Abstract
The patatin-like phospholipase domain-containing protein 3 (PNPLA3) variant is associated with nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH). However, the role of genetic variations of the peroxisome proliferator-activated receptor gamma coactivator-1-alpha gene (PPARGC1A) and glucokinase regulatory (GCKR) gene on NASH in obese patients remains unclear. We studied the effects and interaction of these genetic polymorphisms on NASH in severely obese Taiwanese patients.
The genotypes of PPARGC1A rs8192678, PNPLA3 rs738409, and GCKR rs780094 were determined in 177 severely obese patients who underwent bariatric surgery. NASH was evaluated by liver histopathology.
Of 177 patients, 29 (16.4%), 57 (33.2%), and 91 (51.4%) were in the non-NAFLD, steatosis, and NASH groups, respectively. We found that the PPARGC1A and PNPLA3 variants, but not the GCKR variant, were associated with NASH. The PPARGC1A rs8192678 GA/AA genotype was associated with higher steatosis grade and presence of ballooning degeneration. The PNPLA3 rs738409 GG genotype was associated with higher severity in all histologic features except for fibrosis. In multivariate analysis, both the PPARGC1A rs8192678 GA/AA genotype (odds ratio [OR] 2.32; 95% confidence interval [CI] 1.08–4.98; P = 0.031) and the PNPLA3 rs738409 GG genotype (OR 4.05; 95% CI 1.24–13.23; P = 0.021), and also body mass index were independent risk factors for NASH. Further, there was an additive effect of the PPARGC1A rs8192678 GA/AA genotype and the PNPLA3 rs738409 GG genotype on the presence of NASH (OR 6.83; 95% CI 1.61–29.01; P = 0.009).
The PPARGC1A rs8192678 GA/AA genotype and the PNPLA3 rs738409 GG genotype had an additive effect on NASH in severely obese Taiwanese patients.
INTRODUCTION
Along with the increase in the prevalence of obesity, the disease burden of nonalcoholic fatty liver disease (NAFLD) is increasing in both Western and Eastern countries.1–3 Nonalcoholic steatohepatitis (NASH), an extreme form of NAFLD, is associated with progressive liver disease and can lead to cirrhosis.4 Morbidly obese patients are especially at high risk for NAFLD and NASH. Prevalence rates are as high as 84% to 96% for NAFLD and 25% to 55% for NASH.5
Both genetic and environmental factors are implicated in the development of NASH.6 Using genome-wide association studies (GWAS), Romeo et al first demonstrated that the patatin-like phospholipase domain-containing protein 3 (PNPLA3) rs738409 variant is associated with increased liver fat content, and subsequent studies confirmed the association between this single-nucleotide polymorphism (SNP) and NASH.7,8 Recently, we also reported that the PNPLA3 rs738409 GG genotype increases susceptibility to NASH in severely obese Taiwanese patients.9
Several SNPs have also been associated with NASH. A GWAS revealed that genetic variants in or near neurocan (NCAN), lysophospholipase-like 1 (LYPLAL1), glucokinase regulatory protein (GCKR), and protein phosphatase 1 regulatory subunit 3b (PPP1R3B) were associated with hepatic steatosis, lobular inflammation, or fibrosis in persons with European ancestry,10 but inconsistent results were found in different ethnic groups.11,12 In addition, only the genetic variant in GCKR was found to be associated with ultrasound-defined NAFLD in obese Taiwanese children.13 The peroxisome proliferator-activated receptor γ coactivator-1α gene (PPARGC1A) rs8192678 variant was also found to be inconsistently associated with ultrasound-defined NAFLD.14,15 However, no study reported the association between the PPARGC1A variant and biopsy-proven NASH.
Although the prevalence of NAFLD is high in morbidly obese patients, not all of them develop NASH. The role of genetic disposition in the pathogenesis of NASH remains not well established. Although the association between PNPLA3 and NAFLD is consistent in most studies, the association between other SNPs and NAFLD is inconsistent. Furthermore, studies about the association between these SNPs and NASH are scarce. Because the allele frequency and genotype frequency of homozygous mutants in NCAN, LYPLAL1, and PPP1R3B are very low in the Taiwanese population,13 we examined only the effects of the PPARGC1A variant and the GCKR variant on NASH in severely obese Taiwanese patients. We also attempted to elucidate the effects of these SNPs and their interactions with the PNPLA3 variant on NASH.
METHODS
Eligible Patients
According to the recommendations of the Asia-Pacific consensus,16 obese Taiwanese patients with a body mass index (BMI) ≥37 kg/m2 or a BMI ≥32 kg/m2 with obesity-related comorbidities were considered for bariatric surgery. The patients who underwent bariatric surgery and intraoperative liver biopsy in E-Da Hospital, Kaohsiung, Taiwan, were consecutively enrolled in the present study. The exclusion criteria included age <20 years, alcohol consumption >140 g/week, use of hepatotoxic drugs, presence of liver diseases (such as Wilson disease, α1-antitrypsin deficiency, hemochromatosis, or autoimmune hepatitis), and malignant diseases. All patients were negative for hepatitis B and C viral markers. To avoid the potential influence of medications on NAFLD, we also excluded patients on glucose-lowering drugs. Written informed consent was obtained from each participant, and the study was approved by the E-Da Hospital Ethical Committee.
