Abstract
Choline kinase α (CHKA), the enzyme that converts choline to phosphocholine, has been studied in human carcinogenesis widely. However, the expression and underlying clinicopathological characteristics of CHKA in lung adenocarcinoma remains elusive. In the present study, a tissue microarray of 119 pairs of lung adenocarcinoma samples and corresponding adjacent normal mucosae was used to analysis CHKA expression by immunohistochemistry, and CHKA was observed to exhibit enhanced expression in lung adenocarcinoma tissues. Elevated CHKA expression in lung adenocarcinoma tissues at the gene and protein level was observed. The levels of CHKA expression were closely associated with the poor prognosis status of lung adenocarcinoma patients. Furthermore, certain clinicopathological characteristics such as tumor diameter and differentiation were observed to be significant in those lung adenocarcinoma patients who displayed enhanced CHKA expression. The analysis of CHKA expression could provide a more precise way to predict the prognosis of lung adenocarcinoma patients. Collectively, the present study revealed a novel biomarker in lung adenocarcinoma, and indicated that CHKA may be a promising prognostic marker and therapeutic target for lung adenocarcinoma.
Keywords: CHKA, lung adenocarcinoma, prognostic indicator, metastasis
Introduction
Lung cancer has become a common malignant tumor in the past recent years (1). Along with tobacco smoking and environmental factors, lung adenocarcinoma is one of the most important pathological causes of lung cancer (2), and is the first cause of cancer-associated mortalities in men (3). The occurrence and development of lung adenocarcinoma is a disease process involving multiple genes (4). As the occurrence mechanism has not been clarified thus far, the treatment for lung cancer still lacks specificity.
Choline kinase α (CHKA) is an important enzyme at the crossroads of the main growth factor-triggered survival signaling pathways such as Ras activation, and is constitutively activated in certain human tumor cells and tissues (5). In addition, a large number of studies in cancer cells suggest that CHKA expression and activity are directly associated not only with increased cancer cell proliferation but also with malignancy and metastasis (6), making it a potential prognostic biomarker of certain common cancers such as hepatocellular carcinoma (7) and ovarian cancer (8), as well as a potential novel target for cancer therapy. However, as an important oncogene, its association with lung adenocarcinoma carcinogenesis and development has not been reported thus far.
In the present study, the role of CHKA in lung adenocarcinoma and its correlation with the prognosis of lung adenocarcinoma was investigated. The present findings may provide a prognostic indicator and underlying target for lung adenocarcinoma prevention and therapy.
Materials and methods
Microarray tissue samples
A tissue microarray block containing 119 surgically resected lung adenocarcinoma specimens and paired non-cancerous surrounding tissues was constructed using a tissue microarrayer (Shanghai Outdo Biotech Co. Ltd., Shanghai, China).
Patients and lung adenocarcinoma samples
A total of 39 cancer samples were randomly retrieved from lung adenocarcinoma patients who underwent curative resection at the Provincial Hospital Affiliated to Shandong University (Jinan, China) from September 2010 to December 2012. The procedure of human sample collection was approved by the Ethics Committee of the Provincial Hospital Affiliated to Shandong University. Written informed consent was obtained from the patients.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using RNA Simple Total RNA kit (Tiangen Biotech, Co., Ltd., Beijing, China), according to the manufacturer's protocol. qPCR was performed using a SYBR Green PCR kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an Applied Biosystems™ 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.). The following thermal cycling conditions were used: 2 min at 50°C, 1 min at 94°C, 1 min at 55°C and 1 min at 72°C for 40 cycles, and 5 min at 72°C. The messenger RNA level of specific genes was normalized against 18S using the 2-∆∆Cq method. The sequences of the primers used in the present study are listed in Table I.
Table I.
Sequence of primers for reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence (5′-3′) |
|---|---|
| CHKA forward | CACTTTCCGAGGCTCATCAC |
| CHKA reverse | GGACGAGTTCCACATCAGTGT |
| 18S forward | CGGCTACCACATCCAAGGAA |
| 18S reverse | GCTGGAATTACCGCGGCT |
CHKA, choline kinase α.
