Abstract
Despite the fact that the biological function of cluster of differentiation (CD)133 remains unclear, this glycoprotein is currently used in the identification and isolation of tumor-initiating cells from certain malignant tumors, including pancreatic cancer. In the present study, the involvement of mucin 1 (MUC1) in the signaling pathways of a highly tumorigenic CD133+ cellular subpopulation sorted from the pancreatic cancer cell line HPAF-II was evaluated. The expression of MUC1-cytoplasmic domain (MUC1-CD) and oncogenic signaling transducers (epidermal growth factor receptor, protein kinase C delta, glycogen synthase kinase 3 beta and growth factor receptor-bound protein 2), as well as the association between MUC1 and β-catenin, were characterized in HPAF-II CD133+ and CD133low cell subpopulations and in tumor xenografts generated from these cells. Compared with HPAF CD133low cells, HPAF-II CD133+ cancer cells exhibited increased tumorigenic potential in immunocompromised mice, which was associated with overexpression of MUC1 and with the accordingly altered expression profile of MUC1-associated signaling partners. Additionally, MUC1-CD/β-catenin interactions were increased both in the HPAF-II CD133+ cell subpopulation and derived tumor xenografts compared with HPAF CD133low cells. These results suggest that, in comparison with HPAF CD133low cells, CD133+ cells exhibit higher expression of MUC1, which contributes to their tumorigenic phenotype through increased interaction between MUC1-CD and β-catenin, which in turn modulates oncogenic signaling cascades.
Keywords: pancreatic cancer, MUC1, cancer stem cells, cell signaling
Introduction
The pentaspan membrane glycoprotein prominin-1, also known as cluster of differentiation (CD)133, was initially described as a cell surface antigen specific for hematopoietic stem cells and progenitor cells (1,2). The biological function of CD133 remains unclear. However, it is currently used in the identification and isolation of tumor-initiating cells from certain malignant tumors, whereby it correlates with poor prognosis (3–5). Tumor-initiating cells, also called cancer stem cells (CSCs), are characterized by their self-renewal capacity and the ability to generate cell subpopulations during tumor growth (6–8). Although CD133 is a reasonable marker of various CSCs, several types of cancer arise from cells with different markers (9,10).
Pancreatic cancer is the fifth most lethal cancer in developed countries, with a 5-year survival rate of <6% (11). Early metastasis, late diagnosis and the low effectiveness of the currently available therapies contribute to its high mortality rate (12). A previous study on pancreatic cancer identified that cells expressing CD44, CD24 and epithelial-specific antigen surface markers were associated with an increase in the tumorigenic and self-renewal capacity of tumor cells isolated from primary tumors or low-passage tumor xenografts (8). However, these markers do not identify CSCs within all pancreatic tumors, and other studies revealed that the use of CD133 to isolate tumor-initiating cells yielded populations of cells with enhanced tumorigenic potential, high resistance to standard chemotherapy and a close association with metastatic phenotype (7,13). In addition, a recent study confirmed that enforced expression of CD133 enhanced the aggressive behavior of pancreatic cancer cells (14).
MUC1 is a heavily glycosylated transmembrane glycoprotein expressed at low levels in the apical surfaces of epithelial cells (15). This glycoprotein possesses oncogenic properties, and is overexpressed in >80% of pancreatic tumors, contributing to tumor progression, metastasis and mortality in patients with pancreatic cancer (16–20). The MUC1 gene encodes a protein comprised of a large extracellular domain with a tandem repeat region, a transmembrane domain and a highly conserved cytoplasmic domain (MUC1-CD), which participates in several oncogenic signaling pathways (21). MUC1-CD is highly conserved, and contains seven tyrosine residues and several serine and threonine residues that represent potential docking sites for proteins with Src homology 2 domains and recognition sites for receptor tyrosine kinases and other kinases, including protein kinase C delta (PKCδ), glycogen synthase kinase 3 beta (GSK3β) and ErbB receptors such as epidermal growth factor receptor (EGFR) (22). Furthermore, MUC1-CD contains a serine-rich motif that functions as a β-catenin binding site, and the phosphorylation of MUC1-CD modulates this affinity (23). MUC1-CD/β-catenin interactions enhance the malignant phenotype of tumor cells by regulating the activity of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors, thus modulating the expression of several genes involved in the tumorigenic phenotype, including target genes in the Wnt signaling pathway (24). Recently, a transmembrane cleaved form of MUC1 has been reported to exert an important role in chemoresistance to standard chemotherapy agents (25), and to potentially serve as an accurate marker of pluripotency in human embryonic stem cells (26). The expression of MUC1 in CSCs has been documented by a novel antibody against tumor-associated MUC1 that recognizes a sequence in the tandem repeat region of MUC1, which is different from the sequences recognized by the majority of commercially available antibodies against MUC1 (27).