Clinical and Biochemical Characteristics
Preoperative data, including age, sex, coexisting medical diseases (eg, type 2 diabetes mellitus and hypertension), and anthropometric measurements (weight and height) were collected. The BMI was calculated as the weight in kilograms divided by the height in meters squared. Biochemical data included aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transpeptidase (GGT), fasting blood glucose, fasting insulin, total cholesterol, triglycerides, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and uric acid levels. Homeostasis model assessment of insulin resistance (HOMA-IR) was calculated using glucose and insulin levels in a fasting state. Briefly, the HOMA-IR was calculated as follows: (insulin [mU/mL]) × (glucose [mmol/L])/22.5.17
Histologic Examination
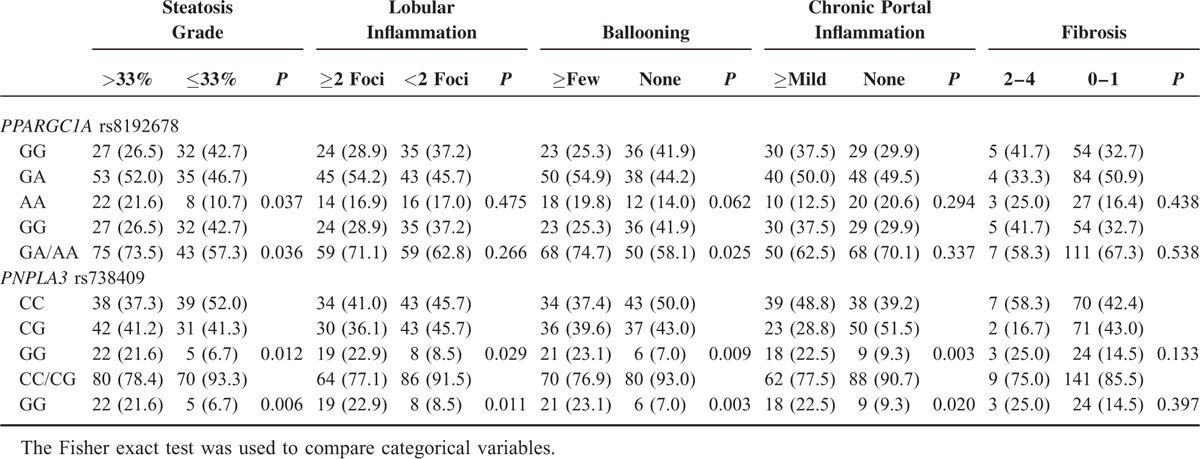
Wedge liver biopsy specimens, about 2 cm × 2 cm, obtained at the beginning of bariatric surgery, were routinely stained with hematoxylin and eosin, and Masson trichrome. All biopsy specimens were evaluated by an experienced pathologist (JC Hwang). According to the histologic results, patients were classified into one of 3 groups, as follows: non-NAFLD; steatosis; and NASH. The diagnosis of NASH was made according to the criteria of Brunt et al, and required the presence of steatosis, ballooning, and lobular inflammation.18,19 Histologic features were assessed semiquantitatively.20 Steatosis was graded on a scale of 0 to 3: 0—steatosis <5%; 1—steatosis 5% to 33%; 2—steatosis >33% to 66%; and 3—steatosis >66%. Lobular inflammation was scored according to the number of foci of inflammatory infiltration per 200× microscopic field, as follows: 0—no inflammation; 1—<2 foci; 2—2 to 4 foci; and 3—>4 foci. Ballooning degeneration was scored between 0 and 2 points as follows: 0—none; 1—few balloon cells; and 2—many balloon cells. Fibrosis was staged as follows: 0—no fibrosis; 1—perisinusoidal or periportal fibrosis; 2—perisinusoidal and portal/periportal fibrosis; 3—bridging fibrosis; and 4—cirrhosis. Portal inflammation was scored between 0 and 2 points as follows: 0—none; 1—mild; and 2—> mild.
DNA Extraction and SNP Genotyping
Genomic DNA was extracted from 3 mL of ethylenediaminetetraacetic acid whole blood samples using the salt fractionation method, which was standardized in the Puregene kit commercialized by Gentra (Research Triangle, NC) and was stored at −80°C. Approximately 10 ng of DNA was used for genotyping by TaqMan polymerase chain reaction (Applied Biosystems, Foster City, CA) according to the manufacturer's directions. After thermal cycling (50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles at 95°C for 15 seconds and at 60°C for 90 seconds), allelic discrimination for rs8192678 (cat.C_1643192_20) in PPARGC1A, rs738409 (cat.C_7241_10) in PNPLA3, and rs780094 (cat.C_2862873_10) in GCKR was performed by measuring the allele-specific fluorescence on an ABI Prism Sequence Detection System ABI 7900HT (Applied Biosystems).