Western blotting
Total protein was extracted using Minute TM Total Protein Extraction Kit for Animal Cultured Cells/Tissues (Invent Biotechnologies, Inc., Plymouth, MN, USA). The lung cancer tissue extract was separated on 12% polyacrylamide-sodium dodecyl sulfate gels, and transferred to polyvinylidene difluoride membranes. The membranes were blocked with Tris-buffered saline (TBS) with 5% milk for 2 h. Subsequently, the membranes were washed with TBS with Tween 20 (TBST) and the membranes were incubated with a rabbit polyclonal anti-CHKA and anti-GAPDH (catalog nos., ab88053 and ab9485; 1 µg/ml; Abcam Trading Co., Ltd., Shanghai, China) at 4°C overnight. Then the membranes were washed with TBST three times and incubated with Dylight 680, goat anti-rabbit IgG (catalog no., A23720; 1:1,000; Abbkine, Inc., Redlands, CA, USA) for 2 h at room temperature. The protein band, specifically bound to the primary antibody, was detected with a LI-COR imaging system (LI-COR Biotechnology, Lincoln, NE, USA).
Immunohistochemistry (IHC) and tissue microarray analysis
IHC analysis of tumor tissue microarray was performed using an anti-CHKA antibody (catalog no., AP8179b; Abgent Biotech Co., Ltd., Suzhou, China). The tissue array was incubated with this primary anti-CHKA antibody at 4°C overnight, followed by incubation with a horseradish peroxidase-conjugated secondary antibody at 37°C for 30 min. Next, the tissue array was incubated with 3,3′-diaminobenzidine and counterstained with hematoxylin for detection. Assessment of tissue microarray staining was based on the percentage of positively stained cells, and staining intensity was measured by Aperio ImageScope software (Leica Microsystems, Inc., Buffalo Grove, IL, USA).
Statistical analysis
Overall survival was defined as the interval between the dates of surgery and mortality. All statistical analyses were performed with SPSS version 18.0 software (SPSS, Inc., Chicago, IL, USA). Survival curves were calculated using the Kaplan-Meier method and compared with the log-rank test. The Cox proportional hazards model was used to determine the independent factors affecting survival based on variables selected by univariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
CHKA expression was upregulated in lung adenocarcinoma
To examine the clinical relevance of expression deregulation of CHKA in lung adenocacinoma patients, two lung cancer tissue microarrays containing 119 lung adenocarcinoma samples were used for IHC staining. It was noticed that the expression of CHKA was significantly elevated in lung adenocacinoma samples compared with peri-tumor tissues (Fig. 1A and B). These results were further confirmed by RT-qPCR and western blot analyses (Fig. 1C and D). This finding prompted us to speculate whether CHKA is important in lung adenocarcinoma.
Figure 1.
