Abstract
MicroRNAs (miRNAs) bind to the 3′-untranslated regions (3′-UTRs) of mRNAs, affecting translation and regulating cell differentiation, tumorigenesis and apoptosis. Genetic polymorphisms in these regions in target genes are able to affect the binding affinity between miRNA and target genes, ultimately affecting the expression of individual miRNAs. In the present case-control study, genotyping of 5 microRNA single nucleotide polymorphisms (SNPs) located at the binding site of the 3′-UTR of RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667), GOLGA7 (rs11337) and KRT81 (rs3660) genes was assessed in order to investigate its role in gastric cancer (GC). The results indicated that the rs4901706 SNP, which is located in the 3′-UTR of C14orf101, was associated with GC development risk, as determined by χ2 analysis (relative risk, 1.630; 95% confidence interval, 1.070–2.483; P=0.022). A Renilla/luciferase reporter assay also indicated the different binding affinity between the SNP of rs4901706 and microRNA. In conclusion, rs4901706 SNP of C14orf101 gene in the microRNA binding site may be used as a valuable biomarker when predicting GC risk.
Keywords: single nucleotide polymorphism, rs4901706, C14orf101, gastric cancer, cancer risk
Introduction
Gastric cancer (GC) is associated with high morbidity and mortality rates. It is fourth most common type of cancer and one of the leading causes of cancer-associated mortality around the world (1–3). Based on the GLOBOCAN 2012 data from the World Health Organization (http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx), ~1 million new cases of stomach cancer have occurred in 2012 (952,000 cases and 6.8% of the total cancer) with 70% of cases occurring in developing countries, particularly in Eastern Asia. Half the world total occurred in Eastern Asia, which made it the fifth most common malignancy in the world. Stomach cancer is also the third leading cause of cancer death in both genders worldwide (723,000 deaths and 8.8% of the total cancer). GC is a multifactorial disease, with environmental and genetic factors contributing to its etiology (4,5). Although advancements have been achieved in the treatment of GC in recent years, GC is still associated with a poor prognosis, particularly due to delayed diagnosis (6).
MicroRNAs (miRNAs) are ~22-nucleotide long molecules that regulate the expression of mRNAs by base pairing to their 3′-untranslated region (3′-UTR), thereby preventing translation (7,8). A great number of studies have indicated that miRNAs are critical in numerous biological processes, including apoptosis, proliferation, insulin secretion, tumorigenesis and cellular differentiation (7,8). In humans, >700 miRNAs have been identified, and these miRNAs regulate the expression of 30% of protein-coding genes (9). In fact, miRNAs target nucleotides in the ‘seed region’ of the 3′-UTR (2–8 nucleotides in the 5′ end) of the mRNA. Perfect complementarity between the miRNA and its target mRNA sequence leads to RNA silencing, and thus, the protein levels are reduced (10,11). It has been demonstrated that single nucleotide polymorphisms (SNPs) in the 3′-UTR alter target gene expression, affecting the risk of cancer development in individuals (12,13).
A study by Yu et al (14) identified 12 SNPs associated with cancer development risk, which were located in the miRNA target site. These SNPs were then genotyped in healthy subjects from the Hebei area in China, and 6 SNPs were excluded (14) due to presenting a minor allele frequency of <5%. The SET8 expression have been proven to be associated with the outcome of gastric cancer (15). The potential value of one of these SNPs, rs16917496, in the SET8 gene was examined in a preliminary study to evaluate the correlation between rs16917496 polymorphisms and SET8 expression in GC patients (unpublished data). Thus, the present study aimed to investigate the remaining 5 miRNA binding-site SNPs in the 3′-UTR of RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667), GOLGA7 (rs11337) and KRT81 (rs3660) in GC patients in order to assess their association with the risk of cancer development.