Statistical Analysis
Descriptive results regarding categorical variables were given as percentages, and continuous variables were presented as the mean and standard deviation (SD). Continuous variables, such as insulin, HOMA-IR, AST, ALT, GGT, and triglycerides were not normally distributed, and they were therefore logarithm-transformed to normality before use in statistical models. The Hardy–Weinberg equilibrium test was employed to assess the discrepancy between the observed and the expected SNP genotypic frequency. Data for continuous and categorical variables were compared using the chi-square test, Fisher exact test, or Student t test as appropriate. Multiple comparisons of means of continuous variables were analyzed by 1-way analysis of variance and were compared post hoc by the Bonferroni test. The association between SNPs and NAS was analyzed using the Cochran–Armitage test for linear trends. The odds ratio (OR) and corresponding 95% confidence intervals (CIs) calculated from logistic regression models were employed to evaluate the association between genotypic distribution (additive, dominant, or recessive model) with NASH. After adjustment for potential confounding variables contributing to the development of NASH, including age, sex, BMI, HOMA-IR, and triglycerides, we assessed the independent effect of the PPARGC1A rs8192678 and PNPLA3 rs738409 genes on NASH using multiple logistic regression models. The potential gene–gene interaction was evaluated by fitting a multiplicative model with cross-product terms representing the interaction between the 2 genes studied. The statistical analyses were performed using SAS (version 9.3; SAS Institute, Inc., Cary, NC).
RESULTS
Clinical and Biochemical Characteristics in Patients With Non-NAFLD, Steatosis, and NASH
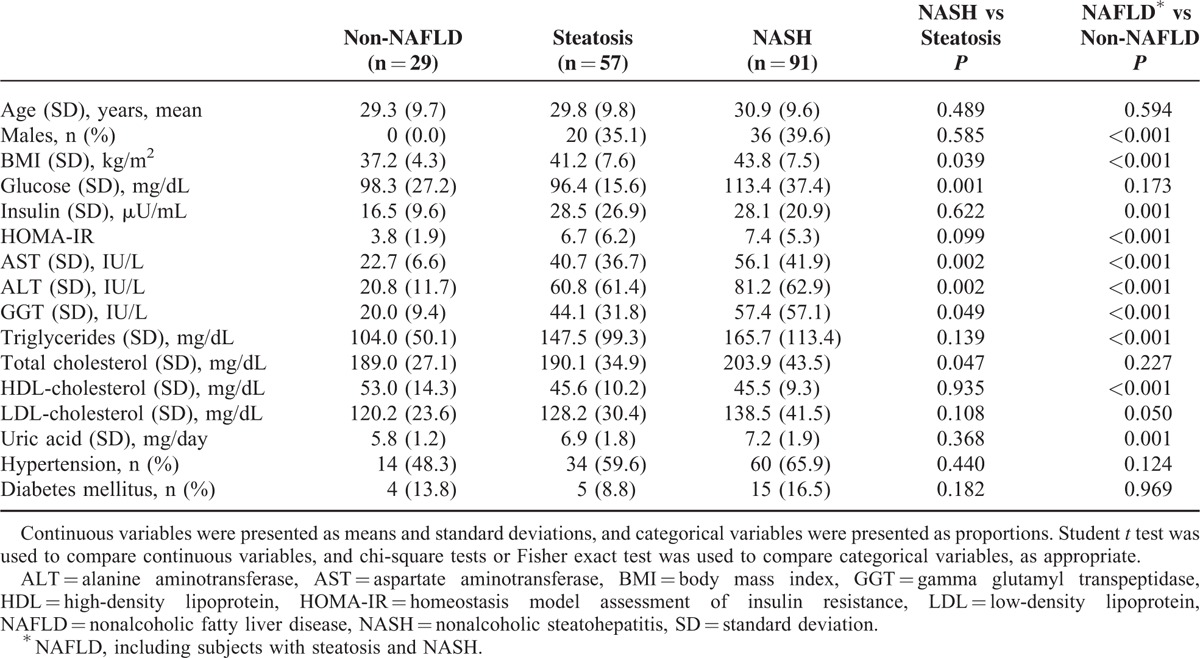
Between November 2007 and August 2009, 243 obese patients were consecutively enrolled. Sixty-two patients were excluded from this study, including 40 patients with hepatitis B and/or hepatitis C, 12 patients because of alcohol consumption, and 10 patients on glucose-lowering drugs. In addition, 4 patients without available PPARGC1A rs8192678 variant results were excluded. Finally, a total of 177 obese patients (56 [31.6%] men and 121 [68.4%] women) were enrolled in the present study (Supplementary Figure 1). The liver histology demonstrated that 29 (16.4%) patients were non-NAFLD, 57 (32.2%) patients were steatosis, and 91 (51.4%) patients were NASH. Compared with non-NAFLD patients, more NAFLD patients were male who had significantly higher BMIs and levels of all biochemical variables except for glucose, total cholesterol, and low-density lipoprotein cholesterol (Table 1). Among NAFLD patients, patients with NASH had significantly higher BMIs and levels of glucose, AST, ALT, GGT, and total cholesterol than those without (Table 1).
TABLE 1.
