Abstract
Recently, kinesin motor proteins have been focused on as targets for cancer therapy. Kinesins are microtubule-based motor proteins that mediate diverse functions within the cell, including the transport of vesicles, organelles, chromosomes and protein complexes, as well as the movement of microtubules. In the current study, the expression of kinesin family member 18A (KIF18A), a member of kinesin superfamily, was investigated in breast cancer using immunohistochemistry, and its effect on breast cancer prognosis was examined. KIF18A expression level was significantly associated with lymph node metastasis (P=0.047). In patients with high levels of KIF18A expression, survival was significantly poorer compared to patients with low levels of KIF18A expression (disease-free survival, P=0.030). Multivariate analysis revealed that venous invasion (hazard ratio, 9.22; 95% confidence interval, 3.90–23.66; P<0.001) and KIF18A expression (hazard ratio, 3.20; 95% confidence interval, 1.34–6.09; P=0.010) were independent predictive factors for lymph node metastasis. KIF18A may be a useful predictive marker for lymph node metastasis in breast cancer, which could facilitate curative adjuvant treatment.
Keywords: kinesin family member 18A, biomarker, breast cancer
Introduction
Breast cancer is one of the most prevalent cancers and is a major public health concern among women. The most recent statistics for Japan document >76,000 cases per year (1), with a mortality rate of >13,000 per year (2). Breast cancer care and research has improved early detection and treatment. However, even after apparently successful localized treatments, there are long-term risks of recurrence and metastasis (3).
Breast cancer is currently classified into subtypes based on immunohistochemical (IHC) classification (4,5). Subtypes are defined by clinicopathological biomarkers, including estrogen receptor (ER), progesterone receptors (PgR), human epidermal growth factor receptor 2 (HER2) and Ki-67; these biomarkers can indicate the optimum treatment and play a key role in breast cancer treatment to improve prognosis (6,7). To facilitate the treatment of cancer, it is important to exploit new biomarkers that can improve the reliability of prognosis prediction, and to develop targeted therapies for breast cancer patients.
Recently, specific kinesin motor proteins have been studied as key proteins that regulate mitotic events and potential targets of therapy (8–11). KIF18A is a member of the kinesin protein superfamily, which is associated with the molecular motor proteins that use ATP hydrolysis to produce force and movement along microtubules (12–17). The basic mechanism for these activities is not well understood. However, recent studies have demonstrated that KIF18A regulates chromosome congression (18) and suppresses kinetochore movements to control mitotic chromosome alignment (19). Chromosome congression relies on the presence of KIF18A, indicating that this motile microtubule depolymerase has a key role in the dynamics of kinetochore microtubules driving chromosome alignment in the pre-anaphase state of the human cell cycle (18–20).
It has been reported that KIF18A is involved in breast, colorectal and hepatocellular cancers, and cholangiocarcinoma (21–24). However, there is essentially no information regarding the clinical relevance of KIF18A in breast cancer patients. Therefore, the present study investigated the clinicopathological significance of KIF18A expression in human breast cancer.
Materials and methods
Patients and specimen collection
Primary breast cancer and paired normal tissues were obtained from the operative specimens of 144 patients, who underwent radical surgery at the Medical Hospital of the Tokyo Medical and Dental University (Tokyo, Japan) between January 2004 and December 2006. Normal tissue was collected from at least 1 cm away from the primary breast cancer site and was histopathologically identified as normal by a pathologist at the Tokyo Medical and Dental University. Written informed consent was obtained from all patients according to the guidelines approved by the Institutional Research Board. Patients ranged in age from 26 to 91 years, with a mean age of 54 years. Patients were excluded if they received anti-hormone therapy, chemotherapy or radiotherapy prior to surgery. Patients with non-invasive breast cancer were also not included in the study. All patients were closely followed up after surgery at regular 3- to 6-month intervals, and the total follow-up periods ranged from 3 months to 7.6 years, with a median of 5.9 years. Following surgery, all patients were clearly classified into a category of breast cancer based on the clinicopathological criteria described by the Japanese Society for Breast Cancer (25). All data, including age, tumor size, lymphatic invasion, vascular invasion, nuclear grade, lymph node metastasis, ER status, PgR status, HER2 score, recurrence, pathological histology and clinical stage, were obtained from the clinical and pathological records. HER2 status was scored using the HER2 expression criteria (26). For primary tumors with a HER2 score of 2+, the IHC results were additionally validated with fluorescence in situ hybridization. All patients were treated with anti-hormonal therapy, chemotherapy and/or radiotherapy subsequent to surgery, according to breast cancer treatment guidelines in Japan, which were based on St. Gallen International Breast Cancer Conference and National Comprehensive Cancer Network recommendations (27).