Materials and methods
Tissue specimen collection and DNA extraction
Blood samples were collected before GC resection from a total of 153 GC patients (112 males and 41 females) who underwent GC resection at the Department of General Surgery at the Fourth Hospital of Hebei Medical University (Shijiazhuang, China) prior to the operation between February 2008 and November 2010. All GC patients were diagnosed preoperatively via histopathological examination (16). The TNM system was used to determine the different lymph node metastasis and clinical stages (17). In addition, blood samples were collected from 233 healthy subjects (169 males, 64 females) with no previous history of cancer. The characteristics of patients and controls were listed in Table I. Patients provided their informed consent for participation in the present study. All procedures were supervised and approved by the Human Tissue Research Committee of the Fourth Hospital of Hebei Medical University (Shijiazhuang, China).
Table I.
Clinical characteristics of gastric cancer patients and healthy controls.
| Characteristic | Cases, n | Controls, n | P-value |
|---|---|---|---|
| Gender | 0.885 | ||
| Male | 112 | 169 | |
| Female | 41 | 64 | |
| Age (years) | 0.209 | ||
| ≤60 | 61 | 108 | |
| >60 | 92 | 125 | |
| Tumor size (diameter) | |||
| ≤6 cm | 76 | ||
| >6 cm | 77 | ||
| Tumor location | |||
| Upper | 55 | ||
| Middle | 47 | ||
| Lower | 51 | ||
| Lymph node metastasis | |||
| N0 | 43 | ||
| N1 | 36 | ||
| N2 | 34 | ||
| N3 | 40 | ||
| Clinical stages | |||
| I+II | 46 | ||
| III+IV | 90 | ||
| Extent of differentiation | |||
| Moderate | 96 | ||
| Poor | 57 | ||
| Pathological subtype | |||
| Diffuse | 100 | ||
| Intestinal | 27 | ||
| Mixed | 26 |
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (A1125; Promega Corp., Madison, WI, USA). Briefly, 100 µl blood was added to the cell lysis solution for red blood cells, followed by lysis of the white blood cells and their nuclei in the Nuclei Lysis Solution with RNase. The cellular proteins were then removed by a salt-precipitation step. Finally, the genomic DNA was concentrated and desalted by isopropanol precipitation.
SNP genotyping of microRNA binding-site SNPs
The miRNA SNPs located at the binding site, including RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667), GOLGA7 (rs11337) and KRT81 (rs3660), were genotyped using the ligation detection reaction method (18), featuring forward and reverse primers, in order to amplify DNA fragments flanking the SNPs. This procedure was performed according to the SNP database of the National Center for Biotechnology Information (Bethesda, MD, USA; http://www.ncbi.nlm.nih.gov/snp/).
Polymerase chain reaction (PCR) was performed with 50 ng genomic DNA, 1 µl of 10 nm primer pairs, 12.5 µl of Master Mix and distilled water to a final volume of 25 µl using a PCR Master Mix kit (K1081; Promega Corp.), according to the manufacturer's instructions, and for 35 thermal cycles. The primer and probe sequences used in PCR are shown in Table II. Ligation was performed with various probes, which were matched to the SNPs. Subsequently, the ABI PRISM 3730xl DNA Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA) was used to separate the ligated products. SNPs were detected and verified based on differences in the length of ligated products. The experiment was performed once if it was successful.
Table II.
Primers and probes used for genotyping of miRNA SNPs.