Clinical and Biochemical Characteristics in Patients With Non-NAFLD, Steatosis, and NASH
PPARGC1A rs8192678 and PNPLA3 rs738409 Variants Associated With Nash
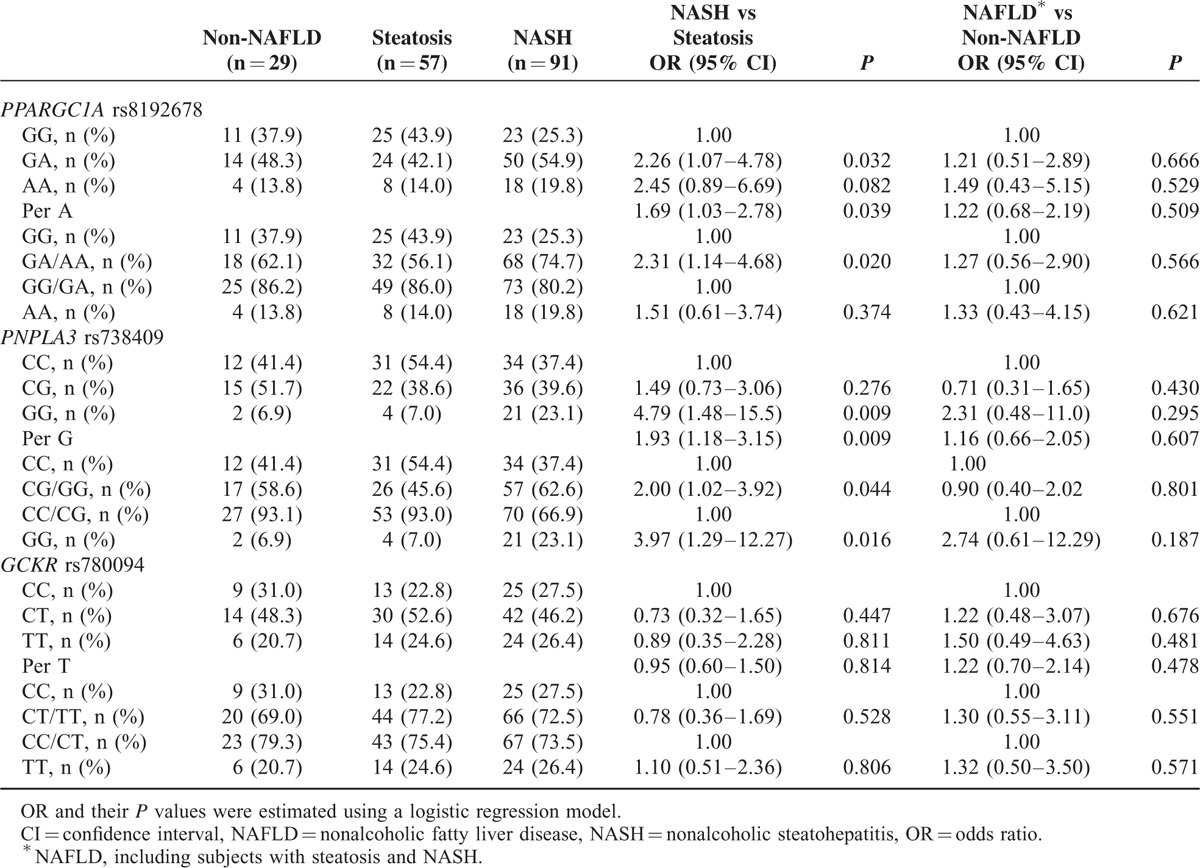
None of the SNPs deviated from the Hardy–Weinberg equilibrium (all P > 0.05). We found that the PPARGC1A and PNPLA3 variants, but not the GCKR variant, were associated with NASH (Table 2). The distribution of the PPARGC1A rs8192678 genotype was GG in 59 patients (33.3%), GA in 88 patients (49.7%), and AA in 30 patients (17.0%). Severely obese patients with the PPARGC1A rs8192678 GA genotype showed a significant association with NASH compared with patients having the PPARGC1A rs8192678 GG genotype (OR 2.26; 95% CI 1.07–4.78; P = 0.032). Marginally significant association was found in patients with the PPARGC1A rs8192678 AA genotype (P = 0.082). In addition, we found that patients with the PPARGC1A rs8192678 GA/AA genotype (dominant model) showed a significant association with NASH compared with patients having the PPARGC1A rs8192678 GG genotype (OR 2.31; 95% CI 1.14–4.68; P = 0.020). However, patients with the PNPLA3 rs738409 GG genotype (recessive model) had a significant association with NASH compared with patients having the PNPLA3 rs738409 CC/CG genotype (OR 3.97; 95% CI 1.29–12.27; P = 0.016).
TABLE 2.
