transferred into the heat map in order to easily understand the situation of the miRNA screening of each sample. Each column represents a sample and each row displays miRNA. Upregulated miRNAs in the sevoflurane group are shown in red, whereas downregulated miRNAs are presented as green (Fig. 1).
Figure 1.

Heat map for the analysis of differentially-expressed miRNA. Each column represents samples and each row represents miRNAs. Red represents upregulated miRNAs and green shows downregulated miRNAs in the sevoflurane group. The first four columns on each figure represent the level of expression in the control group, while the last four columns represent the level of corresponding miRNA expression in the sevoflurane group. The heat map for 53/291 detected miRNAs (18.2%) that were differentially expressed with P<0.01 and signal value >500 are depicted.
Single validation of representative miRNA by qPCR
To confirm the accuracy of the µParaflo microfluidic chip, nine differentially-expressed miRNAs were selected, which have been reported to be associated with brain development, cognition or both (23–31) and were used for qPCR in triplicate. From the results of the qPCR, the expression levels of miR-204, miR-214 and miR-146a-5p were identified to be upregulated the in sevoflurane group and miR-9a-5p, miR-128, miR-138 and miR-17-5p were downregulated. In addition, rno-U87 was used as an internal control due to its stable expression in the rats' hippocampi. The results indicated that the fold change of miR-204, miR-17-5p, miR-128, miR-146a-5p, miR-9a-5p, miR-138 and miR-214 measured by qPCR were in concordance with the µParaflo microarray data (Table III; Fig. 2).
Table III.
Primer sequence for quantitative polymerase chain reaction.
| miRNA | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Length |
|---|---|---|---|
| rno-U87 | ACAATGATGACTTATGTTTTT | GCTCAGTCTTAAGATTCTCT | 21 |
| rno-miR-17-5p | GCCGCCAAAGTGCTTACAGT | CCAGTGCAGGGTCCGAGGT | 20 |
| rno-miR-9a-5p | ACGGCGGTCTTTGGTTATCTA | CAGTGCAGGGTCCGAGGTAT | 21 |
| rno-miR-128-3p | CAGCCGTCACAGTGAACCG | TCCACACCACCTGAGCCG | 20 |
| rno-miR-137-3p | GGCGGCGGTTATTGCTTA | CAGTGCAGGGTCCGAGGTAT | 18 |
| rno-miR-138-5p | GCGAGCTGGTGTTGTGAATC | CAGTGCAGGGTCCGAGGTAT | 20 |
| rno-miR-204-5p | TGCCTTCCCTTTGTCATCCTAT | TCCACACCACCTGAGCCG | 21 |
| rno-miR-214-3p | GCGGACAGCAGGCACAGACAG | CAGTGCAGGGTCCGAGGTATTC | 22 |
| rno-miR-146a-5p | GCGGCGGTGAGAACTGAAT | CAGTGCAGGGTCCGAGGTAT | 20 |
| rno-miR-124-3p | CCGACGGTAAGGCACGC | CGCAGGGTCCGAGGTATTC | 17 |
miRNA, microRNA.
Figure 2.
Correlation of miRNA between microarray and RT-qPCR. (A) Results of relative detectable signal intensity of differentially-expressed miRNAs between the sevoflurane and control groups by microarray. (B) The results of the relative expression level of differentially-expressed miRNAs analyzed by qPCR. (C) Comparison of qPCR observations to microarray results by fold-change of 7-selected miRNAs. #P<0.05, ##P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miRNA predicted targets and relevant bioinformatic analysis
To improve our understanding of the potential role in biosynthesis of these nine differentially-expressed miRNAs, three software packages were used (TargetScan, miRanda and PicTar), the Gene Ontology Database and the DAVID bioinformatics resources for target prediction, GO enrichment analysis and KEGG annotation analysis. A total of 306 putative target genes of the nine differentially-expressed miRNAs in two groups were identified using the three software packages (data not shown). Table IV lists a total of 85 predicted target genes of nine miRNAs. These genes were identified to be associated to brain development and cognitive function, implying that miRNAs are involved in regulating the pathological processes of sevoflurane-induced neurotoxicity by targeting these genes. In addition, the present study revealed that the majority of miRNAs had multiple potential target genes, whereas some unigenes may also be targeted by more than one miRNA.
Table IV.