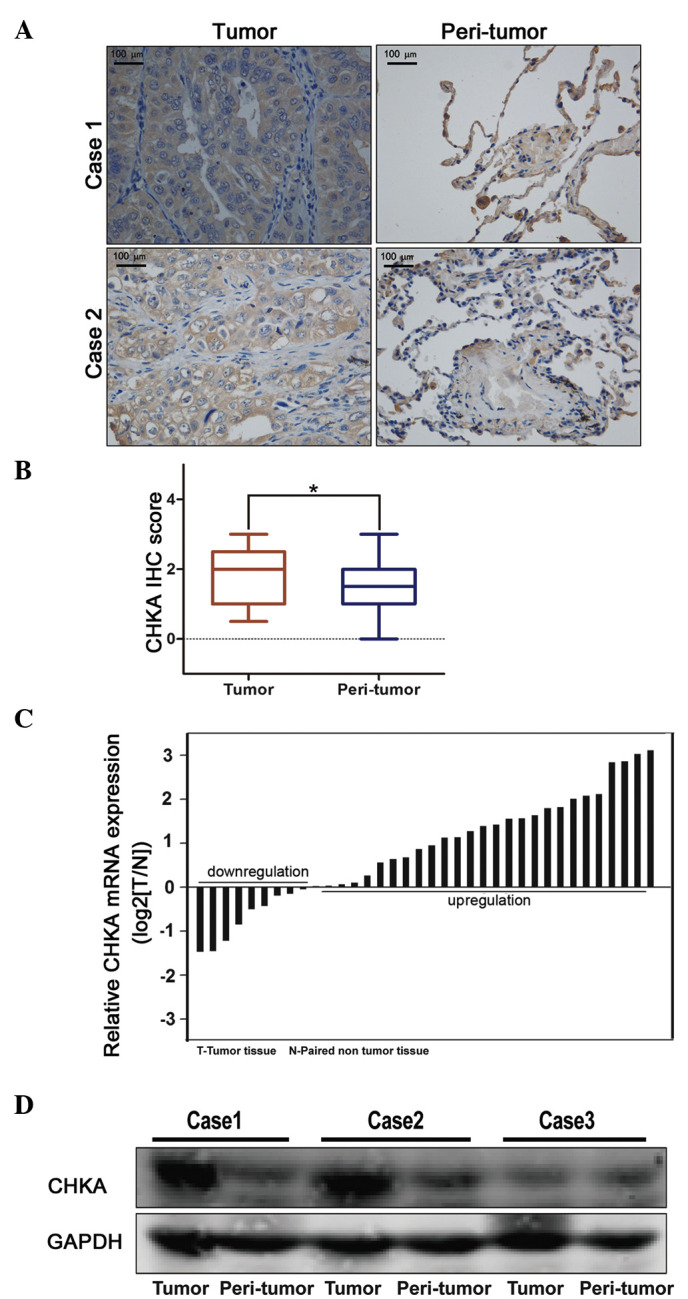
Enhanced expression of CHKA in lung adenocarcinoma. (A and B) Immunohistochemical staining of CHKA expression in tissue microarray (n=119). The expression of CHKA was analyzed in tumor and peri-tumor tissues of lung adenocarcinoma patients. (A) Representative images (magnification, ×200) and (B) quantitative results are shown (*P=0.032). (C) Comparison of CHKA expression in tumor and peri-tumor tissues of lung adenocarcinoma patients by reverse transcription-quantitative polymerase chain reaction. (D) Representative western blot analysis of the expression of CHKA protein in tumor and peri-tumor tissues from three patients with lung adenocarcinoma. CHKA, choline kinase α; IHC, immunohistochemistry; mRNA, messenger RNA; T, tumor; N, non-tumor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Clinical significance of CHKA expression in lung adenocarcinoma
The clinical implication of CHKA expression in lung adenocarcinoma was analyzed. High levels of CHKA were observed to be associated with poor survival in lung adenocarcinoma patients (Fig. 2A). The association between CHKA expression in tumor tissues and the clinicopathological characteristics of the 119 patients was examined (Table II). Correlation regression analysis indicated that prognosis was correlated with several individual parameters, including CHKA expression, tumor diameter, tumor stage and differentiation (Table III). These individual parameters were further analyzed with a multivariate Cox proportional hazards model. The results indicated that CHKA expression, tumor diameter and differentiation were significant and independent factors that affected the survival of lung adenocarcinoma patients (Table III and Fig. 2B). Among these factors, the significant hazard ratio (HR) value for the cumulative survival of CHKA expression levels was 2.644 [95% confidence interval (CI), 1.217–5.746; P=0.05]. Tumor diameter and differentiation also exhibited a significant HR value for cumulative survival. For tumor diameter, the HR value was determined to be 2.462 (95% CI, 1.249–4.853; P=0.01). Poor differentiation displayed the greatest HR value (HR, 2.644; 95% CI, 1.217–5.746; P=0.01). Since poor differentiated lung cancer exhibited the greatest HR value for cumulative survival, the expression level of CHKA in poor- vs. well-differentiated cancer was further analyzed. Patients with poor differentiation tended to have high CHKA expression levels (Fig. 2C). As known, lung cancer patients with poor differentiation are more inclined to bad prognosis compared with high differentiation group (9); therefore, this result suggested that CHKA may act as an oncogene and mediate the poor prognosis and poor differentiation of lung cancer.