Based on the reported associations of MUC1 with CSCs, the present study aimed to investigate the potential contribution of MUC1 to the oncogenic signaling pathways of CD133+ pancreatic cancer cells. The results revealed that MUC1/β-catenin interactions are associated with enhanced tumorigenic properties of CD133+ pancreatic cancer cells.
Materials and methods
Cell culture
The human pancreatic cell line HPAF-II was obtained from the American Type Culture Collection (Manassas, VA, USA), and was cultured in RPMI 1640 medium Gibco; Thermo Fisher Scientific Inc.,Waltham, MA USA containing GlutaMAXTMI (Gibco; Thermo Fisher Scientific, Inc.) and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 50 mg/ml gentamicin (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were grown at 37°C with 5% CO2 in a humidified atmosphere.
CD133 cell-surface expression analysis by flow cytometry
The expression levels of CD133 in the HPAF-II cell line were assessed by flow cytometry with an anti-CD133/2-phycoerythrin (PE) monoclonal antibody (MAb) [#130-080-901; mouse immunoglobulin (IgG)1; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany]. A mouse IgG1 MAb served as a control (#130-092-212; Miltenyi Biotec GmbH).
To perform flow cytometry analysis, cells were trypsinized when 80% of confluence was reached. For each analysis, 5×105 cells were used. Cells were incubated with a mouse IgG1 MAb solution (1:80) for 10 min at 4°C, and next resuspended in an anti-CD133/2-PE antibody solution (1:10) for 10 min at 4°C in the dark. Upon incubation, the cells were washed with 0.1% PBS two times, and resuspended in 500 µl magnetic-activated cell sorting (MACS) buffer [phosphate-buffered saline (PBS) supplemented with 0.5% bovine serum albumin (BSA) and 2 mM ethylenediaminetetraacetic acid (EDTA)], prior to be analyzed in a FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). A cell suspension that was only incubated with mouse IgG1 MAb was used as a control. Analysis of the results was performed using FlowJo version 7.2.5 software (FlowJo, LLC, Ashland, OR, USA).
MACS
Cell subpopulations (CD133− and CD133+) were isolated using a MACS system and microbeads coupled to anti-CD133/1 MAb (Miltenyi Biotec GmbH).
Magnetic separation was performed using the MidiMACSTM magnetic separation kit (Miltenyi Biotec GmbH), according to the manufacturer's protocol with minor alterations. Briefly, 1×108 cells were washed twice with 0.1% PBS and passed through a pre-separation filter (30 µm) in order to remove cell clumps. Subsequently, the cell suspension was incubated with human IgG FcR Blocking Reagent (1:3 in MACS buffer; Miltenyi Biotec, GmbH) and CD133 microbeads (1:5; Miltenyi Biotec, GmbH) for 30 min at 4°C. Following the incubation step, cells were washed twice with 0.1% PBS, and the pellet was resuspended in 500 µl MACS buffer (PBS supplemented with 0.5% BSA and 2 mM EDTA). The cell suspension was then transferred to an LS column (Milteny Biotec GmbH) previously hydrated with 3 ml buffer, and placed in a magnetic support. The total effluent was collected as the CD133− fraction, and the column was then washed three times with 3 ml buffer. Next, the column was removed from the magnet and, with the aid of a plunger, 5 ml MACS buffer were used to flush the microbeads-labeled cells out of the column. The effluent was collected as the CD133+ fraction.
The CD133− cell subpopulation was subsequently passed through the LS column, and washed three times with 1 ml MACS buffer to further deplete the remaining CD133+ cells. Both CD133+ and CD133− fractions were centrifuged (300 × g; Centrifuge 5810R; Eppendorf, Hamburg, Germany), and the pellets were resuspended in culture medium and maintained at 37°C in a humidified atmosphere containing 5% CO2.
In vivo tumorigenic assay
The present in vivo tumorigenic assay was approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center (Omaha, USA; protocol 98-088-03FC). Three groups of non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice (n=5/group) were subcutaneously injected in the right dorsal flanks with 3,500 HPAF-II cells [wild-type (wt), CD133low or CD133+]. Mice were bred and maintained under pathogen-free conditions, which included: A 12 h light/12 h dark cycle, 6 AM/6 PM; water bag accessible at all times; Nestlets (Animal Specialties and Provisions, LLC, Quakertown, PA, USA) or NestPaks (WF Fisher and Son, Inc., Somerville, NJ, USA) for enrichment; 18–23°C with 40–60% humidity; and Standard Chow food, similar to LabDiet 5010 (protein 23%; fat content not less than 4.5%). Animals were observed twice a day by trained veterinary staff and once a day by laboratory staff from the Eppley Institute for Research in Cancer and Allied Disease (Omaha, NE, USA). Mice were euthanized 4 weeks following cell injection, which was the time point when it was necessary to euthanize the first mouse due to the initial signs of suffering. The maximum tumor size achieved was 263.8 mm3. Animals were sacrificed with the aid of CO2. Following 5 min without signs of heartbeat or respiration, the animals were subjected to cervical dislocation to ensure mortality. Tumors were collected, fixed in 10% formalin (Thermo Fisher Scientific Inc.) and embedded in paraffin (Thermo Fisher Scientific Inc.) prior to sectioning (Shandon™ Finesse™ 325 microtome; Thermo Fisher Scientific Inc.). Growth of internal tumors was evaluated by direct examination (palpation), or by careful observation of animal behavior and estimation of post-procedure pain, discomfort, distress or morbidity. Anesthesia, when required, was induced by intraperitoneal administration of ketamine hydrochloride (100 mg/ml; injectable-RL 3760; NDC-0409-2051-05; Hospira, Inc., Lake Forest, IL, USA) and xylazine hydrochloride (20 mg/ml; injectable-AnaSed NADA; 139–236; Lloyd, Inc., Shenandoah, IA, USA).