Predicted miRNA target genes associated with brain development and cognitive function.
| miRNA | Count | Target genes |
|---|---|---|
| rno-miR-9a-5p | 1 | Arid3b |
| rno-miR-17-5p | 7 | Cct6a Limk1 Mmaa Klf9 Lzic Plekhm1 Mtmr3 |
| rno-miR-128-3p | 8 | Stra6 Fam83f Gosr1 Kcnk2 Leng1 Mrpl13 Slc39a13 Surf6 |
| rno-miR-146a-5p | 4 | Chmp3 Efha1 Hapln1 Rhov |
| rno-miR-138-5p | 12 | Sh2b2 Olig1 Vamp4 Gosr1 Ftsjd2 Derl1 Cav3 Nup210 Plxnb2 Pdrg1 Nrarp Ankrd54 |
| rno-miR-204-5p | 12 | Arid3b Comtd1 Cyp4a8 Dnmt3a Ephb6 Fam134c Mcoln1 Pitx1 Pvalb Slc1a1 Tnpo3 Xirp2 |
| rno-miR-214-3p | 23 | Adora2a Nodal Slc7a5 Emx1 Sox8 Grin1 S100b Dll1 Dvl1 Mapk8ip2 Cplx1 Apba1 Prps2 Abt1 Ephb1 Kcnj10 Lgi4 Uhmk1 Pld2 Cntnap1 Plxdc1 Txn2 Vsx2 |
| rno-miR-125b-5p | 13 | C1qtnf1 Psd4 Sstr3 Cdk16 Suv420h2 Map3k11 Zfhx2 Sgsm2 Phf7 Foxo4 Slc26a10 Tex19 Cdk16 |
| rno-miR-124-3p | 5 | Parp16 Frzb Ftsjd1 Tlx1 Emx1 |
miRNA, microRNA.
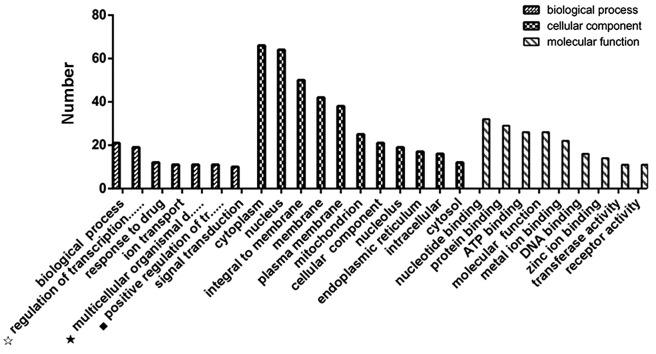
With the use of GO (Gene ontology; http://amigo.geneontology.org/amigo), these putative miRNA targets can be used as an input in order to perform the GO functional enrichment. From the enrichment results, the main 27 GO terms are shown in Fig. 3. These results could demonstrate that the target genes of nine miRNAs in neonatal rats' hippocampus are involved in a wide variety of pathophysiological processes after sevoflurane exposure. In order to explore the possible mechanism of neurotoxicity induced by sevoflurane, 19 GO terms were selected that were thought to be associated to brain development and cognition among the ~3,200 GO terms of the nine miRNAs (Table V). The most significant GOs associated with the nine cognition-related miRNAs included the neuronal cell body (GO:0043025), dendrite (GO:0030425) and synapse (GO:0045202).
Figure 3.
Categories and distribution of the gene ontology terms of predicted targets for nine miRNAs. In the biological process, significant target genes were distributed in the regulation of transcription, DNA-dependency, response to drug, ion transport, multicellular organismal development, positive regulation of transcription from RNA, polymerase II promoter and signal transduction. In the field of the cellular component, genes are enriched in the cytoplasm, nucleus, integral to membrane, membrane, plasma membrane, mitochondrion, nucleolus, endoplasmic reticulum, intracellular part and the cytosol. For the molecular function, the major significant genes are enriched in nucleotide, protein, ATP, metal ion, DNA, zinc ion binding, and transferase and receptor activity. ☆Regulation of transcription, DNA-dependency, ★multicellular organismal development. ◆Positive regulation of transcription from RNA polymerase II promoter. miRNA, microRNA; ATP, adenosine triphosphate.
Table V.
GO functional enrichment for target genes involved in brain development and cognitive function.
| GO term | Count | Target genes |
|---|---|---|
| GO:0007268-synaptic transmission | 3 | Slc17a7, Lrp6, Apba1 |
| GO:0007420-brain development | 2 | Nodal, Emx1 |
| GO:0007611-learning or memory | 2 | Grin1, S100b |
| GO:0007616-long-term memory | 2 | Grin1, Slc17a7 |
| GO:0014069-postsynaptic density | 3 | Grin1, Adora2a, Mapk8ip2 |
| GO:0030182-neuron differentiation | 2 | Tlx1, Emx1 |
| GO:0031175-neuron projection development | 3 | Uhmk1, Pld2, Cntnap1 |
| GO:0043025-neuronal cell body | 8 | Lrp6, Adora2a, S100b Plxdc1, Mapk8ip2, Cplx1, Txn2, Dvl1 |
| GO:0045202-synapse | 6 | Slc17a7, Rims2, Lrp6, Grin1, Cplx1, Dvl1 |
| GO:0048167-regulation of synaptic plasticity | 2 | Grin1, Adora2a |
| GO:0048168-regulation of neuronal synaptic plasticity | 2 | Grin1, S100b |
| GO:0048169-regulation of long-term neuronal synaptic plasticity | 2 | Grin1, Kcnj10 |
| GO:0060079-regulation of excitatory postsynaptic membrane potential | 4 | Slc17a7, Grin1, Adora2a, Mapk8ip2 |
| GO:0007417-central nervous system development | 2 | Tlx1, Acvr1b |
| GO:0021987-cerebral cortex development | 3 | Lrp6, Grin1, Emx1 |
| GO:0030425-dendrite | 8 | Clip2, Ephb1, Grin1, Adora2a, Txn2, Plxdc1, Cplx1, Dvl1 |
| GO:0007399-nervous system development | 4 | Sh2b2, Nodal, Slc7a5, Dll1 |
| GO:0043525-positive regulation of neuron apoptotic process | 2 | Map3k11, Casp9 |
| GO:0035249-synaptic transmission, glutamatergic | 3 | Slc17a7, Grin1, Adora2a |
GO, gene ontology.