Figure 2.
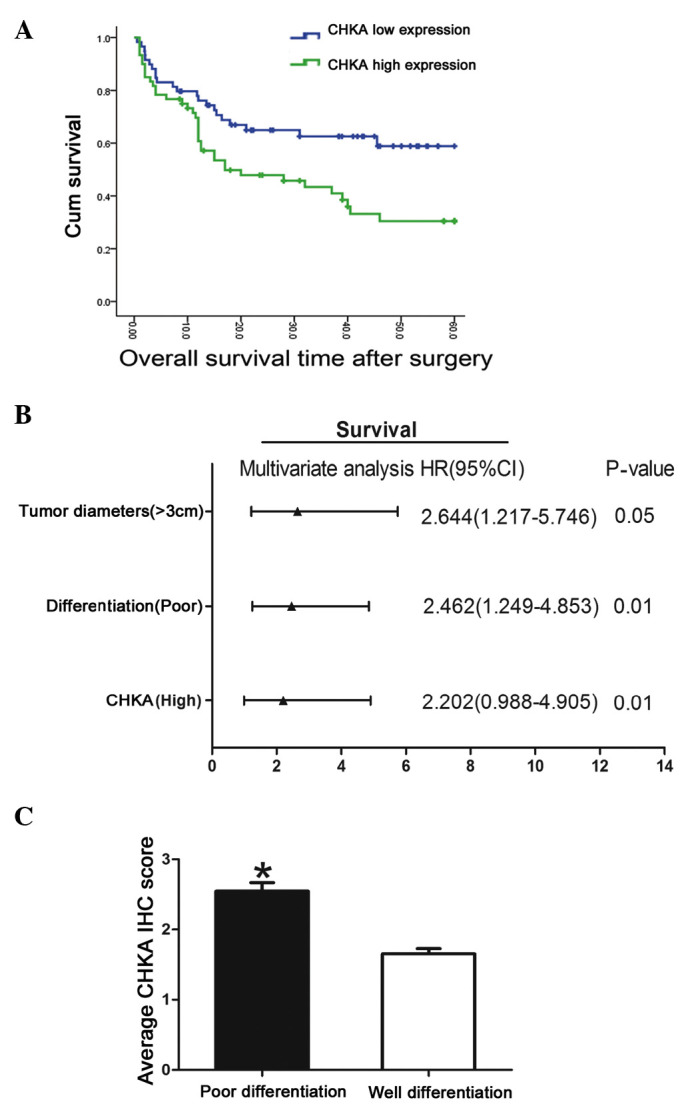
Clinical Significance of CHKA expression in lung adenocarcinoma. (A) Lung adenocarcinoma patients were divided into a CHKA ‘high’ group (whose fold-change of relative expression was higher than the median) and ‘low’ group (whose fold-change of relative expression was lower than the median), and the overall survival rates of the 119 lung adenocarcinoma patients were compared between the CHKA ‘high’ group and the CHKA ‘low’ group. (B) Multivariate analysis of hazard ratios for overall survival of lung adenocarcinoma patients in tissue microarray. (C) Comparison of CHKA expression between lung adenocarcinoma patients whose differentiation status is poor or well. *P<0.05. Patients with poor differentiation exhibited higher CHKA expression than those with well-differentiated tumors. Cum, cumulative; CHKA, choline kinase α; HR, hazard ratio; CI, confidence interval; IHC, immunohistochemistry.
Table II.