Immunohistochemistry
IHC studies for KIF18A expression were performed on formalin-fixed, paraffin-embedded breast cancer tissues. After tissue sections (4 µm) were deparaffinized over 5 × 10 min incubations in xylene, the sections were rehydrated, and antigen retrieval was performed by incubation in antigen activation liquid (pH 9.0) in a microwave processor at 98°C for 30 min. Endogenous peroxidase activity was blocked using a solution of 3% hydrogen peroxide in absolute methanol for 15 min. A polyclonal rabbit anti-KIF18A antibody (catalog no., A301-080A; Bethyl Labotatories, Montgomery, TX, USA) was applied at a dilution of 1:150 and incubated overnight at 4°C. The Histofine Simple Stain MAX PO kit (Nichirei Corporation, Tokyo, Japan) was used according to the manufacturer's instructions to block non-specific binding and to detect bound primary antibody. The color was developed by diaminobenzidine (Nichirei Corporation) for 10 min at room temperature. The sections were then counterstained with Mayer's hematoxylin. Negative control staining was conducted by substituting non-immune rabbit serum and phosphate-buffered saline for primary antibodies. An expert pathologist selected and evaluated five representative fields at 400x magnification from each slide to produce digital photographs for measuring of the staining intensity of KIF18A. KIF18A expression was quantified using ImageJ software (Java 1.6.0_30 (32-bit); http://rsb.info.nih.gov/ij/index.html), which calculates the staining intensity of an area by converting RGB pixels to brightness values (11). The 5 most typically stained areas within the tumor were selected for calculating the average value.
Statistical analysis
Data from the IHC analysis was analyzed by JMP 10 software for Windows (version 10.0.1; SAS Institute, Cary, NC, USA). Differences between groups were determined using the χ2 test and analysis of variance. Disease-free survival (DFS) rates were calculated actuarially according to the Kaplan-Meier analysis and compared with the generalized log-rank test. Variables with a value of P<0.05 in univariate analysis were used in a subsequent multivariate analysis using nominal logistic regression. P<0.05 was considered as to indicate statistical significance.
Results
KIF18A expression in breast cancer tissues
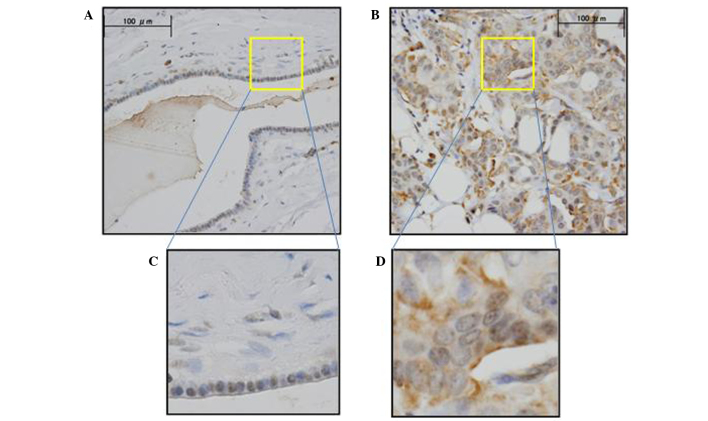
KIF18A expression was assessed by IHC analysis in the primary breast cancer tissue and normal breast tissue samples. IHC analysis with anti-KIF18A antibody verified that KIF18A was highly expressed in cancer cells compared to normal cells (Fig. 1). The cancer and normal cell types exhibited differential staining patterns: Positive IHC staining was observed in the nucleus of normal and cancer cells; positive IHC staining of the cytoplasm was observed predominantly in cancer cells and slightly in normal cells. ImageJ software was used to quantitatively evaluate the staining intensity of KIF18A in 144 breast cancer samples. Following assay optimization, a cutoff level of expression was determined as 30.02 (the average expression level of KIF18A in tumor): Breast cancer specimens <30.02 were assigned to the low expression group (n=83; 57.6%), whereas those with values ≥30.02 were assigned to the high expression group (n=61; 42.4%).