In order to evaluate the biological pathway involved in sevoflurane induced neurotoxicity, the DAVID gene annotation tool (http://david.abcc.ncifcrf.gov/) was also used to perform KEGG pathway annotation of the miRNA targets (Table VI). The main enriched pathway were metabolic, with 18 annotated genes representing 22.2%, followed by the MAPK signaling pathway (7 target genes for 8.6%) and Wnt signaling pathway (5 target genes for 0.2%) (Table VI). KEGG pathway analysis revealed some of the biological processes that may be involved in sevoflurane-induced neurotoxicity, and provided useful insights to investigate further the miRNAs target in the neurotoxic effects of sevoflurane.
Table VI.
KEGG pathway annotations for the nine miRNAs targets.
| Pathway ID | Pathway description | Gene number | P-value |
|---|---|---|---|
| 270 | Cysteine and methionine metabolism | 3 | 0.0157a |
| 4114 | Oocyte meiosis | 5 | 0.0332a |
| 4130 | SNARE interactions in vesicular transport | 3 | 0.0374a |
| 4320 | Dorso-ventral axis formation | 2 | 0.0457a |
| 5014 | Amyotrophic lateral sclerosis | 3 | 0.0601 |
| 3320 | PPAR signaling pathway | 3 | 0.0878 |
| 4360 | Axon guidance | 5 | 0.1004 |
| 4310 | Wnt signaling pathway | 5 | 0.1073 |
| 564 | Glycerophospholipid metabolism | 4 | 0.1074 |
| 5010 | Alzheimer's disease | 4 | 0.116 |
| 4010 | MAPK signaling pathway | 7 | 0.1989 |
| 5012 | Parkinson's disease | 2 | 0.2719 |
| 5016 | Huntington's disease | 3 | 0.3061 |
| 4720 | Long-term potentiation | 2 | 0.3299 |
| 4722 | Neurotrophin signaling pathway | 3 | 0.4341 |
| 1100 | Metabolic pathways | 18 | 0.4969 |
| 4120 | Ubiquitin mediated proteolysis | 1 | 0.8667 |
| 4080 | Neuroactive ligand-receptor interaction | 2 | 0.8735 |
P<0.05. KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; SNARE, soluble NSF attachment protein receptor; PPAR, peroxisome proliferator-activated receptor; MAPK. mitogen-activated protein kinase.
Discussion
Volatile anesthetics have an effect on cerebral blood flow and the metabolism (34,35), voltage-gated sodium channels (36), signaling via γ-aminobutyric acid receptors (37) and N-methyl-d-aspartate receptors (38), and these effects occur during anesthesia. In the present study, the hippocampal miRNAs expression profiles in neonatal rats were analyzed at 24 h after 1 minimum alveolar concentration sevoflurane exposure for 4 h. The changes in miRNA expression in neonatal rat hippocampi occurred for longer periods following the withdrawal of anesthetic gases. Therefore, these changes may be associated to the neurotoxicity induced by sevoflurane, but not due to the anesthetic effects of volatile anesthetics.
In the present study, one minimum alveolar concentration (2.4%) of sevoflurane was administered because this dose was as effective and was the same as the clinical doses used (39). A similar dose was also used in previous studies (40,41). None of these pups appeared to undergo hypoxemia or hypocapnia during the sevoflurane exposure for 4 h, as verified by their arterial blood analysis. Furthermore, there were no differences observed in the results of the arterial blood gas analysis of the sevoflurane-exposed group compared to the control. These results rule out the contribution of hypercapnia and hypoxemia to the miRNAs expression in rat pups' hippocampus.