Association between CHKA expression and clinicopathological characteristics of lung adenocarcinoma patients.
| CHKA expression, N (%) | ||||
|---|---|---|---|---|
| Characteristics | N | Low N=59 | High N=60 | P-value |
| Age | 0.650 | |||
| ≤65 years | 63 | 30 (47.6) | 33 (52.4) | |
| >65 years | 56 | 29 (51.8) | 27 (48.2) | |
| Gender | 0.230 | |||
| Male | 61 | 27 (44.3) | 34 (55.7) | |
| Female | 58 | 32 (55.2) | 26 (44.8) | |
| Smoking | 0.770 | |||
| Yes | 69 | 35 (50.7) | 34 (49.3) | |
| No | 50 | 24 (48.0) | 26 (52.0) | |
| Tumor diametera | 0.020 | |||
| ≤3 cm | 54 | 33 (61.1) | 21 (38.9) | |
| >3 cm | 65 | 26 (40.0) | 39 (60.0) | |
| Lymph metastasis | 0.530 | |||
| Yes | 72 | 34 (47.2) | 38 (52.8) | |
| No | 47 | 25 (53.2) | 22 (46.8) | |
| Differentiationa | 0.001 | |||
| Well | 64 | 43 (67.2) | 21 (32.8) | |
| Poor | 55 | 16 (29.1) | 39 (70.9) | |
| TNM stagea | 0.050 | |||
| T1/2 | 66 | 38 (57.6) | 28 (42.4) | |
| T3/4 | 53 | 21 (39.6) | 32 (60.4) | |
Significant parameters. TNM, tumor-node-metastasis; CHKA, choline kinase α
Table III.
Univariate and multivariate analyses of the primary cohort.
| Variable | N | Time, monthsb | Univariate P-value | Multivariate P-value |
|---|---|---|---|---|
| CHKA expressiona | 0.003 | 0.05 | ||
| High | 60 | 27.117±6.103 | ||
| Low | 59 | 38.186±4.296 | ||
| Lymph metastasis | 0.159 | 0.25 | ||
| Yes | 26 | 43.094±3.829 | ||
| No | 33 | 37.750±16.277 | ||
| Tumor diametera | 0.022 | 0.01 | ||
| ≤3 cm | 54 | 40.074±3.839 | ||
| >3 cm | 65 | 23.389±8.494 | ||
| TNM stage | 0.050 | 0.07 | ||
| 1/2 | 72 | 41.194±3.986 | ||
| 3/4 | 47 | 29.989±4.901 | ||
| Smoking | 0.368 | 0.42 | ||
| Yes | 69 | 35.225±4.776 | ||
| No | 50 | 31.007±6.400 | ||
| Age | 0.482 | 0.53 | ||
| ≤65 years | 63 | 34.000±4.786 | ||
| >65 years | 56 | 30.604±6.271 | ||
| Differentiationa | 0.000 | 0.01 | ||
| Poor | 22 | 21.409±11.274 | ||
| Well | 97 | 36.743±3.636 |
Significant parameters. bResults are presented as the mean ± standard error. TNM, tumor-node-metastasis; CHKA, choline kinase α.
Discussion
Lung cancer is one of the malignant tumors that severely threat people's health and life (10). It has a high malignant degree and a high mortality rate, and its 5-year survival rate is just 10–15% (11–13). In recent years, its morbidity and mortality rate have markedly increased (14). The occurrence and development of lung cancer is a disease process involving multiple genes (15). As the occurrence mechanism has not been clarified thus far, the treatment for lung cancer still lacks specificity (16). The biological characteristics of lung cancer are complicated, with different types of tumors being driven by different genes and exhibiting expression of different proteins (17).
CHKA has been identified as the initial enzyme, playing a regulatory role, in the biosynthesis of phosphatidylcholine, which occurs via the cytidine diphosphate-choline pathway (18). Besides its regular function (19), CHKA was reported to be important in cancer carcinogenesis and development (8,20). Granata et al reported that CHKA could act as a potential druggable target for ovarian cancer by regulating cell aggressiveness and drug sensitivity (21). In addition, the study by de la Cueva et al revealed that a CHKA inhibitor combined with fluorouracil displayed a synergistic antitumoral effect in colorectal cancer therapy (22). Furthermore, previous studies on the human triple negative breast cancer cell line MDA-MB-231 demonstrated that CHKA knockdown significantly decreased the proliferation and malignance of cancer cells, supporting CHKA downregulation as a cancer-specific treatment (23).