Association of PPARGC1A rs8192678, PNPLA3 rs738409, and GCKR rs780094 With NAFLD and NASH
Clinical and Biochemical Characteristics Stratified by PPARGC1A rs8192678 Genotypes or PNPLA3 rs738409 Genotypes
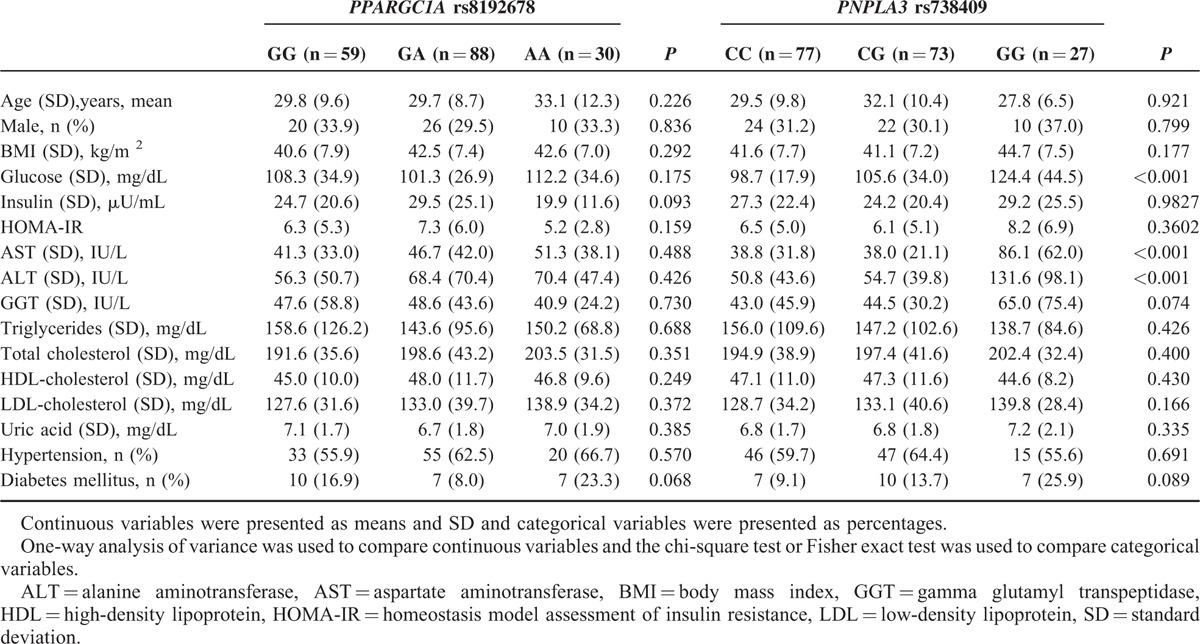
We further investigated the association between genotypes, and clinical and biochemical characteristics (Table 3). No significant difference was found among the 3 PPARGC1A rs8192678 genotypes. However, patients with the PNPLA3 rs738409 GG genotype had significantly higher levels of glucose, AST, and ALT compared with patients having the PNPLA3 rs738409 CG or CC genotypes.
TABLE 3.
Clinical and Biochemical Characteristics Stratified by PPARGC1A rs8192678 Genotypes or PNPLA3 rs738409 Genotypes
PPARGC1A rs8192678 and PNPLA3 rs738409 Genotypes Associated With Histologic Severity
All histologic features except for chronic portal inflammation were correlated with the diagnostic categories (P < 0.001 for all) (Table 4). We further investigated the association between genotypes and histologic severity (Table 5). Patients with PPARGC1A rs8192678 GA/AA genotype had higher steatosis grade and presence of ballooning degeneration compared with patients having the PPARGC1A rs8192678 GG genotype. Compared with patients with the PNPLA3 rs738409 CG/CC genotype, patients with the PNPLA3 rs738409 GG genotype had higher severity in all histologic features except for fibrosis.
TABLE 4.
Histologic Features in Patients With Non-NAFLD, Steatosis, and NASH
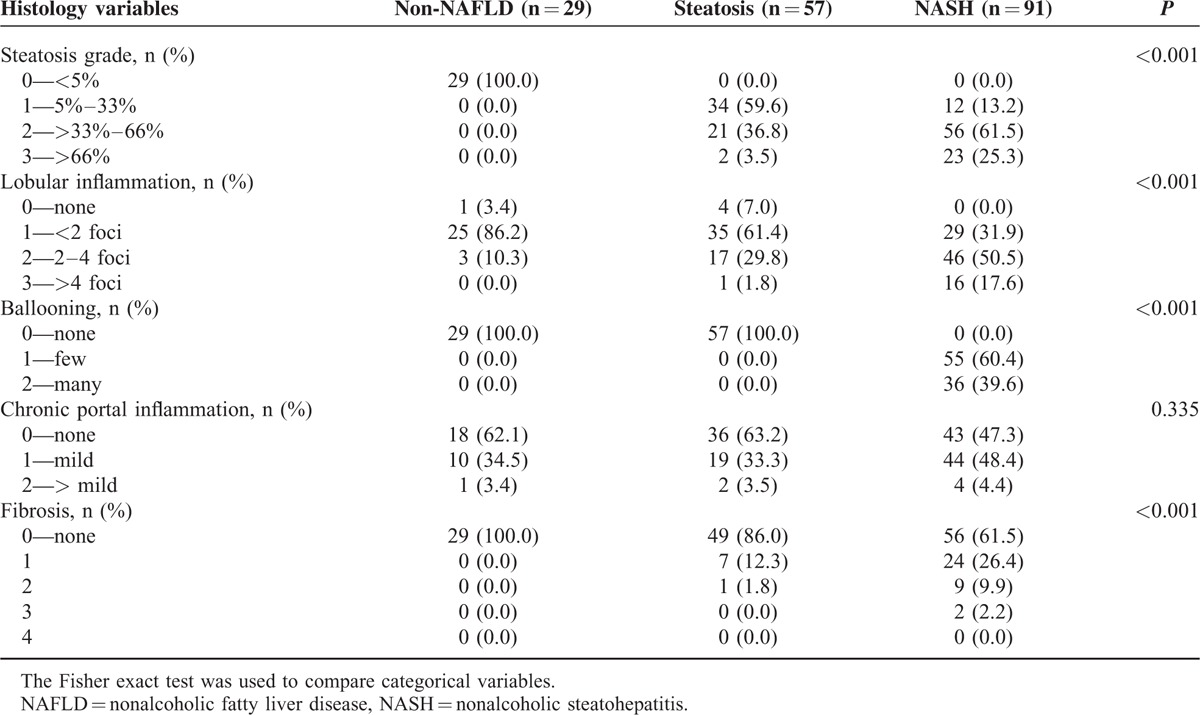
TABLE 5.