The microfluidic chip assay is an efficient method to generate miRNA expression profiles. This method has previously been used to study the functional linkage between miRNAs and physiological or pathological processes (42,43). The present study used the microfluidic chip assay technique on neonatal rat hippocampi after sevoflurane exposure, which revealed an abnormal miRNA expression pattern. Next, several potential miRNA targets and their possible underlying molecular mechanisms were discussed. In the present study, 53 differentially-expressed miRNAs, including miR-124, miR-128, miR-214, miR-146a-5p, miR-9a-5p, miR138, miR137, miR-17-5p (44) and miR-204 are known to be involved in brain development and cognitive function (45). For example, multiple lines of evidence indicated that miR-124 was abundant in developing the mammalian brain and are important in hippocampal demyelination and memory dysfunction (46). In addition, it was revealed that melatonin can rescue the expression of synaptic proteins and the impairment of dendritic spines by activating the EPACs/miR-124/Egr1 signaling pathway (47). Furthermore, it has been demonstrated that when miR-128, a brain enriched miRNA, is overexpressed it causes morphological changes in SH-SY5Y neuroblastoma cells, implying that miRNA-128 was involved in neuronal differentiation (48). For miR-214, recent observations indicate that desflurane, a new inhaled anesthetic, could accelerate neuronal cytotoxicity of Aβ by downregulating miR-214 (49). Furthermore, it has been revealed that miR-146a was upregulated in brain white matter lesions from multiple sclerosis patients and in a mouse model as well (50). In addition, Kong et al (51) found that miRNA-9a may be involved in the pathogenesis of AD by having an effect on the metabolism of amino acids in the brain. Miska et al demonstrated that the levels of mir-138 expression increase with age in a developing brain of a rat reaching a peak in juvenile and adult rats (52). Furthermore, mir-138 was also revealed to be a negative regulator of dendritic spine size, which may possibly occur by tuning the activity of antagonistic signaling that are known to regulate the actin cytoskeleton in spines (53). miR-137 is capable of regulating synaptic plasticity and neurogenesis, and recently it has been confirmed as an important susceptibility-related gene for schizophrenia (54). Finally, these observations indicate that dysregulated expression of miRNAs may be involved in the mechanism of sevoflurane-induced neurotoxicity.
Since miRNAs primarily control target mRNA, they can regulate the basic activities of cells and tissues. Therefore, the prediction of the target genes should be significant in elucidating the biological function of the miRNAs. For instance, it has been demonstrated that downregulation of miR-214 was associated with the neurotoxicity of Aβ induced by desflurane (48). Among its target genes, Grin1, S100b, Slc17a7, Dvl1 and Adora2a had been reported to participate in some neurological disorders (55–59). The Grin1 gene, that is a subunit of N-methyl-D-aspartate receptors, is important in the excitatory neurotransmission, synaptic plasticity and brain development (60). Furthermore, it is suggested to be a candidate susceptibility gene for neuropsychiatric disorders, including bipolar disorder, schizophrenia and attention deficit and hyperactivity disorder (61). In addition, an increasing number of studies focused on the role of S100β in cognitive function. In previous large-scale investigations, researchers regarded the S100β protein as the marker to evaluate the neurocognitive outcome following cardiac surgery (55,62,63). It is likely that these target genes are important in sevoflurane-induced neurotoxicity. However, a simple prediction of miRNA using bioinformatics software is not enough. In the future, pertinent miRNA transfection and a luciferase gene assay will be performed to verify the authenticity and accuracy of the predicted target genes.
Following the GO analysis, the involvement of predicted targets of miRNAs in a variety of biological processes was shown. A positive regulation of the neuron apoptotic process, dendrite, nervous system development, regulation of synaptic plasticity, neuron differentiation and synaptic transmission, which are correlated to brain development and cognition, were also enriched as seen from the predicted targets, indicating that these targets may be important in the mechanism of sevoflurane-induced neurotoxicity.
In an attempt to further characterize the molecular response to sevoflurane in the rat pups' hippocampus, nine differentially-expressed miRNAs were classified in order to identify the potential molecular signaling pathway according to the KEGG functional annotations. Few candidate miRNAs (miR-124, miR-128, miR-214, miR-146a-5p, miR-9a-5p, miR138, miR137, miR-17-5p41 and miR-204) and few target genes, identified using the three predicted tools, are thought to be the main reason that there were only four pathways with a significant difference. Nevertheless, the present study also identified that within the MAPK and Wnt signaling pathways a number of targets existed that are known to be closely associated with synaptic dysfunction, hippocampal neurogenesis and behavioral deficits in rats. For the MAPK signaling pathway, sevoflurane may lead to neuroapoptosis by suppressing the ERK1/2 MAPK signaling in a developing brain (64,65). For the Wnt signaling pathway, its importance of maintaining and protecting neural connections throughout the lifespan has been recognized gradually and numerous studies regarded it as a potential strategy for neuroprotection or potential regulator of hippocampal synapses that could impact learning and memory (66,67). In addition, further study of the MAPK and Wnt signaling pathways may be helpful to elucidate the mechanism of neurotoxicity induced by sevoflurane. The present study proposes that these KEGG results will provide new insights into the neurotoxicity and neuroprotection of volatile anesthetics on infants.