Figure 1.
Representative KIF18A immunohistochemistry images of breast cancer tissues. Positive staining was observed in cancer cells, but very limited in normal cells. (A) Normal breast, magnification ×400. (B) Breast cancer, magnification ×400. Enlarged views of (C) normal breast and (D) breast cancer tissues, magnification ×1,000.
Clinicopathological significance of KIF18A expression in breast cancer tissue
The clinicopathological factors analyzed in relation to KIF18A expression in breast cancer tissue are shown in Table I. The incidence of lymph node metastasis was significantly higher (P=0.047) in the high-expression group than in the low-expression group. Conversely, no significant differences were observed with regard to age, menopause status, tumor stage, lymphovascular invasion, nuclear grade, hormone status, HER2 status or recurrence.
Table I.
Clinicopathological significance of the KIF18A expression ratio in breast cancer.
| KIF18A expression ratio | |||||
|---|---|---|---|---|---|
| Low | High | ||||
| Clinicopathological factor | n | % | n | % | P-value |
| Age, years (mean ± SD) | 54.6±12.9 | 54.0±13.5 | 0.814 | ||
| Menopause status | 0.876 | ||||
| Pre | 37 | 44.6 | 28 | 45.9 | |
| Post | 46 | 55.4 | 33 | 54.1 | |
| Tumor stage | 0.110 | ||||
| T1 | 34 | 40.9 | 33 | 54.1 | |
| T2-3 | 49 | 59.1 | 28 | 45.9 | |
| Lymph node metastasis | 0.047a | ||||
| Absent | 62 | 74.7 | 36 | 59.1 | |
| Present | 21 | 25.3 | 25 | 40.9 | |
| Lymphatic invasion | 0.649 | ||||
| Absent | 44 | 53.0 | 30 | 49.2 | |
| Present | 39 | 47.0 | 31 | 50.8 | |
| Venous invasion | 0.929 | ||||
| Absent | 47 | 56.6 | 35 | 57.4 | |
| Present | 36 | 43.4 | 26 | 42.6 | |
| Nuclear grade | 0.185 | ||||
| Grade 1 | 47 | 56.6 | 40 | 65.6 | |
| Grade 2 | 15 | 18.1 | 13 | 21.3 | |
| Grade 3 | 21 | 25.3 | 8 | 13.1 | |
| Nuclear atypia | 0.528 | ||||
| Score 1 | 5 | 6.0 | 6 | 9.8 | |
| Score 2 | 66 | 79.5 | 49 | 80.4 | |
| Score 3 | 12 | 14.5 | 6 | 9.8 | |
| Mitotic counts | 0.256 | ||||
| Score 1 | 48 | 57.8 | 41 | 67.2 | |
| Score 2 | 17 | 20.5 | 13 | 21.3 | |
| Score 3 | 18 | 21.7 | 7 | 11.5 | |
| Estrogen receptor | 0.961 | ||||
| Absent | 12 | 14.5 | 9 | 14.8 | |
| Present | 71 | 85.5 | 52 | 85.2 | |
| Progesterone receptor | 0.629 | ||||
| Absent | 22 | 26.5 | 14 | 22.9 | |
| Present | 61 | 73.5 | 47 | 77.1 | |
| HER2 score | 0.221 | ||||
| 0–1 | 69 | 83.1 | 55 | 90.2 | |
| 2–3 | 14 | 16.9 | 6 | 9.8 | |
| Recurrence | 0.534 | ||||
| Absent | 69 | 83.1 | 53 | 16.9 | |
| Present | 14 | 86.9 | 8 | 13.1 | |
P<0.05, statistical significance. KIF18a, kinesin family member 18A; SD, standard deviation; HER2, human epidermal growth factor receptor 2.