PPARGC1A rs8192678 and PNPLA3 rs738409 Genotypes Associated With Histologic Severity
Interaction Between the PPARGC1A rs8192678 Variant and the PNPLA3 rs738409 Variant on NASH in NAFLD Patients
After adjustment for potential confounders contributing to the development of NASH, including age, sex, BMI, HOMA-IR, and triglycerides, we determined that both the PPARGC1A rs8192678 GA/AA genotype (OR 2.32; 95% CI 1.08–4.98; P = 0.031) and the PNPLA3 rs738409 GG genotype (OR 4.05; 95% CI 1.24–13.23 P = 0.021) were independent risk factors for NASH (Table 6). As shown in Table 7, there was an additive significant effect of the combined PPARGC1A rs8192678 GA/AA and PNPLA3 rs738409 CC/CG genotypes on the presence of NASH (OR 2.72; 95% CI 1.21–6.09; P = 0.015). Of note, among severely obese patients, a much higher risk of NASH was observed with the combined PPARGC1A rs8192678 GA/AA and the PNPLA3 rs738409 GG genotypes (OR 6.83; 95% CI 1.61–29.01; P = 0.009). No such effect was observed for the combined PPARGC1A rs8192678 GG and PNPLA3 rs738409 GG genotypes (Table 7).
TABLE 6.
The PPARGC1A rs8192678 Variant and PNPLA3 rs738409 Variant Predict the Risk of NASH in NAFLD Patients
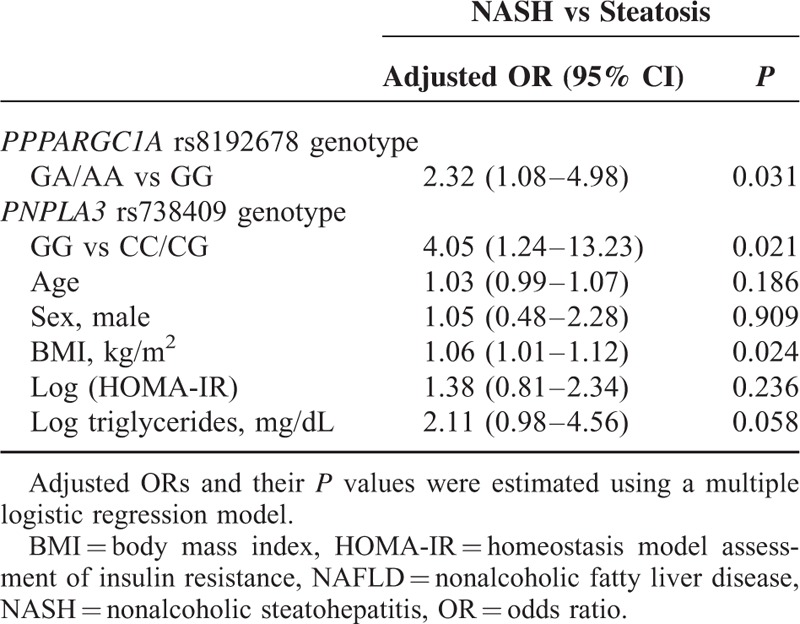
TABLE 7.
Interaction Between the PPARGC1A rs8192678 Variant and the PNPLA3 rs738409 Variant on NASH in NAFLD Patients
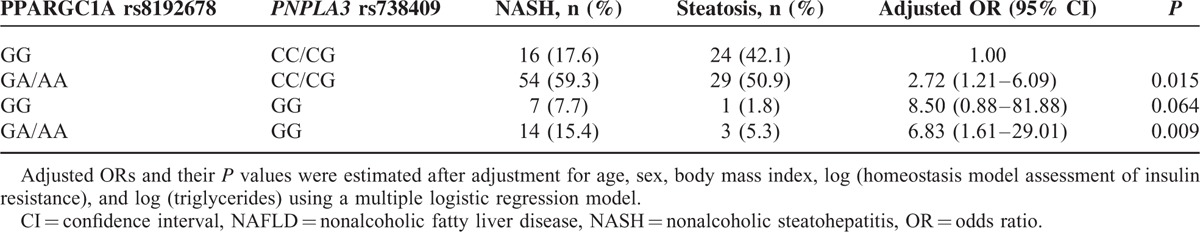
DISCUSSION
In the current study, we reported that the PPARGC1A and PNPLA3 variants, but not the GCKR variant, were associated with NASH in severely obese Taiwanese patients. Patients with the PPARGC1A rs8192678 GA/AA genotype showed increased susceptibility for NASH, which seemed to be independent of the PNPLA3 rs738409 GG genotype. The combination of the PPARGC1A rs8192678 GA/AA genotype and the PNPLA3 rs738409 GG genotype increased the risk of NASH (OR 6.83; 95% CI 1.61–29.01; P = 0.009).