Nevertheless, the present study included a number of limitations: i) The experimental times under anesthesia were only 4 h, and thus it is required to verify time-dependent changes of miRNA expression; ii) although a great deal of studies have validated that 2.4% sevoflurane treatment could impair the learning and memory function of neonatal rats (35,68,69), the present study could not confirm the cause-effect association between altered expression of miRNAs and sevoflurane neurotoxicity due to the lack of specific animal models of sevoflurane-induced neurotoxicity; iii) despite the fact that a microfluidics chip is a new and mature sequencing technology, its results still have a risk for false positive rates (FPRs). To reduce the FPR, certain single miRNAs were selected to perform validation. However, the number of single miRNA validation is limited, therefore the risk of FPR still remains; and iv) finally in order to explore the bioinformatic background of miRNAs in a more concentrated manner, nine differentially-expressed miRNAs among ~141 were selected to undertake the bioinformatics analysis. Therefore the KEGG functional annotation of nine miRNAs was not evident and it may be that certain other significant signaling pathways were missed; v) while bioinformatics is a useful tool for identifying potential targets of a specific miRNA, its reliability is still a matter of concern. The present study neither performed functional in vivo or in vitro tests to confirm the predicted targets, nor did it validate the roles of these candidates in regulating the signaling pathways that were identified in the analysis. Thus, further studies are required to elucidate the role of differentially-expressed miRNA in sevoflurane-induced neurotoxicity.
In conclusion, the results of the present study demonstrated an aberrant miRNA expression pattern in the neonatal rats' hippocampus following sevoflurane exposure. Bioinformatics analysis of these differentially-expressed miRNAs offer insights into the mechanism of neurotoxicity induced by sevoflurane and sheds light on novel strategies that can be used to prevent and treat sevoflurane neurotoxicity in a developing brain. However, further studies are required to verify the potential functions of these miRNAs.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81371195), Natural Science Foundation of Hubei Province (grant no. 2013CFB278), Excellent Youth Foundation of Hubei Scientific Committee (grant no. 2014CFA046) and Wuhan Foundation for the key members of young doctors.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: Rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184:2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity - a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014;369:1652. doi: 10.1098/rstb.2013.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 5.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrié S, Anderson TA. Volatile anesthetics for status asthmaticus in pediatric patients: A comprehensive review and case series. Paediatr Anaesth. 2015;25:460–467. doi: 10.1111/pan.12577. [DOI] [PubMed] [Google Scholar]
- 8.Jevtovic-Todorovic V, Hartman RE, Izumi Y, Benshoff ND, Dikranian K, Zorumski CF, Olney JW, Wozniak DF. Early exposure to common anesthetic agents causes widespread neurodegeneration in the developing rat brain and persistent learning deficits. J Neurosci. 2003;23:876–882. doi: 10.1523/JNEUROSCI.23-03-00876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilder RT, Flick RP, Sprung J, Katusic SK, Barbaresi WJ, Mickelson C, Gleich SJ, Schroeder DR, Weaver AL, Warner DO. Early exposure to anesthesia and learning disabilities in a population-based birth cohort. Anesthesiology. 2009;110:796–804. doi: 10.1097/01.anes.0000344728.34332.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Li S, Niu X, Wang P, Wang J, Zhang M. Protective effect of FTY720 against sevoflurane-induced developmental neurotoxicity in rats. Cell Biochem Biophys. 2013;67:591–598. doi: 10.1007/s12013-013-9546-3. [DOI] [PubMed] [Google Scholar]
- 11.Fredriksson A, Ponten E, Gordh T, Eriksson P. Neonatal exposure to a combination of N-methyl-D-aspartate and gamma-aminobutyric acid type A receptor anesthetic agents potentiates apoptotic neurodegeneration and persistent behavioral deficits. Anesthesiology. 2007;107:427–436. doi: 10.1097/01.anes.0000278892.62305.9c. [DOI] [PubMed] [Google Scholar]