DFS analysis
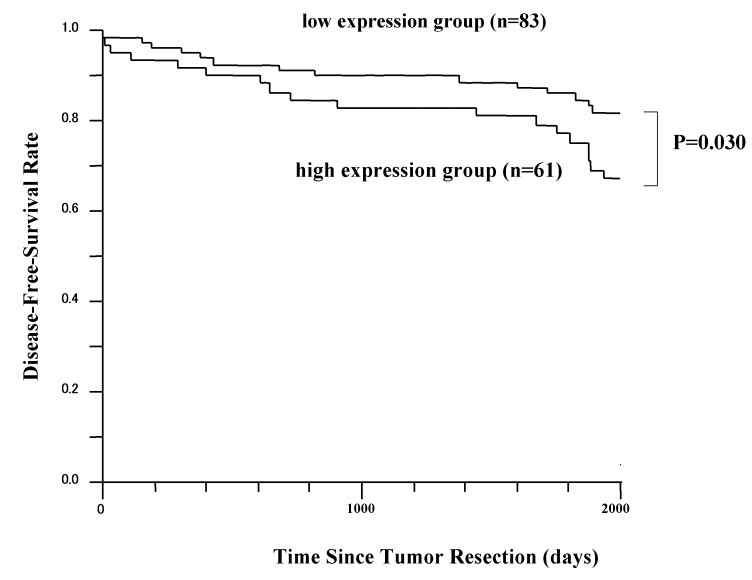
The 5-year DFS rates in patients with high KIF18A expression and patients with low KIF18A expression are presented in Fig. 2. The difference in DFS time between these two groups was statistically significant (P=0.030; log-rank test). However, the overall survival difference between these two groups was not statistically significant (data not shown). Patients received ≥1 postoperative therapy (anti-hormonal treatment, chemotherapy or radiotherapy).
Figure 2.
Disease-free survival rate of patients with breast cancer grouped according to KIF18A expression status of the tumor. Patients with high KIF18A expression (n=61) had a significantly poorer prognosis than those with low KIF18A expression (n=83). KIF18A, kinesin family member 18A.
Univariate and multivariate analysis
Univariate and multivariate logistic regression analyses were performed for factors affecting lymph node metastasis (Table II). Univariate analysis revealed a significant associated between lymph node metastasis and the following factors: Tumor size (P=0.009), lymphatic invasion (P<0.001), venous invasion (P<0.001), recurrence (P=0.004) and KIF18A expression (P=0.047). Multivariate analysis of these parameters revealed that venous invasion (hazard ratio, 9.22; 95% confidence interval, 3.90–23.66; P<0.001) and KIF18A expression (hazard ratio, 3.20; 95% confidence interval, 1.34–6.09; P=0.010) were independent predictive factors for lymph node metastasis.
Table II.
Univariate and multivariate analyses of clinicopathological factors affecting lymph node metastasis.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (<50/≥51) | 0.97 | 0.48–1.97 | 0.93 | – | – | – |
| T stage (T1/T2-3) | 2.69 | 1.30–5.79 | <0.01 | 1.91 | 0.77–4.87 | 0.164 |
| LI (absent/present) | 19.51 | 7.57–61.01 | <0.01 | – | – | – |
| VI (absent/present)) | 9.97 | 4.48–23.92 | <0.01 | 9.22 | 3.90–23.66 | <0.001 |
| ER (absent/present) | 1.60 | 0.58–5.16 | 0.39 | – | – | – |
| PgR (absent/present) | 0.92 | 0.41–2.10 | 0.84 | – | – | – |
| HER2 (absent/present) | 1.51 | 0.55–3.95 | 0.41 | – | – | – |
| Recurrence | 3.90 | 1.54–10.27 | <0.01 | 2.54 | 0.81–8.33 | 0.113 |
| KIF18A (low/high) | 2.05 | 1.01–4.21 | 0.047 | 3.20 | 1.34–8.09 | 0.010 |
LI, lymphatic invasion; VI, venous invasion; ER, estrogen receptor; PgR, progesterone receptor; HER2, human epidermal growth factor receptor 2; KIF18A, kinesin family member 18A; HR, hazard ratio; CI, confidence interval.
Discussion
In recent years, cancer therapy research has focused on proteins involved in the regulatory events of mitosis (8–11). Infiltrating growth of cancer cells, which is associated with abnormal, uncontrolled proliferation, requires the biological activation of numerous proteins that serve central roles. Mitotic inhibitor drugs, which include taxanes and vinca alkaloids, act to target microtubules and have yielded various levels of success in the treatment of various types of carcinomas (28). Several next-generation mitotic drug targets have been developed, and small molecule inhibitors that have been identified are already under investigation in clinical trials (11).
KIF18A is a member of the kinesin 8 family and has been demonstrated to play important roles in chromosome alignment during mitosis (18–20). Through several in vitro assays, it has been revealed that upregulation of KIF18A may affect the biological characteristics of cancer cells (21–24).