Immunohistochemistry (IHC)
Tumor xenografts were paraffin-embedded and sectioned at 4-µm thickness. IHC staining to detect CD133 protein expression in tumor xenografts was performed using the Dako EnVision System (Dako, Glostrup, Denmark). Antigen retrieval was performed in an IHC-Tek™ Epitope Retrieval Steamer Set (IHC World, LLC, Woodstock, MD, USA) for 40 min with 10 mM citrate buffer pH 6.0 (Thermo Fisher Scientific Inc.), following deparaffinization in xylene (Thermo Fisher Scientific Inc.) and rehydration. The slides were cooled for 20 min at room temperature, and endogenous peroxidase was blocked with 3% H2O2 (Merck Millipore, Darmstadt, Germany) for 5 min. Primary antibody incubation was performed for 1 h at room temperature with a mouse anti-human CD133/1 MAb (1:25; clone AC133; Miltenyi Biotech GmbH). Slides were next washed in Tris-buffered saline with Tween 20 (Grisp, Porto, Portugal), and incubated with Dako REAL EnVision-horseradish peroxidase (HRP) secondary antibody (Dako) for 30 min at room temperature. For signal detection, the slides were incubated for 5 min with 3,3′-diaminobenzidine chromogen (Dako). Next, tissues were counterstained with hematoxylin (Richard-Allan Scientific™; Thermo Fisher Scientific Inc.) for 3 min, dehydrated, cleared, mounted with Histomount medium (Richard-Allan Scientific™; Thermo Fisher Scientific Inc.) and cover slipped. Hematoxylin and eosin (Richard-Allan Scientific™; Thermo Fisher Scientific Inc.) staining was performed upon antigen retrieval following a standard protocol (28).
Protein extraction and western blot analysis
The expression levels of MUC1-CD and oncogenic signaling proteins were evaluated by western blotting. Unsorted HPAF-II cells and sorted CD133low and CD133+ cell subpopulations were cultured to 80–90% confluence. Upon washing twice with PBS, lysis buffer [10 mM Tris pH 7.4, 150 mM NaCl, 0.1% (v/v) sodium dodecyl sulfate (SDS; Bio-Rad Laboratories, Inc., Hercules, CA, USA), 1 mM phenylmethylsulfonyl fluoride and 1% (v/v) Triton X-100] was added, and cells were scraped. Cell lysates were incubated on ice for 1 h and centrifuged (14,000 × g; Centrifuge 5417R; Eppendorf) for 30 min at 4°C to collect the supernatants. Protein content was assessed wit a bicinchoninic acid protein assay kit (Bio-Rad Laboratories, Inc.), as described in the manufacturer's protocol.
Protein extracts were analyzed by 10% SDS-polyacrylamide gel electrophoresis (Invitrogen; Thermo Fisher Scientific, Inc.), transferred to a polyvinylidene fluoride membrane (GE Healthcare Life Sciences, Chalfont, UK) and incubated overnight at 4°C with anti-MUC-1 Armenian hamster MAb (1:300; catalogue no. Ab-5; Thermo Fisher Scientific, Inc.), anti-EGFR mouse MAb (1:200; catalogue no. sc-81449; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-PKCδ rabbit MAb (1:200; catalogue no. sc-213; Santa Cruz Biotechnology, Inc.), anti-GSK3β mouse MAb (1:200; catalogue no. sc-53931; Santa Cruz Biotechnology), anti-growth factor receptor-bound protein 2 (GRB2) mouse MAb (1:200; catalogue no. sc-8034; Santa Cruz Biotechnology, Inc.), anti-β-catenin MAb (1:1,000; catalogue no., 610153; BD Biosciences) and anti-β-actin MAb (1:2,000; catalogue no. sc-69879; Santa Cruz Biotechnology, Inc.) in 5% non-fat milk diluted in PBS containing 0.1% Tween 20 (Sigma-Aldrich, St. Louis, MO, USA). Next, membranes were washed three times with PBS containing 0.1% Tween 20, and incubated with the corresponding goat anti-Armenian hamster (catalogue no. sc-2443), anti-mouse (catalogue no. sc-2005) or anti-rabbit (catalogue no. sc-2004) peroxidase conjugated antibody (1:2,000; Santa Cruz Biotechnology, Inc.) in 5% non-fat milk diluted in PBS containing 0.1% Tween 20. Proteins were visualized using an enhanced chemiluminescence detection kit (Bio-Rad Laboratories, Inc.).