- 12.Satomoto M, Satoh Y, Terui K, Miyao H, Takishima K, Ito M, Imaki J. Neonatal exposure to sevoflurane induces abnormal social behaviors and deficits in fear conditioning in mice. Anesthesiology. 2009;110:628–637. doi: 10.1097/ALN.0b013e3181974fa2. [DOI] [PubMed] [Google Scholar]
- 13.Stratmann G, Sall JW, May LD, Bell JS, Magnusson KR, Rau V, Visrodia KH, Alvi RS, Ku B, Lee MT. Isoflurane differentially affects neurogenesis and long-term neurocognitive function in 60-day-old and 7-day-old rats. Anesthesiology. 2009;110:834–848. doi: 10.1097/ALN.0b013e31819c463d. [DOI] [PubMed] [Google Scholar]
- 14.Kodama M, Satoh Y, Otsubo Y, Araki Y, Yonamine R, Masui K, Kazama T. Neonatal desflurane exposure induces more robust neuroapoptosis than do isoflurane and sevoflurane and impairs working memory. Anesthesiology. 2011;115:979–991. doi: 10.1097/ALN.0b013e318234228b. [DOI] [PubMed] [Google Scholar]
- 15.Gentry KR, Steele LM, Sedensky MM, Morgan PG. Early developmental exposure to volatile anesthetics causes behavioral defects in Caenorhabditis elegans. Anesth Analg. 2013;116:185–189. doi: 10.1213/ANE.0b013e31826d37c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H, Dong Y, Xu Z, Crosby G, Culley DJ, Zhang Y, Xie Z. Sevoflurane anesthesia in pregnant mice induces neurotoxicity in fetal and offspring mice. Anesthesiology. 2013;118:516–526. doi: 10.1097/ALN.0b013e3182834d5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satomoto M, Satoh Y, Terui K, Miyao H, Takishima K, Ito M, Imaki J. Neonatal exposure to sevoflurane induces abnormal social behaviors and deficits in fear conditioning in mice. Anesthesiology. 2009;110:628–637. doi: 10.1097/ALN.0b013e3181974fa2. [DOI] [PubMed] [Google Scholar]
- 18.Wang WY, Wang H, Luo Y, Jia LJ, Zhao JN, Zhang HH, Ma ZW, Xue QS, Yu BW. The effects of metabotropic glutamate receptor 7 allosteric agonist N,N'-dibenzhydrylethane-1,2-diamine dihydrochloride on developmental sevoflurane neurotoxicity: Role of extracellular signal-regulated kinase 1 and 2 mitogen-activated protein kinase signaling pathway. Neuroscience. 2012;205:167–177. doi: 10.1016/j.neuroscience.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Cheng Y, Liu G, Tian X, Tu X, Wang J. Chronic exposure of gestation rat to sevoflurane impairs offspring brain development. Neurol Sci. 2012;33:535–544. doi: 10.1007/s10072-011-0762-6. [DOI] [PubMed] [Google Scholar]
- 20.Kalenka A, Hinkelbein J, Feldmann RE, Jr, Kuschinsky W, Waschke KF, Maurer MH. The effects of sevoflurane anesthesia on rat brain proteins: A proteomic time-course analysis. Anesth Analg. 2007;104:1129–1135. doi: 10.1213/01.ane.0000260799.37107.e6. [DOI] [PubMed] [Google Scholar]
- 21.Pan Z, Lu XF, Shao C, Zhang C, Yang J, Ma T, Zhang LC, Cao JL. The effects of sevoflurane anesthesia on rat hippocampus: A genomic expression analysis. Brain Res. 2011;1381:124–133. doi: 10.1016/j.brainres.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, Kato I. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: A possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, Lukiw WJ. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011;499:109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Bian S, Zhang J, Zhang H, Tang B, Sun T. The Silencing Effect of microRNA miR-17 on p21 Maintains the Neural Progenitor Pool in the Developing Cerebral Cortex. Front Neurol. 2014;5:132. doi: 10.3389/fneur.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilje P, Gidlöf O, Rundgren M, Cronberg T, Al-Mashat M, Olde B, Friberg H, Erlinge D. The brain-enriched microRNA miR-124 in plasma predicts neurological outcome after cardiac arrest. Crit Care. 2014;18:R40. doi: 10.1186/cc13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ignacio C, Mooney SM, Middleton FA. Effects of Acute Prenatal Exposure to Ethanol on microRNA Expression are Ameliorated by Social Enrichment. Front Pediatr. 2014;2:103. doi: 10.3389/fped.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong Y, Wu J, Yuan L. MicroRNA expression analysis of adult-onset Drosophila Alzheimer's disease model. Curr Alzheimer Res. 2014;11:882–891. doi: 10.2174/1567205011666141001121416. [DOI] [PubMed] [Google Scholar]
- 30.Schröder J, Ansaloni S, Schilling M, Liu T, Radke J, Jaedicke M, Schjeide BM, Mashychev A, Tegeler C, Radbruch H, et al. MicroRNA-138 is a potential regulator of memory performance in humans. Front Hum Neurosci. 2014;8:501. doi: 10.3389/fnhum.2014.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, Li K. miR-137: A new player in schizophrenia. Int J Mol Sci. 2014;15:3262–3271. doi: 10.3390/ijms15023262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Pan Y, Cheng B, Chen J, Bai B. Identification of Conserved and Novel microRNAs in Cerebral Ischemia-Reperfusion Injury of Rat Using Deep Sequencing. J Mol Neurosci. 2014;54:671–683. doi: 10.1007/s12031-014-0383-7. [DOI] [PubMed] [Google Scholar]