Reported associations between the PPARGC1A rs8192678 variant and ultrasound-defined NAFLD have been inconsistent. Hui et al14 reported no association between the PPARGC1A rs8192678 variant and ultrasound-defined NAFLD in Chinese adults. In contrast, Lin et al15 suggested an association between the PPARGC1A rs8192678 variant and ultrasound-defined NAFLD in obese Taiwanese children. These contradictory conclusions may be due to the different ultrasonographic criteria for the definition of NAFLD and the age ranges of subjects in these studies.15 In the current study, we reported the first association study of PPARGC1A rs8192678 variant with the presence of NASH by using the diagnostic gold standard for NASH, liver histopathology, to avoid the potential image bias in severely obese Asian patients.
Proxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) encoded by the PPARGC1A gene is a transcriptional coactivator of the nuclear receptor PPARγ that exerts powerful effects on hepatic glucose and fat metabolism and may be involved in the development of NAFLD.21 The role of PGC-1α in protecting against hepatic steatosis has been found in both in vivo and in vitro studies.21,22 In addition, Chen et al22 also reported in an in vivo study that the PPARGC1A rs8192678 mutation can impair the activities of PGC-1α in decreasing fat deposition in cultured hepatocytes and can result in steatosis.
Mitochondrial dysfunction that promotes reactive oxygen species production and lipid peroxidation plays a key role in the pathophysiology of NASH.23 PGC-1α orchestrates mitochondrial biogenesis and functional capacity, and abnormal PGC-1α activities may result in mitochondrial dysfunction and thereby may lead to the development of NASH.24 However, although the PPARGC1A rs8192678 mutation may affect the activities of PGC-1α, further studies are needed to elucidate whether the PPARGC1A rs8192678 mutation contributes to the development of NASH.
Moreover, we also found an additive effect of the PPARGC1A rs8192678 variant and the PNPLA3 rs738409 variant on NASH in severely obese patients. The mechanism by which the PNPLA3 rs738409 variant results in steatosis was hypothesized to involve promoting triglyceride accumulation by limiting triglyceride hydrolysis.25–29 Although the mechanism by which the PNPLA3 rs738409 variant results in NASH is still elusive, the additive effect of these 2 SNPs on NASH noted in the current study suggests that there might be different mechanisms for the PPARGC1A rs8192678 and PNPLA3 rs738409 variants on the susceptibility of NASH.
Significant associations between the GCKR rs780094 variant and NAFLD were found in several studies, including a meta-analysis.30 However, studies about the association between the GCKR rs780094 variant and NASH are scarce and inconsistent. Although Tan et al11 reported a significant association between the GCKR rs780094 variant and NASH, Gorden et al31 reported that the GCKR rs780094 variant did not increase the risk for NASH in morbidly obese patients. In agreement with a previous study,31 we also found that the GCKR rs780094 variant was not associated with NASH in severely obese Taiwanese patients. However, further studies that enroll more patients from diverse ethnic groups are needed to confirm this finding.
The novelty of the present study is that we used biopsy-proven NASH to confirm that the PPARGC1A rs8192678 variant is associated with NASH. However, our study has some limitations. First, there might exist selection bias in the current study, because we enrolled only patients undergoing bariatric surgery, and overall they were young and predominantly female. Further studies are needed to elucidate whether our findings can be applied either to the general population and specific ethnic populations. Second, the association between NASH and the PPARGC1A rs8192678 AA genotype was only marginally significant (OR 2.45; 95% CI 0.89–6.69; P = 0.082). This could result from a limited number of patients with the PPARGC1A rs8192678 AA genotype. Therefore, further studies with larger sample sizes are needed to clarify the issue.
In conclusion, we reported that the PPARGC1A rs8192678 GA/AA genotype and the PNPLA3 rs738409 GG genotype had an additive effect on NASH in severely obese Taiwanese patients.
Supplementary Material
Acknowledgment
We would like to thank Hua-Ling Yang for genetic testing.
Footnotes
Abbreviations: ALT = alanine aminotransferase, AST = aspartate aminotransferase, BMI = body mass index, GCKR = glucokinase regulatory protein, GGT = gamma-glutamyl transpeptidase, GWAS = genome-wide association study, HOMA-IR = homeostasis model assessment of insulin resistance, NAFLD = nonalcoholic fatty liver disease, NASH = nonalcoholic steatohepatitis, PGC-1α = proxisome proliferator-activated receptor γ coactivator-1α, PNPLA3 = patatin-like phospholipase domain-containing protein 3, PPARγ = proxisome proliferator-activated receptor γ, PPARGC1A = peroxisome proliferator–activated receptor γ coactivator-1α gene, PPP1R3B = protein phosphatase 1 regulatory subunit 3b, SNP = single-nucleotide polymorphism.