In the present study, CHKA expression was observed to be increased in lung adenocarcinoma, and this elevated expression was associated with malignant clinicopathological characteristics. It was observed that the tumor survival rate substantially differed between patients with high and low expression levels of CHKA in tumor tissues. Multivariate analysis revealed that CHKA expression was an independent and significant risk factor affecting the survival rate subsequent to curative resection, with the greatest HR value identified for survival. Importantly, CHKA overexpression displayed enhanced accuracy in predicting poor outcome in patients with lung adenocarcinoma at early stages. Furthermore, CHKA expression exhibited a close correlation with poor differentiation in lung adenocarcinoma; thus, further analysis of CHKA expression would help to assess the prognosis of lung adenocarcinoma patients.
In conclusion, the present data provided a new insight into the prognosis of lung adenocarcinoma and the underlying target therapy of patients. Therefore, the authors propose that CHKA functions as a proto-oncogene and contributes to malignant progression.
Acknowledgements
The authors would like to thank Dr Qi Wang (Department of Respiratory Medicine, Provincial Hospital Affliated to Shandong University, Jinan, China) for his technical assistance. The present study was supported by a grant from the Technology Development Schedule of Shandong Province (Jinan, China; grant number 2010G0020227).
References
- 1.Quoix E, Lemarié E. Epidemiological novelties in lung cancer. Rev Mal Respir. 2011;28:1048–1058. doi: 10.1016/j.rmr.2010.12.015. (In French) [DOI] [PubMed] [Google Scholar]
- 2.Sasaki H, Maekawa M, Tatematsu T, Okuda K, Moriyama S, Yano M, Fujii Y. Increased BRAF copy number in lung adenocarcinoma. Oncol Lett. 2015;9:709–712. doi: 10.3892/ol.2014.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shields PG. Molecular epidemiology of smoking and lung cancer. Oncogene. 2002;21:6870–6876. doi: 10.1038/sj.onc.1205832. [DOI] [PubMed] [Google Scholar]
- 4.Woo HI, Kim JA, Jung HA, Kim KK, Lee JY, Sun JM, Ahn JS, Park K, Lee SY, Ahn MJ. Correlation of genetic polymorphisms with clinical outcomes in pemetrexed-treated advanced lung adenocarcinoma patients. Pharmacogenomics. 2015;16:383–391. doi: 10.2217/pgs.15.14. [DOI] [PubMed] [Google Scholar]
- 5.Janardhan S, Srivani P, Sastry GN. Choline kinase: An important target for cancer. Curr Med Chem. 2006;13:1169–1186. doi: 10.2174/092986706776360923. [DOI] [PubMed] [Google Scholar]
- 6.Clem BF, Clem AL, Yalcin A, Goswami U, Arumugam S, Telang S, Trent JO, Chesney J. A novel small molecule antagonist of choline kinase-α that simultaneously suppresses MAPK and PI3K/AKT signaling. Oncogene. 2011;30:3370–3380. doi: 10.1038/onc.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwee SA, Hernandez B, Chan O, Wong L. Choline kinase alpha and hexokinase-2 protein expression in hepatocellular carcinoma: Association with survival. PLoS One. 2012;7:e46591. doi: 10.1371/journal.pone.0046591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granata A, Nicoletti R, Perego P, Iorio E, Krishnamachary B, Benigni F, Ricci A, Podo F, Bhujwalla ZM, Canevari S, et al. Global metabolic profile identifies choline kinase alpha as a key regulator of glutathione-dependent antioxidant cell defense in ovarian carcinoma. Oncotarget. 2015;6:11216–11230. doi: 10.18632/oncotarget.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, Shen J, Wang L, Shi Z, Wang M, Jiang BH, Shu Y. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2015;69:301–305. doi: 10.1016/j.biopha.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 11.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: From targets to treatments. Cancer Treat Rev. 2015;41:361–375. doi: 10.1016/j.ctrv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Riquet M, Mordant P, Pricopi C, Legras A, Foucault C, Dujon A, Arame A, Le Pimpec-Barthes F. A review of 250 ten-year survivors after pneumonectomy for non-small-cell lung cancer. Eur J Cardiothorac Surg. 2014;45:876–881. doi: 10.1093/ejcts/ezt494. [DOI] [PubMed] [Google Scholar]