- 34.Mielck F, Stephan H, Buhre W, Weyland A, Sonntag H. Effects of 1 MAC desflurane on cerebral metabolism, blood flow and carbon dioxide reactivity in humans. Br J Anaesth. 1998;81:155–160. doi: 10.1093/bja/81.2.155. [DOI] [PubMed] [Google Scholar]
- 35.Yonamine R, Satoh Y, Kodama M, Araki Y, Kazama T. Coadministration of hydrogen gas as part of the carrier gas mixture suppresses neuronal apoptosis and subsequent behavioral deficits caused by neonatal exposure to sevoflurane in mice. Anesthesiology. 2013;118:105–113. doi: 10.1097/ALN.0b013e318275146d. [DOI] [PubMed] [Google Scholar]
- 36.Ratnakumari L, Vysotskaya TN, Duch DS, Hemmings HC., Jr Differential effects of anesthetic and nonanesthetic cyclobutanes on neuronal voltage-gated sodium channels. Anesthesiology. 2000;92:529–541. doi: 10.1097/00000542-200002000-00037. [DOI] [PubMed] [Google Scholar]
- 37.Gyulai FE, Mintun MA, Firestone LL. Dose-dependent enhancement of in vivo GABA (A)-benzodiazepine receptor binding by isoflurane. Anesthesiology. 2001;95:585–593. doi: 10.1097/00000542-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 38.Ming Z, Knapp DJ, Mueller RA, Breese GR, Criswell HE. Differential modulation of GABA- and NMDA-gated currents by ethanol and isoflurane in cultured rat cerebral cortical neurons. Brain Res. 2001;920:117–124. doi: 10.1016/S0006-8993(01)03044-X. [DOI] [PubMed] [Google Scholar]
- 39.Obal D, Preckel B, Scharbatke H, Müllenheim J, Höterkes F, Thämer V, Schlack W. One MAC of sevoflurane provides protection against reperfusion injury in the rat heart in vivo. Br J Anaesth. 2001;87:905–911. doi: 10.1093/bja/87.6.905. [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa M, Tanaka S, Arai M, Genda Y, Sakamoto A. Differences in microRNA changes of healthy rat liver between sevoflurane and propofol anesthesia. Anesthesiology. 2012;117:1245–1252. doi: 10.1097/ALN.0b013e3182746676. [DOI] [PubMed] [Google Scholar]
- 41.Goto G, Hori Y, Ishikawa M, Tanaka S, Sakamoto A. Changes in the gene expression levels of microRNAs in the rat hippocampus by sevoflurane and propofol anesthesia. Mol Med Rep. 2014;9:1715–1722. doi: 10.3892/mmr.2014.2038. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Zheng Y, Wang G, Li H. Identification of microRNA and bioinformatics target gene analysis in beef cattle intramuscular fat and subcutaneous fat. Mol Biosyst. 2013;9:2154–2162. doi: 10.1039/c3mb70084d. [DOI] [PubMed] [Google Scholar]
- 43.Katchy A, Williams C. Profiling of estrogen-regulated microRNAs in breast cancer cells. J Vis Exp. 2014;e51285 doi: 10.3791/51285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaalund SS, Venø MT, Bak M, Møller RS, Laursen H, Madsen F, Broholm H, Quistorff B, Uldall P, Tommerup N, et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia. 2014;55:2017–2027. doi: 10.1111/epi.12839. [DOI] [PubMed] [Google Scholar]
- 46.Dutta R, Chomyk AM, Chang A, Ribaudo MV, Deckard SA, Doud MK, Edberg DD, Bai B, Li M, Baranzini SE, et al. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann Neurol. 2013;73:637–645. doi: 10.1002/ana.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Wang ZH, Wu YY, Tang H, Tan L, Wang X, Gao XY, Xiong YS, Liu D, Wang JZ. Melatonin attenuates scopolamine-induced memory/synaptic disorder by rescuing EPACs/miR-124/Egr1 pathway. Mol Neurobiol. 2013;47:373–381. doi: 10.1007/s12035-012-8355-9. [DOI] [PubMed] [Google Scholar]
- 48.Guidi M, Muiños-Gimeno M, Kagerbauer B, Marti E, Estivill X, Espinosa-Parrilla Y. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol Biol. 2010;11:95. doi: 10.1186/1471-2199-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, Zhang Y. Desflurane accelerates neuronal cytotoxicity of Aβ by downregulating miR-214. Neurosci Lett. 2013;554:28–33. doi: 10.1016/j.neulet.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 50.Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, Singh N, Nagarkatti M, Nagarkatti P. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci. 2014;11:810–818. doi: 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong Y, Wu J, Yuan L. MicroRNA Expression Analysis of Adult-Onset Drosophila Alzheimer's Disease Model. Curr Alzheimer Res. 2014;11:882–891. doi: 10.2174/1567205011666141001121416. [DOI] [PubMed] [Google Scholar]