Immunoprecipitation assay
The interaction between MUC1-CD and β-catenin in the HPAF-II cell line was evaluated by immunoprecipitation. Proteins from cell lysates (750 µg) were incubated overnight at 4°C with protein G-agarose beads (Sigma-Aldrich) previously linked to anti-MUC1 Ab-5 MAb and normal Armenian hamster IgG (eBioscience, Inc., San Diego, CA, USA). Following three washes, the immune complexes were dissociated from the beads with reducing NuPAGE® buffer (Invitrogen; Thermo Fisher Scientific, Inc.). The immunoprecipitates and cell lysates were separated in 12% Tris-glycine gels (Invitrogen; Thermo Fisher Scientific, Inc.), and immunoblotted following the aforementioned procedure.
In situ proximity ligation assay (PLA)
PLA was used to assess the close proximity (and putative interaction) between MUC1-CD and β-catenin in tumor xenografts. PLAs were performed using Duolink® In Situ Detection Reagents Brightfield (Olink Bioscience, Uppsala, Sweden), according to the manufacturer's protocol. Antigen retrieval was performed in an IHC-Tek™ Epitope Retrieval Steamer Set for 40 min with 10 mM citrate buffer pH 6.0, following deparaffinization and rehydration. Subsequently, the slides were incubated at 37°C for 30 min with a blocking solution (Olink Bioscience) in a humidity chamber.
For β-catenin staining, the mouse primary antibody was used under the same conditions as the ones above described for IHC, and a secondary anti-mouse antibody conjugated with Duolink® In Situ PLA® Probe Anti-Mouse MINUS (Olink Bioscience) was added, followed by 1-h incubation at 37°C in a humidity chamber.
For MUC1 staining, anti-MUC1 Ab-5 primary antibody directly conjugated with DuolinkII Probemarker Plus (Olink Bioscience) was used. The conjugation of the antibody with the probe was performed following the manufacturer's protocol, and hybridization was conducted for 1 h at 37°C in a humidity chamber. Following the ligation of the probes for 30 min at 37°C, amplification of the signal was performed for 120 min at 37°C; both steps occurred in a humidity chamber. To detect the signal, the slides were incubated with HRP-labeled probes and chromogen (catalogue no., DUO92012-30RXN; Olink Bioscience). Subsequently, tissues were counterstained with hematoxylin, dehydrated, cleared and mounted with Histomount medium.
Results
Isolation of a CD133+ cell subpopulation from the HPAF-II cell line
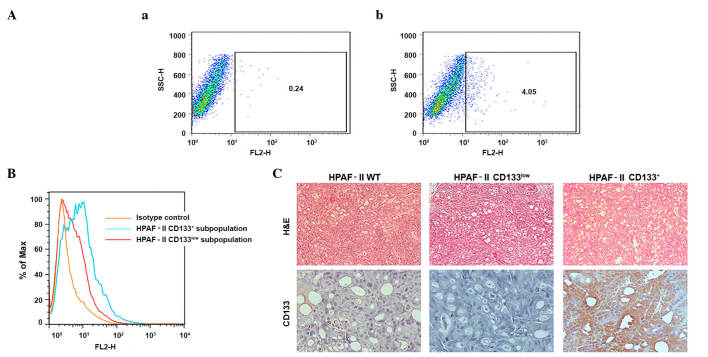
Low-passage/highly tumorigenic samples of the HPAF-II cell line (104 cells produced tumors in 100% of animals; data not shown) were evaluated for CD133 expression levels by flow cytometry. The results indicated that low-passage HPAF-II cells contained ~4% CD133+ cells (Fig. 1A). These cells were isolated using MACS, and both CD133+ and CD133− subpopulations were cultured. To evaluate the enrichment obtained with the sorting methodology used, CD133 was again measured by flow cytometry in the above two cell subpopulations prior to injection into immunodeficient mice (Fig. 1B). The results revealed that the CD133+ subpopulation was highly enriched in CD133+ cells. However, the putative CD133− subpopulation retained a very low percentage of cells expressing CD133, and was therefore called CD133low. Repeated selection did not improve the performance of this procedure (data not shown).
Figure 1.