| Gene | rs NCBI | Primer sequence | Probe sequence |
|---|---|---|---|
| RYR3 | rs1044129 | F: ATGGAGTAATGCTTTATGGTC | S1: TTTAGGTGAATCTCCTCAAATACAA |
| (A/G) | R: CAGTCACAGAGTGGTTGTAGA | S2: TTTTTTAGGTGAATCTCCTCAAATACAG | |
| S3: TGAAGTGCCCACTGCAATAAAGTAA | |||
| KIAA0423 | rs1053667 | F: CATGAAATCTGAGTCACATGG | S1: TTTTAGTAATCATGTTTTAATGTAGAACC |
| (C/T) | R: GCTGAGAAATGAGACATACCA | S2: TTTTTTTAGTAATCATGTTTTAATGTAGAA | |
| S3: TCAAACAGGATGGAACATCAGTGGATTT | |||
| C14orf101 | rs4901706 | F: AAACTAAGTCATCTCCCAGATA | S1: TTTTTTTTTTAATGGGGTATTCAGTGACTAAGA |
| (A/G) | R: GTCATCTGGTGAAAGACTGGA | S2: TTTTTTTTTTTTTAATGGGGTATTCAGTGACTAAGG | |
| S3: TCTGCTATTTATGCAAAATTCTGTTTTTTTT | |||
| GOLGA7 | rs11337 | F: CGCTGTATTTGGGAGAGAGTT | S1: TTTTTTTTTTTTACCATTAAAAGTTTCACTGTCAGAG |
| (G/T) | R: CAGGCTGTAAAGTAACAAATGAG | S2: TTTTTTTTTTTTTTTACCATTAAAAGTTTCACTGTCAGAT | |
| S3: ATATTGTAGGTGCTAATACTGGATTTTTTTTTTT | |||
| KRT81 | rs3660 | F: GTTAGGCACCCCAACTCAAGT | S1: TTTTTATTTTTGAGAAAAGTCCTGCTCAC |
| (G/C) | R: GCCAGCGGACTTCTTTCTAGG | S2: TTTTTTTTATTTTTGAGAAAAGTCCTGCTCAT | |
| S3: TTGCACTATTCTATAGAAACTACAATTT |
F represents the forward primer. R represents reverse primer. S1 and S2 represent probes that match to different alleles of the SNP. S3 represents probes downstream of the SNP. miR, microRNA; SNP, single nucleotide polymorphism; NCBI, National Center for Biotechnology Information.
Renilla/luciferase reporter assays
A total of 4 oligonucleotides were synthesized based on dbSNP, the NCBI database of genetic variation, which consisted of the following parts (from 5′ to 3′): A XhoI sticky end (5 bp), a fragment from the 3′-UTR of the C14orf101 gene containing the GG or AA genotype (rs4901706; 47 bp), and a NotI sticky end (2 bp). The following sequences were used: GG-containing sense, 5′-TCGAGAGTGCTCAGCTACTTCTCCTCCACTTTGAAAGACCCCTCCCAGATCTGC-3′, and antisense, 5′-GGCCGCAGATCTGGGAGGGGTCTTTCAAAGTGGAGGAGAAGTAGCTGAGCACTC-3′; AA-containing sense, 5′-TCGAGAGTGCTCAGCTACTTCTCCTGCACTTTGAAAGACCCCTCCCAGATCTGC-3′, and antisense, 5′-GGCCGCAGATCTGGGAGGGGTCTTTCAAAGTGCAGGAGAAGTAGCTGAGCACTC-3′. The four oligonucleotides were incubated for 5 min with 1X NEBuffer 2 (New England Biolabs, Ipswich, MA, USA) in a heating block at 95°C. Next, the temperature was gradually reduced until it reached room temperature. The psiCheck-2 vector featuring the Renilla/luciferase and controlled firefly luciferase genes was linearized by digestion with NotI and XhoI (New England Biolabs), and the vector was then purified using a 1% agarose gel electrophoresis. The oligonucleotides were ligated in the linearized psiCheck-2 vector (Promega Corp.) into the cloning sites (NotI and XhoI), which were downstream of the Renilla luciferase reporter gene with T4 DNA ligase (Promega Corp.). Subsequently, the ligated vectors were transformed in Escherichia coli-competent cells, as follows: Competent cells were taken from −80°C freezer and thawed on ice for 20 min. 1 µl of ligated vector was mixed into 100 µl of competent cells in a microcentrifuge. The competent cell/DNA mixture was then placed on ice for 20–30 min and each transformation tube was heat shocked by placing the tube into a 42°C water bath for 90 sec. The tubes were placed back on the ice for 2 min and 500 µl of Luria-Bertani (LB) medium was added. The cells were grown at 37°C in a shaking incubator for 45 min. All of the transformations were plated onto a 10 cm LB agar plate and the plates were incubated at 37°C overnight. A total of 3 bacterial colonies were selected and the bacterial culture was incubated at 37°C for 12–18 h in a shaking incubator. The bacterial fluid samples were sequenced by the methods of Sanger dideoxy (Sangon Biotech, Shanghai, China), and sequencing was used to identify and select positive clones.