Funding: This study was supported by research grants from the National Science Council (NSC102–2314-B-650–002-MY2), the E-Da Hospital, Taiwan (EDAHI102004 and EDAHI103004), and the Kaohsiung Medical University (“Aim for the Top Universities Grant, grant No. KMU-TP104D12”).
The authors have conflicts of interest to disclose.
REFERENCES
- 1.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 2004; 40:1387–1395. [DOI] [PubMed] [Google Scholar]
- 2.Chitturi S, Wong VW, Farrell G. Nonalcoholic fatty liver in Asia: firmly entrenched and rapidly gaining ground. J Gastroenterol Hepatol 2011; 26 suppl 1:163–172. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh MH, Ho CK, Hou NJ, et al. Abnormal liver function test results are related to metabolic syndrome and BMI in Taiwanese adults without chronic hepatitis B or C. Int J Obes (Lond) 2009; 33:1309–1317. [DOI] [PubMed] [Google Scholar]
- 4.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med 2002; 346:1221–1231. [DOI] [PubMed] [Google Scholar]
- 5.Clark JM. The epidemiology of nonalcoholic fatty liver disease in adults. J Clin Gastroenterol 2006; 40 suppl 1:S5–10. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science 2011; 332:1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008; 40:1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148 M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 2011; 53:1883–1894. [DOI] [PubMed] [Google Scholar]
- 9.Tai CM, Huang CK, Tu HP, et al. PNPLA3 genotype increases susceptibility of nonalcoholic steatohepatitis among obese patients with nonalcoholic fatty liver disease. Surg Obes Relat Dis 2014; 11:888–894. [DOI] [PubMed] [Google Scholar]
- 10.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 2011; 7:e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan HL, Zain SM, Mohamed R, et al. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J Gastroenterol 2014; 49:1056–1064. [DOI] [PubMed] [Google Scholar]
- 12.León-Mimila P, Vega-Badillo J, Gutiérrez-Vidal R, et al. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp Mol Pathol 2015; 98:178–183. [DOI] [PubMed] [Google Scholar]
- 13.Lin YC, Chang PF, Chang MH, et al. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am J Clin Nutr 2014; 99:869–874. [DOI] [PubMed] [Google Scholar]
- 14.Hui Y, Yu-Yuan L, Yu-Qiang N, et al. Effect of peroxisome proliferator-activated receptors-gamma and co-activator-1alpha genetic polymorphisms on plasma adiponectin levels and susceptibility of non-alcoholic fatty liver disease in Chinese people. Liver Int 2008; 28:385–392. [DOI] [PubMed] [Google Scholar]
- 15.Lin YC, Chang PF, Chang MH, et al. A common variant in the peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene is associated with nonalcoholic fatty liver disease in obese children. Am J Clin Nutr 2013; 97:326–331. [DOI] [PubMed] [Google Scholar]
- 16.Lee WJ, Wang W. Bariatric surgery: Asia-Pacific perspective. Obes Surg 2005; 15:751–757. [DOI] [PubMed] [Google Scholar]
- 17.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care 2004; 27:1487–1495. [DOI] [PubMed] [Google Scholar]
- 18.Brunt EM, Janney CG, Di Bisceglie AM, et al. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol 1999; 94:2467–2474. [DOI] [PubMed] [Google Scholar]
- 19.Kleiner DE, Brunt EM. Nonalcoholic fatty liver disease: pathologic patterns and biopsy evaluation in clinical research. Semin Liver Dis 2012; 32:3–13. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005; 41:1313–1321. [DOI] [PubMed] [Google Scholar]
- 21.Estall JL, Kahn M, Cooper MP, et al. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 2009; 58:1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Mu P, He S, et al. Gly482Ser mutation impairs the effects of peroxisome proliferator-activated receptor gamma coactivator-1alpha on decreasing fat deposition and stimulating phosphoenolpyruvate carboxykinase expression in hepatocytes. Nutr Res 2013; 33:332–339. [DOI] [PubMed] [Google Scholar]
- 23.Begriche K, Igoudjil A, Pessayre D, et al. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion 2006; 6:1–28. [DOI] [PubMed] [Google Scholar]
- 24.Nassir F, Ibdah JA. Role of mitochondria in nonalcoholic fatty liver disease. Int J Mol Sci 2014; 15:8713–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S, McPhaul C, Li JZ, et al. A sequence variation (I148 M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010; 285:6706–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148 M in mouse liver causes hepatic steatosis. J Clin Invest 2012; 122:4130–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumari M, Schoiswohl G, Chitraju C, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab 2012; 15:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Xu J, Xu F, et al. The SRE motif in the human PNPLA3 promoter (-97 to -88bp) mediates transactivational effects of SREBP-1c. J Cell Physiol 2015; 230:2224–2232. [DOI] [PubMed] [Google Scholar]
- 29.Sookoian S, Pirola CJ. PNPLA3, the triacylglycerol synthesis/hydrolysis/storage dilemma and nonalcoholic fatty liver disease. World J Gastroenterol 2012; 18:6018–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zain SM, Mohamed Z, Mohamed R. A common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol 2015; 30:21–27. [DOI] [PubMed] [Google Scholar]
- 31.Gorden A, Yang R, Yerges-Armstrong LM, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered 2013; 75:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