Validation of the CSC model. (A) Identification of a CSC subpopulation (CD133+ cells) in the HPAF-II pancreatic cancer cell line and evaluation of CD133 expression in cell subpopulations sorted by flow cytometry. (a) Isotype stained cells were used as controls. (b) HPAF-II cells stained with CD133/2-phycoerythrin monoclonal antibody. (B) Enrichment of HPAF-II CD133+ subpopulation isolated by magnetic-activated cell sorting represented on a frequency distribution histogram. The HPAF-II CD133+ subpopulation exhibits 8.89% of CD133+ cells, while the HPAF-II CD133low subpopulation exhibited 3.07% of CD133+ cells, representing an enriched and a depleted population, respectively. (C) CD133 expression in HPAF-II tumor xenografts determined by immunohistochemistry (magnification, ×400). Hemotoxylin and eosin staining was used to reveal the morphology of the tumors (magnification, ×100). CSC, cancer stem cell; CD, cluster of differentiation; H&E, hematoxylin and eosin; SSC-H, measures cell granularity or internal complexity; FH2, phycoerythrin detection; % of Max, % of maximum (normalization method).
The sorted cells were evaluated for tumorigenicity and tumor phenotype. The CD133+ enriched subpopulation exhibited increased tumorigenic potential when injected subcutaneously into NOD/SCID mice, since higher number of tumors were obtained from these cells (Table I), and tumor growth was initiated at earlier time points (3 weeks), compared with the CD133low population (Table I). IHC analysis demonstrated that the xenografts recapitulated the HPAF-II CD133 subpopulation expression levels. Tumors derived from the HPAF-II CD133+ subpopulation retained high CD133 expression levels, with limited negative cells, while the HPAF-II CD133low xenografts were largely negative for expression of CD133. The parental HPAF-II wt-derived xenografts displayed a small percentage of CD133+ cells, similar to the original cell line (Fig. 1C).
Table I.
In vivo tumorigenic assay.
| Time (weeks)a | ||||
|---|---|---|---|---|
| Cell subpopulation | 1 | 2 | 3 | 4 |
| HPAF-II wt | 0/5 | 0/5 | 0/5 | 1/5 |
| HPAF-II CD133low | 0/5 | 0/5 | 0/5 | 1/5 |
| HPAF-II CD133+ | 0/5 | 0/5 | 2/5 | 4/5 |
Data represent the number of animals with tumors vs. the total number of animals injected with different cell subpopulations. CD, cluster of differentiation; wt, wild-type.
Expression of MUC1 in the CD133+ cell subpopulation
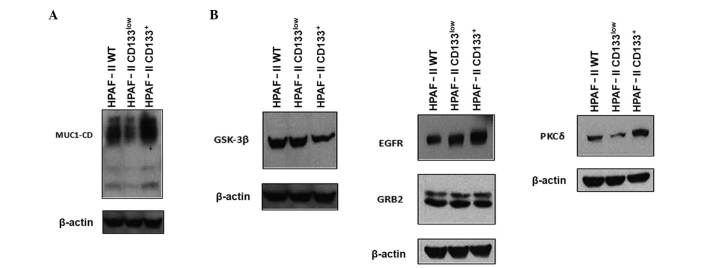
In order to evaluate the relevance of MUC1 glycoprotein in CD133+ cell biology, the expression levels of MUC1 were analyzed in CD133low and CD133+ subpopulations by immunoblotting (Fig. 2A). HPAF-II CD133+ cells were highly enriched in MUC1 expression, compared with HPAF-II wt and HPAF-II CD133low cells. In addition, MUC1 expression levels in the HPAF-II CD133low cell subpopulation were lower than in HPAF-II wt cells (Fig. 2A).
Figure 2.
Expression of MUC1 and signaling partners. (A) Expression of MUC1 in HPAF-II wt, HPAF-II CD133low and HPAF-II CD133+ cells was evaluated by western blotting. β-actin was used as a loading control. (B) Expression of MUC1 signaling partners (epidermal growth factor receptor, growth factor receptor-bound protein 2, protein kinase C delta and glycogen synthase kinase 3 beta) in HPAF-II wt, HPAF-II CD133low and HPAF-II CD133+ cells was evaluated by western blotting. β-actin was used as a loading control. MUC1, mucin 1; MUC1-CD, mucin 1 cytoplasmic domain; EGFR, epidermal growth factor receptor; PKCδ, protein kinase C delta; GSK3β, glycogen synthase kinase 3 beta; GRB2, growth factor receptor-bound protein 2; CD, cluster of differentiation; wt, wild-type.
Expression of MUC1 signaling partners in CD133+ cells
MUC1 function in oncogenic pathways depends on the phosphorylation of its CD by several kinases, including EGFR, PKCδ and GSK3β (21). Since it is not possible to assess the phosphorylation status of MUC1-CD due to the unavailability of antibodies sensitive to phosphorylation, the expression of selected kinases in CD133+ cells was evaluated by immunoblot analysis. The results indicated that CD133+ cells were enriched in EGFR and PKCδ expression, whereas CD133low cells were enriched in GSK3β expression. The protein expression levels of GRB2 were equivalent in HPAF-II wt, HPAF-II CD133+ and HPAF-II CD133low cells (Fig. 2B).