A human GC cell line (SGC 7901) purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China) was seeded in 48-well plates and transfected with the modified psiCheck-2 vector (800 ng) using Lipo 2000 (11668-027; Invitrogen, CA, USA) at 37°C for 4–6 h with the GG or AA genotype. At 48 h after transfection, the Renilla luciferase activity was assessed using the Dual-Lucy Assay kit (Vigorous Instrument Co., Ltd., Beijing, China) and a luminometer BioFix Lumi-10 (Macherey-Nagel, Dusseldorf, Germany). The transfection efficiency was normalized against the firefly luciferase activity.
Statistical analysis
The χ2 test was performed to analyze dichotomous variables, including the presence or absence of an individual SNP in the patients and healthy controls. The odds ratio (OR) and 95% confidence interval (CI) were calculated using an unconditional logistic regression model. In addition, Student's t-test was used to compare the differential expression levels between genotypic groups in the Renilla/luciferase reporter assays. SPSS version 18.0 software (SPSS, Inc., Chicago, IL, USA) was used to perform statistical analyses. Statistically significant differences were defined when the P-value was <0.05.
Results
Patient characteristics
A total of 153 patients and 233 healthy controls were included in the present study. The clinical characteristics of patients including gender, age, tumor size and location, metastasis, clinical stage, differentiation status and pathological types were listed in Table I. The characteristics of the control, including gender and age were also listed in Table I. The distribution frequency was not identified to be different between patients and controls with respect to their age and gender (P=0.885 and 0.209) respectively. In addition, the mean age between patients and controls was not statistically different. An analysis for these patients and the controls was subsequently performed.
Association of C14orf101 SNP with risk of GC
In the present study, miRNA binding-site SNPs were genotyped in the patients and healthy controls, including C14orf101 (rs4901706), RYR3 (rs1044129), GOLGA7 (rs11337), KRT81 (rs3660) and KIAA0423 (rs1053667) to evaluate their association with the risk for cancer. The cancer risk association was evaluated for the distribution frequency of rs4901706 (GG vs. AG+AA), rs1044129 (AA vs. AG+GG), rs11337 (GG vs. GT+TT), rs3660 (GG vs. CG+CC) and rs1053667 (TT vs. CT+CC) between patients and controls using the χ2 test. As shown in Table III, the rs1044129 AA carriers were 47 in GC patients and 67 in the controls whereas the AG+GG carriers were 106 in GC patients and 166 in the controls; the rs1053667 TT carriers were 113 in GC patients and 161 in the controls whereas the CT+TT carriers were 40 in GC patients and 72 in the controls. In addition, the rs11337 GG carriers were 92 in GC patients and 141 in the controls while the GT+TT carriers were 61 in GC patients and 92 in the controls; Finally, the rs3660 GG carriers were 97 in GC patients and 145 in the controls while CG+CC carriers were 56 in GC patients and 88 in the controls. The rs 1044129 (relative risk, 1.099; 95% CI, 0.704–1.715; P=0.679), rs11337 (relative risk, 0.984; 95% CI, 0.649–1.493; P=0.940), rs3660 (relative risk, 1.023; 95% CI, 0.662–1.583; P=0.917) and rs1053667 (relative risk, 1.263; 95% CI, 0.801–1.992; P=0.314) were not associated with cancer risk by the analysis of the present study. The GG carriers were 100 in GC patients and 125 in the controls whereas the AG+AA were 53 in GC patients and 108 in the controls. It was also noted that the GG genotype was susceptible to GC carcinogenesis (relative risk, 1.630; 95% CI, 1.070–2.483; P=0.022). The data of the present study demonstrated that the rs4901706 SNP of C14orf101 was a predictive marker for GC risk.
Table III.