- 52.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, Li K. miR-137: A new player in schizophrenia. Int J Mol Sci. 2014;15:3262–3271. doi: 10.3390/ijms15023262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rothermundt M, Peters M, Prehn JH, Arolt V. S100B in brain damage and neurodegeneration. Microsc Res Tech. 2003;60:614–632. doi: 10.1002/jemt.10303. [DOI] [PubMed] [Google Scholar]
- 56.Ciani L, Boyle KA, Dickins E, Sahores M, Anane D, Lopes DM, Gibb AJ, Salinas PC. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin-dependent protein kinase II. Proc Natl Acad Sci USA. 2011;108:10732–10737. doi: 10.1073/pnas.1018132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkinson MB, Dias C, Magida J, Mazei-Robison M, Lobo M, Kennedy P, Dietz D, Covington H, III, Russo S, Neve R, et al. A novel role of the WNT-dishevelled-GSK3β signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J Neurosci. 2011;31:9084–9092. doi: 10.1523/JNEUROSCI.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Yu T, Huang X, Cao Y, Li X, Liu B, Yang F, Li W, Zhao X, Feng G, et al. SLC17A7 gene may be the indicator of selective serotonin reuptake inhibitor treatment response in the Chinese Han population. J Clin Psychopharmacol. 2014;34:331–336. doi: 10.1097/JCP.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 59.Ohata S, Nakatani J, Herranz-Pérez V, Cheng J, Belinson H, Inubushi T, Snider WD, García-Verdugo JM, Wynshaw-Boris A, Alvarez-Buylla A. Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron. 2014;83:558–571. doi: 10.1016/j.neuron.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasan MT, Hernández-González S, Dogbevia G, Treviño M, Bertocchi I, Gruart A, Delgado-García JM. Role of motor cortex NMDA receptors in learning-dependent synaptic plasticity of behaving mice. Nat Commun. 2013;4:2258. doi: 10.1038/ncomms3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qin S, Zhao X, Pan Y, Liu J, Feng G, Fu J, Bao J, Zhang Z, He L. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur J Hum Genet. 2005;13:807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- 62.Abu-Sultaneh S, Hehir DA, Murkowski K, Ghanayem NS, Liedel J, Hoffmann RG, Cao Y, Mitchell ME, Jeromin A, Tweddell JS. Changes in cerebral oxygen saturation correlate with S100B in infants undergoing cardiac surgery with cardiopulmonary bypass. Pediatr Crit Care Med. 2014;15:219–228. doi: 10.1097/PCC.0000000000000055. [DOI] [PubMed] [Google Scholar]
- 63.Harilall Y, Adam JK, Biccard BM, Reddi A. The effect of optimising cerebral tissue oxygen saturation on markers of neurological injury during coronary artery bypass graft surgery. Heart Lung Circ. 2014;23:68–74. doi: 10.1016/j.hlc.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 64.Eyre H, Baune BT. Neuroplastic changes in depression: A role for the immune system. Psychoneuroendocrinology. 2012;37:1397–1416. doi: 10.1016/j.psyneuen.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 65.Tong L, Prieto GA, Kramár EA, Smith ED, Cribbs DH, Lynch G, Cotman CW. Brain-derived neurotrophic factor-dependent synaptic plasticity is suppressed by interleukin-1β via p38 mitogen-activated protein kinase. J Neurosci. 2012;32:17714–17724. doi: 10.1523/JNEUROSCI.1253-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avila ME, Sepúlveda FJ, Burgos CF, Moraga-Cid G, Parodi J, Moon RT, Aguayo LG, Opazo C, De Ferrari GV. Canonical Wnt3a modulates intracellular calcium and enhances excitatory neurotransmission in hippocampal neurons. J Biol Chem. 2010;285:18939–18947. doi: 10.1074/jbc.M110.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ciani L, Boyle KA, Dickins E, Sahores M, Anane D, Lopes DM, Gibb AJ, Salinas PC. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin-dependent protein kinase II. Proc Natl Acad Sci USA. 2011;108:10732–10737. doi: 10.1073/pnas.1018132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shih J, May LD, Gonzalez HE, Lee EW, Alvi RS, Sall JW, Rau V, Bickler PE, Lalchandani GR, Yusupova M, et al. Delayed environmental enrichment reverses sevoflurane-induced memory impairment in rats. Anesthesiology. 2012;116:586–602. doi: 10.1097/ALN.0b013e318247564d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang SQ, Fang F, Xue ZG, Cang J, Zhang XG. Neonatal sevoflurane anesthesia induces long-term memory impairment and decreases hippocampal PSD-95 expression without neuronal loss. Eur Rev Med Pharmacol Sci. 2013;17:941–950. [PubMed] [Google Scholar]