MUC1 and β-catenin interaction in the HPAF-II cell line and tumor xenografts
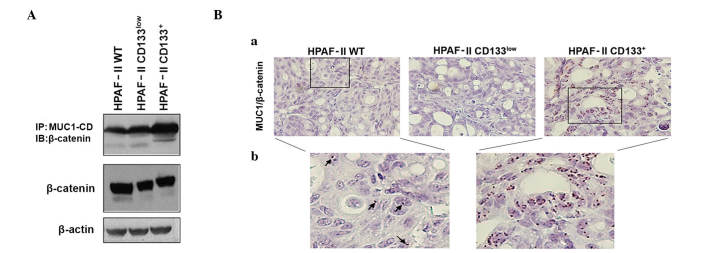
MUC1-CD contains docking sites for β-catenin, and the interactions between MUC1 and β-catenin are known to contribute to the malignant phenotype of tumor cells by modifying the expression of target genes in the Wnt signaling pathway (24,29). To assess if MUC1/β-catenin interaction was potentiated in CD133+ cells, MUC1 was immunoprecipitated from cell lysates of HPAF-II wt, HPAF-II CD133+ and HPAF-II CD133low cells, followed by β-catenin immunoblotting. An enrichment in MUC1/β-catenin interaction was observed in HPAF-II CD133+ cells. β-catenin expression levels were similar in all conditions (Fig. 3A).
Figure 3.
Evaluation of MUC1/β-catenin interaction in HPAF-II cells and xenografts. (A) β-catenin expression and its interaction with MUC1-CD in HPAF-II wt, HPAF-II CD133low and HPAF-II CD133+ cells was evaluated by immunoprecipitation and western blot analysis. β-actin was used as a loading control. (B) In situ PLA in tumor xenografts was used to evaluate the interaction between MUC1 and β-catenin; magnification, (a) ×400; (b) ×650 (Duolink in situ Detection Reagents Brightfield staining). Brown dots correspond to proximity/interaction between MUC1 and β-catenin. Arrows indicate the PLA signals in HPAF-II wt tumors. IP, immunoprecipitation; IB, immunoblotting; CD, cluster of differentiation; wt, wild-type; MUC1, mucin 1; MUC1-CD, mucin 1 cytoplasmic domain; PLA, proximity ligation assay.
MUC1/β-catenin interactions in tumor xenografts were confirmed by in situ PLA. Significant interactions between MUC1 and β-catenin were observed in HPAF-II wt and HPAF-II CD133+-derived xenografts, but almost no interactions were observed in HPAF-II CD133low-derived tumors. The most abundant interactions were observed in the CD133+ tumors (Fig. 3B). In all cases, the interaction signals were predominantly observed in the nuclei of the cells.
Discussion
In the present study, the involvement of CD133 and MUC1 in the highly tumorigenic low-passage pancreatic cancer cell line HPAF-II, which was derived from the ascites of a pancreatic cancer patient, was investigated (30). Using a well-established CD133 selection method, an isolated CD133+ cell subpopulation was demonstrated to exhibit features associated with CSCs (enhanced tumorigenicity) and concomitant enriched expression of MUC1.
CSCs are known to aberrantly activate canonical signaling pathways (31–33). Recently, a MUC1 spliced form was reported to be associated with the differentiation status of stem cells (34). In the present study, the expression of MUC1 and oncogenic signaling transducers (EGFR, PKCδ, GSK3β and GRB2), as well as the MUC1/β-catenin association, were characterized in pancreatic cancer cells that expressed CD133. MUC1-CD, EGFR and PKCδ expression levels were increased in the HPAF-II CD133+ subpopulation, while GSK3β expression was decreased, and no significant differences were observed regarding GRB2 and β-catenin expression levels. These results clearly demonstrate that pancreatic HPAF-II CD133+ cells have a distinct expression profile, which includes MUC1 and its associated signaling partners, compared with the subpopulation of cells that do not express the stem cell surface marker CD133.
MUC1-CD contains docking sites for molecules such as β-catenin, and the association of these proteins is modulated by motifs that may be phosphorylated by several kinases, namely EGFR, PKCδ, GSK3β and GRB2 (35). The phosphorylation of MUC1-CD influences its interaction with β-catenin, which directly binds at the MUC1-CD motif SAGNGGSSLS (22,23). In the present study, increased interactions between MUC1-CD and β-catenin were observed in the HPAF-II CD133+ subpopulation, which was correlated with enhanced expression of EGFR and PKCδ, and decreased expression of GSK3β (24,36–42). It is known that MUC1-CD phosphorylation by EGFR and PKCδ promotes interactions between β-catenin and MUC1, while phosphorylation by GSK3β leads to a decrease in this association (39–41). It was observed in the present study that EGFR and PKCδ expression were upregulated, while GSK3β expression was downregulated, in the HPAF-II CD133+ subpopulation, compared with the HPAF-II CD133low subpopulation, which likely explains the observed increase in MUC1-CD/β-catenin interactions in the CD133+ subpopulation, despite the fact that the steady-state levels of β-catenin remained unchanged in these cells.