Distribution frequency of miRNA-binding SNPs between GC patients and healthy controls.
| Variant | Gene | Genotype | Cases | Controls | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| rs1044129 | RYR3 | AA | 47 | 67 | 0.679 | 1.099 | 0.704–1.715 |
| AG+GG | 106 | 166 | |||||
| rs1053667 | KIAA0423 | TT | 113 | 161 | 0.314 | 1.263 | 0.801–1.992 |
| CT+CC | 40 | 72 | |||||
| rs4901706 | C14orf101 | GG | 100 | 125 | 0.022 | 1.630 | 1.070–2.483 |
| AG+AA | 53 | 108 | |||||
| rs11337 | GOLGA7 | GG | 92 | 141 | 0.940 | 0.984 | 0.649–1.493 |
| GT+TT | 61 | 92 | |||||
| rs3660 | KRT81 | GG | 97 | 145 | 0.917 | 1.023 | 0.662–1.583 |
| CG+CC | 56 | 88 |
miRNA, microRNA; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
rs4901706 SNP in C14orf101 affects protein translation in GC cells
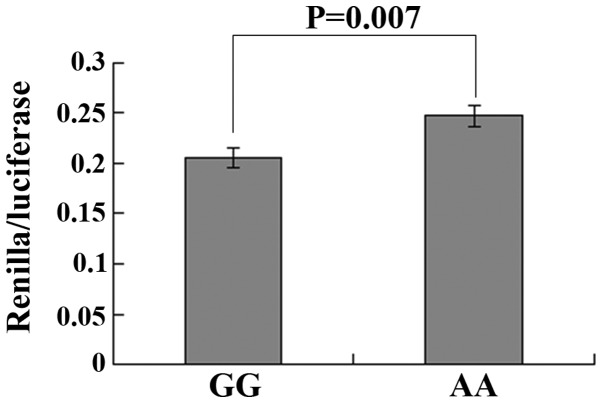
A vector named psiCheck-2 was constructed containing the rs4901706 AA or GG genotypes downstream of the Renilla/luciferase reporter gene in order to assess their functional effect on C14orf101 expression. The vector was transfected into the GC cell line SGC 7901. As shown in Fig. 1, a significant reduction of Renilla/luciferase activity was observed in GG genotypes compared with that of the AA genotypes (P=0.007). These data demonstrated that rs4901706 SNP may change the binding affinity between the rs4901706 genotype at the 3′UTR of C14orf101 and the corresponding miRNA.
Figure 1.
Renilla luciferase assay performed on the GG and AA genotypes at 48 h after transfection with modified psiCheck-2 vector transfected in SGC 7901 cell lines. The data are presented as the ratio of the Renilla luciferase activity against the firefly luciferase activity.
Discussion
In the present study, the SNPs of miRNA binding sites, including RYR3 (rs1044129), C14orf101 (rs4901706), KIAA0423 (rs1053667), GOLGA7 (rs11337) and KRT81 (rs3660), were examined for their ability to predict GC cancer risk. The results revealed that the rs4901706 SNP of C14orf101 gene is a possible risk biomarker for GC. To the best of our knowledge, the current study is the first to investigate the miRNA binding-site SNPs at the 3′-UTR of C14orf101, which exhibit the ability to predict GC risk. The miR-SNPs defined as the SNP at the miRNA binding site, the miRNAs and miRNAs processing machinery genes, are critically involved in disease phenotypes (19,20). We have identified gastric cancer associated miR-SNP of miRNAs processing machinery genes in previous study (21). Yu et al (14) performed a genome-wide analysis of SNPs located in the miRNA-binding sites of the 3′-UTR of various human genes and revealed that some miRNA-binding SNP distribution frequencies between the human cancer EST libraries and the dbSNP database was significantly different. Furthermore, they identified that twelve miRNA-binding SNPs displayed an aberrant allele frequency in human cancers using human cancer specimens against the dbSNP database for case-control association studies. Finally, they deduced that SNPs located in miRNA-binding sites was potentially associated with cancer by affecting miRNA target expression and function (14). Consistent with these previous findings, the frequent allele G of rs4901706 was found to be associated with GC risk in the present study.
MiRNAs appear to be critical in the response of patients to various treatments, particularly cancer treatment (22). In addition, the rs4901706 SNP of the C14orf101 gene has been previously demonstrated to be associated with cancer risk, since it serves as an miRNA binding site; however, the miRNA binding to this site that modulates the C14orf101 expression was not specified (14,23). In a previous study, we demonstrated that this SNP was also associated with survival in patients with non-Hodgkin lymphoma G allele carriers that exhibited a long life span (24). The data demonstrated that the GG genotype of C14orf101 would modify both the carcinogenesis and outcome of the cancer.