In the current study, the expression of KIF18A in breast cancer tissues and the association between KIF18A expression and clinicopathological factors in breast cancer were explored using IHC analysis. The results revealed that KIF18A protein expression was significantly higher in breast cancer tissues than in normal breast tissues (21). This suggests that breast cancer cells may take advantage of KIF18A overexpression to control mitotic chromosome alignment and increase their rate of repetitive cell division.
The present study also revealed that KIF18A overexpression in breast cancer was associated with lymph node metastasis and poor prognosis. The group with high KIF18A expression had a poorer prognosis compared with that of the low-expression group in terms of DFS. KIF18A overexpression was prevalent in breast cancer cells and was also associated with prognostic factors and shorter survival time; these results may suggest that the overexpression of this mitotic protein is associated with aggressive primary tumors. To the best of our knowledge, this is the first study to demonstrate the clinical relevance of KIF18A in invasive breast cancer and its relation to disease outcome.
The axillary lymph node status is the most consistent prognostic factor used in adjuvant therapy decision-making (29). Currently, the sentinel node biopsy is a common surgical procedure to determine the stage of the cancer and select an appropriate treatment plan (30,31). In multivariate analysis, KIF18A overexpression in breast cancer was determined to be an independent and significant predictive factor for lymph node metastasis. Based on these findings, in cases where low KIF18A expression is identified prior to breast surgery, it may be possible to avoid performing the sentinel node biopsy in selected patients with clinically and radiologically normal axilla.
In summary, this is the first report of clinicopathological analysis of KIF18A in breast cancer patients. KIF18A expression was correlated with lymph node metastasis and was an independent predictive factor for the lymph node metastasis and DFS. Kaplan-Meier analyses revealed that the DFS rate was significantly lower in the high KIF18A expression group. These findings suggest that KIF18A may be a useful predictive biomarker of lymph node metastasis, which could aid in the development of optimum adjuvant treatments.
Acknowledgements
The authors would like to thank Ms. Yoko Takagi from the Department of Surgical Oncology, Graduate School of Medicine and Dental Science, Tokyo Medical and Dental University (Tokyo, Japan) for providing technical support.
References
- 1.Matsuda A, Matsuda T, Shibata A, Katanoda K, Sobue T, Nishimoto H. Japan Cancer Surveillance Research Group: Cancer incidence and incidence rates in Japan in 2008: A study of 25 population-based cancer registries for the monitoring of cancer incidence in Japan (MCIJ) project. Jpn J Clin Oncol. 2014;44:388–396. doi: 10.1093/jjco/hyu003. [DOI] [PubMed] [Google Scholar]
- 2.Katanoda K, Hori M, Matsuda T, Shibata A, Nishino Y, Hattori M, Soda M, Ioka A, Sobue T, Nishimoto H. An updated report on the trends in cancer incidence and mortality in Japan. Jpn J Clin Oncol. 2015;45:390–401. doi: 10.1093/jjco/hyv002. [DOI] [PubMed] [Google Scholar]
- 3.Peto R, Boreham J, Clarke M, Davies C, Beral V. UK and USA breast cancer deaths down 25% in year 2000 at ages 20–69 years. Lancet. 2000;355:1822. doi: 10.1016/S0140-6736(00)02277-7. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Harbeck N, Thomssen C, Gnant M. St. Gallen 2013: Brief preliminary summary of the consensus discussion. Breast Care (Basel) 2013;8:102–109. doi: 10.1159/000351193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Panel members: Strategies for subtypes-dealing with the diversity of breast cancer: Highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falck AK, Bendahl PO, Chebil G, Olsson H, Fernö M, Rydén L. Biomarker expression and St Gallen molecular subtype classification in primary tumours, synchronous lymph node metastases and asynchronous relapses in primary breast cancer patients with 10 years' follow-up. Breast Cancer Res Treat. 2013;140:93–104. doi: 10.1007/s10549-013-2617-8. [DOI] [PubMed] [Google Scholar]