- 13.Wao H, Mhaskar R, Kumar A, Miladinovic B, Djulbegovic B. Survival of patients with non-small cell lung cancer without treatment: A systematic review and meta-analysis. Syst Rev. 2013;2:10. doi: 10.1186/2046-4053-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vyas R, Haque S, Mital SP, Srinivasan S. Analysis of incidence of lung cancer in various states of the USA. International Medicine Engineering and Informatics. 2015;7:36–45. doi: 10.1504/IJMEI.2015.066241. [DOI] [Google Scholar]
- 15.Quintans JS, Antoniolli AR, Onofre FM, Onofre AS. Detection of lung cancer using multiple genetic markers - a systematic review. Diagn Cytopathol. 2013;41:834–842. doi: 10.1002/dc.22978. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling S, Jensen K, Herth FJ, Thomas M, Schnabel PA, Herpel E. Lack of prognostic significance of neuroendocrine differentiation and stem cell antigen co-expression in resected early-stage non-small cell lung cancer. Anticancer Res. 2013;33:981–990. [PubMed] [Google Scholar]
- 17.Korpanty GJ, Graham DM, Vincent MD, Leighl NB. Biomarkers that currently affect clinical practice in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol. 2014;4:204. doi: 10.3389/fonc.2014.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton CC, Aitchison AJ, Gehrig K, Ridgway ND. A mechanism for suppression of the CDP-choline pathway during apoptosis. J Lipid Res. 2013;54:3373–3384. doi: 10.1194/jlr.M041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serran-Aguilera L, Nuti R, López-Cara LC, Ríos-Marco P, Carrasco MP, Marco C, Entrena A, Macchiarulo A, Hurtado-Guerrero R. Choline kinase active site provides features for designing versatile inhibitors. Curr Top Med Chem. 2014;14:2684–2693. doi: 10.2174/1568026614666141216093337. [DOI] [PubMed] [Google Scholar]
- 20.Lacal JC, Campos JM. Preclinical characterization of RSM-932A, a novel anticancer drug targeting the human choline kinase alpha, an enzyme involved in increased lipid metabolism of cancer cells. Mol Cancer Ther. 2015;14:31–39. doi: 10.1158/1535-7163.MCT-14-0531. [DOI] [PubMed] [Google Scholar]
- 21.Granata A, Nicoletti R, Tinaglia V, De Cecco L, Pisanu ME, Ricci A, Podo F, Canevari S, Iorio E, Bagnoli M, Mezzanzanica D. Choline kinase-alpha by regulating cell aggressiveness and drug sensitivity is a potential druggable target for ovarian cancer. Br J Cancer. 2014;110:330–340. doi: 10.1038/bjc.2013.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Cueva A, Ramírez de Molina A, Alvarez-Ayerza N, Ramos MA, Cebrián A, Del Pulgar TG, Lacal JC. Combined 5-FU and ChoKα inhibitors as a new alternative therapy of colorectal cancer: Evidence in human tumor-derived cell lines and mouse xenografts. PLoS One. 2013;8:e64961. doi: 10.1371/journal.pone.0064961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gadiya M, Mori N, Cao MD, Mironchik Y, Kakkad S, Gribbestad IS, Glunde K, Krishnamachary B, Bhujwalla ZM. Phospholipase D1 and choline kinase-α are interactive targets in breast cancer. Cancer Biol Ther. 2014;15:593–601. doi: 10.4161/cbt.28165. [DOI] [PMC free article] [PubMed] [Google Scholar]