The interactions between MUC1-CD/β-catenin influence several tumorigenic processes. Binding of MUC1-CD to β-catenin suppresses the capacity of β-catenin to interact with E-cadherin at adherens junctions, resulting in the loss of cell-cell adhesion, thus playing a relevant role in tumor invasion (43). The MUC1-CD/β-catenin complex contributes to β-catenin stabilization by blocking its GSK3β-mediated phosphorylation and consequently its degradation in the proteasome (24). Furthermore, MUC1-CD/β-catenin is translocated to the nucleus, where it may enhance the activity of β-catenin in association with TCF/LEF transcription factors, thus promoting cell proliferation and survival through upregulation of the transcription of Wnt target genes (36–38,42). Recently, this process has been associated with a metastatic gene expression signature and an epithelial-to-mesenchymal transition phenotype of tumor cells (44). In the present study, the results of in situ PLA for tumor xenografts revealed that CD133+ tumors exhibited frequent MUC1-CD/β-catenin interactions, with the MUC1-CD/β-catenin complex being mainly present in the cellular nuclei, where it presumably binds to transcription factors and activates the transcription of genes involved in cell proliferation and survival.
In summary, the present study has demonstrated for the first time that pancreatic CD133+ cells display enhanced expression of MUC1 and its associated signaling partners. CD133 and MUC1 expression are associated with an aggressive tumor phenotype, partly through the production of enhanced MUC1-CD/β-catenin interactions, and this may partly explain the behavior of pancreatic CSCs.
References
- 1.Yin AH, Miraglia S, Zanjani ED, AlmeidaPorada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 2.Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. A novel five-transmembrane hematopoietic stem cell antigen: Isolation, characterization, and molecular cloning. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 3.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 4.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: Promise of targeted therapy. Gastroenterology. 2010;138:2151–2162. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Song X, Chen Z, Li X, Li M, Liu H, Li J. CD133 expression and the prognosis of colorectal cancer: A systematic review and meta-analysis. PLoS One. 2013;8:e56380. doi: 10.1371/journal.pone.0056380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 9.Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, Corbeil D. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005;319:15–26. doi: 10.1007/s00441-004-1018-z. [DOI] [PubMed] [Google Scholar]
- 10.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133, metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 12.Collins A, Bloomston M. Diagnosis and management of pancreatic cancer. Minerva Gastroenterol Dietol. 2009;55:445–454. [PMC free article] [PubMed] [Google Scholar]
- 13.Moriyama T, Ohuchida K, Mizumoto K, Cui L, Ikenaga N, Sato N, Tanaka M. Enhanced cell migration and invasion of CD133+ pancreatic cancer cells cocultured with pancreatic stromal cells. Cancer. 2010;116:3357–3368. doi: 10.1002/cncr.25121. [DOI] [PubMed] [Google Scholar]
- 14.Nomura A, Banerjee S, Chugh R, Dudeja V, Yamamoto M, Vickers SM, Saluja AK. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget. 2015;6:8313–8322. doi: 10.18632/oncotarget.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/A:1011379725811. [DOI] [PubMed] [Google Scholar]
- 16.Costa NR, Paulo P, Caffrey T, Hollingsworth MA, Santos-Silva F. Impact of MUC1 mucin downregulation in the phenotypic characteristics of MKN45 gastric carcinoma cell line. PLoS One. 2011;6:e26970. doi: 10.1371/journal.pone.0026970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonckheere N, Van Seuningen I. The membrane-bound mucins: From cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie. 2010;92:1–11. doi: 10.1016/j.biochi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carraway KL, III, Funes M, Workman HC, Sweeney C. Contribution of membrane mucins to tumor progression through modulation of cellular growth signaling pathways. Curr Top Dev Biol. 2007;78:1–22. doi: 10.1016/S0070-2153(06)78001-2. [DOI] [PubMed] [Google Scholar]
- 20.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh AP, Chauhan SC, Bafna S, Johansson SL, Smith LM, Moniaux N, Lin MF, Batra SK. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate. 2006;66:421–429. doi: 10.1002/pros.20372. [DOI] [PubMed] [Google Scholar]