C14orf101, also known as transmembrane protein 260 (TMEM260), is conserved in chimpanzees, rhesus macaque monkeys, canines, cattle, mice, rats, chickens and zebrafish. However, a functional study of this gene has not been reported to date. The results of the current study suggest that the miRNA-binding SNPs of C14orf101 have an effect on cancer development. A Renilla/luciferase reporter assay highlighted the different binding affinity between the SNP of rs4901706 and miRNA binding to the rs4901706 site. Furthermore, the A to G transition of rs4901706 in the 3′-UTR of C14orf101 may result in the destruction of the A:T bond at the miRNA binding site, so as to alter the affinity of C14orf101 in binding with miRNAs. This may modulate the expression of the gene, thereby initiating GC carcinogenesis. However, the current study should be replicated in other populations and in laboratory-based functional studies in order to validate the results. The miRNA binding to C14orf101 should be identified, and the change in the expression of C14orf101 on proliferation, invasion and apoptosis of gastric cancer cells should be evaluated in future studies.
In conclusion, SNPs in the C14orf101 miRNA binding site appeared to represent a biomarker for cancer risk. rs4901706 SNP was a potential maker for early diagnosis of gastric cancer and valuable for gastric cancer prevention. Further analyzing the genetic polymorphisms of miRNA binding sites may result in better identification of patients susceptible to GC development. The miRNA binding to C14orf101 should be identified as well as the role of the expressional change of C14orf101 on proliferation, invasion and apoptosis of gastric cancer cells in future studies.
Acknowledgements
This study was supported by a grant from the Key Basic Research Program of Hebei (grant no. 14967713D).
References
- 1.Parkin DM. International variation. Oncogene. 2004;23:6329–6340. doi: 10.1038/sj.onc.1207726. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. doi: 10.1016/S0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 3.Stadtländer CT, Waterbor JW. Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis. 1999;20:2195–2208. doi: 10.1093/carcin/20.12.2195. [DOI] [PubMed] [Google Scholar]
- 4.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman D, Burley VJ. Gastric cancer: Global pattern of the disease and an overview of environmental risk factors. Best Pract Res Clin Gastroenterol. 2006;20:633–649. doi: 10.1016/j.bpg.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Xia J, Guo X, Yan J, Deng K. The role of miR-148a in gastric cancer. J Cancer Res Clin Oncol. 2014;140:1451–1456. doi: 10.1007/s00432-014-1649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicateds that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/S1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 12.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, Hemminki K, Lenner P, Försti A. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–1399. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi XL, Guo ZJ, Wang XL, Liu XL, Shi GF. SET8 expression is associated with overall survival in gastric cancer. Genet Mol Res. 2015;14:15609–15615. doi: 10.4238/2015.December.1.12. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko S, Yoshimura T. Time trend analysis of gastric cancer incidence in Japan by histological types, 1975–1989. Br J Cancer. 2001;84:400–405. doi: 10.1054/bjoc.2000.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 18.Yi P, Lu W, Guo J, Liu Q, Chen Z, Han J, Li L. Development of a PCR/ligase detection reaction/nanogold-based universal array approach for the detection of low-abundant DNA point mutations. Cell Biochem Biophys. 2011;61:629–636. doi: 10.1007/s12013-011-9248-7. [DOI] [PubMed] [Google Scholar]
- 19.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Wang Y, Zhao Y, Guo Z. Single-nucleotide polymorphisms of microRNA processing machinery genes are associated with risk for gastric cancer. Onco Targets Ther. 2015;8:567–571. doi: 10.2147/OTT.S79150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B, Liu C, Diao L, Wang C, Guo Z. A polymorphism at the microRNA binding site in the 3′ untranslated region of C14orf101 is associated with non-Hodgkin lymphoma overall survival. Cancer Genet. 2014;207:141–146. doi: 10.1016/j.cancergen.2014.03.007. [DOI] [PubMed] [Google Scholar]