- 8.Marcus AI, Peters U, Thomas SL, Garrett S, Zelnak A, Kapoor TM, Giannakakou P. Mitotic kinesin inhibitors induce mitotic arrest and cell death in taxol-resistant and -sensitive cancer cells. J Biol Chem. 2005;280:11569–11577. doi: 10.1074/jbc.M413471200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miglarese MR, Carlson RO. Development of new cancer therapeutic agents targeting mitosis. Expert Opin Investig Drugs. 2006;15:1411–1425. doi: 10.1517/13543784.15.11.1411. [DOI] [PubMed] [Google Scholar]
- 10.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 11.Kaestner P, Bastians H. Mitotic drug targets. J Cell Biochem. 2010;111:258–265. doi: 10.1002/jcb.22721. [DOI] [PubMed] [Google Scholar]
- 12.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 13.Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/S0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 15.Karcher RL, Deacon SW, Gelfand VI. Motor-cargo interactions: The key to transport specificity. Trends in Cell Biol. 2002;12:21–27. doi: 10.1016/S0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- 16.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/S0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 17.Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: Refocusing on microtubule dynamics. Mol Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 19.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner MK, Odde DJ, Bloom K. Kinesin-8 molecular motors: Putting the brakes on chromosome oscillations. Trends Cell Biol. 2008;18:307–310. doi: 10.1016/j.tcb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Zhu C, Chen H, Li L, Guo L, Jiang W, Lu SH. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 2010;31:1676–1684. doi: 10.1093/carcin/bgq134. [DOI] [PubMed] [Google Scholar]
- 22.Nagahara M, Nishida N, Iwatsuki M, Ishimaru S, Mimori K, Tanaka F, Nakagawa T, Sato T, Sugihara K, Hoon DS, Mori M. Kinesin 18A expression: Clinical relevance to colorectal cancer progression. Int J Cancer. 2011;129:2543–2552. doi: 10.1002/ijc.25916. [DOI] [PubMed] [Google Scholar]
- 23.Rucksaken R, Khoontawad J, Roytrakul S, Pinlaor P, Hiraku Y, Wongkham C, Pairojkul C, Boonmars T, Pinlaor S. Proteomic analysis to identify plasma orosomucoid 2 and kinesin 18A as potential biomarkers of cholangiocarcinoma. Cancer Biomark. 2012;12:81–95. doi: 10.3233/CBM-130296. [DOI] [PubMed] [Google Scholar]
- 24.Liao W, Huang G, Liao Y, Yang J, Chen Q, Xiao S, Jin J, He S, Wang C. High KIF18A expression correlates with unfavorable prognosis in primary hepatocellular carcinoma. Oncotarget. 2014;5:10271–10279. doi: 10.18632/oncotarget.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukai H, Aihara T, Yamamoto Y, Takahashi M, Toyama T, Sagara Y, Yamaguchi H, Akabane H, Tsurutani J, Hara F, et al. The Japanese Breast Cancer Society Clinical Practice Guideline for systemic treatment of breast cancer. Breast Cancer. 2015;22:5–15. doi: 10.1007/s12282-014-0563-x. [DOI] [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, et al. American Society of Clinical Oncology; College of American Pathologists: American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 27.Komoike Y, Inokuchi M, Itoh T, Kitamura K, Kutomi G, Sakai T, Jinno H, Wada N, Ohsumi S, Mukai H. Japanese Breast Cancer Society: Japan Breast Cancer Society clinical practice guideline for surgical treatment of breast cancer. Breast Cancer. 2015;22:37–48. doi: 10.1007/s12282-014-0558-7. [DOI] [PubMed] [Google Scholar]
- 28.Gradishar WJ, Anderson BO, Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A, Giordano SH, Goldstein LJ, et al. National Comprehensive Cancer Network Breast Cancer Panel: Breast cancer version 3.2014. J Natl Compr Canc Netw. 2014;12:542–590. doi: 10.6004/jnccn.2014.0058. [DOI] [PubMed] [Google Scholar]
- 29.Atalay C. New concepts in axillary management of breast cancer. World J Clin Oncol. 2014;5:895–900. doi: 10.5306/wjco.v5.i5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giuliano AE, Dale PS, Turner RR, Morton DL, Evans SW, Krasne DL. Improved axillary staging of breast cancer with sentinel lymphadenectomy. Ann Surg. 1995;222:394–399. doi: 10.1097/00000658-199509000-00016. discussion 399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ollila DW, Brennan MB, Giuliano AE. The role of intraoperative lymphatic mapping and sentinel lymphadenectomy in the management of patients with breast cancer. Adv Surg. 1999;32:349–364. [PubMed] [Google Scholar]