- 22.Carson DD. The cytoplasmic tail of MUC1: A very busy place. Sci Signal. 2008;1:pe35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 24.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 25.Fessler SP, Wotkowicz MT, Mahanta SK, Bamdad C. MUC1* is a determinant of trastuzumab (Herceptin) resistance in breast cancer cells. Breast Cancer Res Treat. 2009;118:113–124. doi: 10.1007/s10549-009-0412-3. [DOI] [PubMed] [Google Scholar]
- 26.Hikita ST, Kosik KS, Clegg DO, Bamdad C. MUC1* mediates the growth of human pluripotent stem cells. PLoS One. 2008;3:e3312. doi: 10.1371/journal.pone.0003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curry JM, Thompson KJ, Rao SG, Besmer DM, Murphy AM, Grdzelishvili VZ, Ahrens WA, McKillop IH, Sindram D, Iannitti DA, et al. The use of a novel MUC1 antibody to identify cancer stem cells and circulating MUC1 in mice and patients with pancreatic cancer. J Surg Oncol. 2013;107:713–722. doi: 10.1002/jso.23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suvarna SK, Layton C, Bancroft JD, editors. Bancroft's Theory and Practice of Histological Techniques. 7th. Churchill Livingstone; London: 2013. The hematoxylins and eosin; pp. 178–179. [Google Scholar]
- 29.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: Diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 30.Metzgar RS, Gaillard MT, Levine SJ, Tuck FL, Bossen EH, Borowitz MJ. Antigens of human pancreatic adenocarcinoma cells defined by murine monoclonal antibodies. Cancer Res. 1982;42:601–608. [PubMed] [Google Scholar]
- 31.Vathipadiekal V, Saxena D, Mok SC, Hauschka PV, Ozbun L, Birrer MJ. Identification of a potential ovarian cancer stem cell gene expression profile from advanced stage papillary serous ovarian cancer. PLoS One. 2012;7:e29079. doi: 10.1371/journal.pone.0029079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S, Hwang SG, Kang SG, Suh Y, Park MJ, Lee SJ. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31:4655–4666. doi: 10.1038/onc.2011.634. [DOI] [PubMed] [Google Scholar]
- 33.Rohner A, Spilker ME, Lam JL, Pascual B, Bartkowski D, Li QJ, Yang AH, Stevens G, Xu M, Wells PA, et al. Effective targeting of Hedgehog signaling in a medulloblastoma model with PF-5274857, a potent and selective Smoothened antagonist that penetrates the blood-brain barrier. Mol Cancer Ther. 2012;11:57–65. doi: 10.1158/1535-7163.MCT-11-0691. [DOI] [PubMed] [Google Scholar]
- 34.Horn G, Gaziel A, Wreschner DH, Smorodinsky NI, Ehrlich M. ERK and PI3K regulate different aspects of the epithelial to mesenchymal transition of mammary tumor cells induced by truncated MUC1. Exp Cell Res. 2009;315:1490–1504. doi: 10.1016/j.yexcr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Vos HL, de Vries Y, Hilkens J. The mouse episialin (Muc1) gene and its promoter: Rapid evolution of the repetitive domain in the protein. Biochem Biophys Res Commun. 1991;181:121–130. doi: 10.1016/S0006-291X(05)81390-7. [DOI] [PubMed] [Google Scholar]
- 36.Baldus SE, Mönig SP, Huxel S, Landsberg S, Hanisch FG, Engelmann K, Schneider PM, Thiele J, Hölscher AH, Dienes HP. MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin Cancer Res. 2004;10:2790–2796. doi: 10.1158/1078-0432.CCR-03-0163. [DOI] [PubMed] [Google Scholar]
- 37.Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Ren J, Chen D, Li Y, Kharbanda S, Kufe D. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther. 2003;2:702–706. doi: 10.4161/cbt.2.6.610. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Kufe D. The human DF3/MUC1 carcinoma-associated antigen signals nuclear localization of the catenin p120(ctn) Biochem Biophys Res Commun. 2001;281:440–443. doi: 10.1006/bbrc.2001.4383. [DOI] [PubMed] [Google Scholar]
- 40.Ren J, Li Y, Kufe D. Protein kinase C delta regulates function of the DF3/MUC1 carcinoma antigen in beta-catenin signaling. J Biol Chem. 2002;277:17616–17622. doi: 10.1074/jbc.M200436200. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 42.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278:38029–38039. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 43.Yuan Z, Wong S, Borrelli A, Chung MA. Down-regulation of MUC1 in cancer cells inhibits cell migration by promoting E-cadherin/catenin complex formation. Biochem Biophys Res Commun. 2007;362:740–746. doi: 10.1016/j.bbrc.2007.08.074. [DOI] [PubMed] [Google Scholar]
- 44.Gnemmi V, Bouillez A, Gaudelot K, Hémon B, Ringot B, Pottier N, Glowacki F, Villers A, Vindrieux D, Cauffiez C, et al. MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/β-catenin pathway and interaction with SNAIL promoter. Cancer Lett. 2014;346:225–236. doi: 10.1016/j.canlet.2013.12.029. [DOI] [PubMed] [Google Scholar]