Abstract
Despite recent technological advances in quantifying antibody drug conjugate (ADC) species, such as total antibody, conjugated antibody, conjugated drug, and payload drug in circulation, the correlation of their exposures with the efficacy of ADC outcomes in vivo remains challenging. Here, the chemical structures and concentrations of intratumor catabolites were investigated to better understand the drivers of ADC in vivo efficacy. Anti-CD22 disulfide-linked pyrrolobenzodiazepine (PBD-dimer) conjugates containing methyl- and cyclobutyl-substituted disulfide linkers exhibited strong efficacy in a WSU-DLCL2 xenograft mouse model, whereas an ADC derived from a cyclopropyl linker was inactive. Total ADC antibody concentrations and drug-to-antibody ratios (DAR) in circulation were similar between the cyclobutyl-containing ADC and the cyclopropyl-containing ADC; however, the former afforded the release of the PBD-dimer payload in the tumor, but the latter only generated a nonimmolating thiol-containing catabolite that did not bind to DNA. These results suggest that intratumor catabolite analysis rather than systemic pharmacokinetic analysis may be used to better explain and predict ADC in vivo efficacy. These are good examples to demonstrate that the chemical nature and concentration of intratumor catabolites depend on the linker type used for drug conjugation, and the potency of the released drug moiety ultimately determines the ADC in vivo efficacy.
Introduction
Antibody drug conjugates (ADC) have a complex structure that combines an antibody with a small-molecule drug (often cytotoxin) through a chemical linker (Senter and Sievers, 2012; Chari et al., 2014; Polakis, 2016). Figure 1A shows a simplified diagram of such an ADC with a drug-to-antibody ratio (DAR) of 2 that can undergo deconjugation to generate DAR1 and DAR0 species as well as a cytotoxic drug. Great efforts have been made to characterize the heterogeneous and dynamic mixtures of these ADC species in circulation (Xu et al., 2011; Alley and Anderson, 2013), each of which could have its own pharmacokinetic and biologic activity profile. However, the best ADC species in circulation to use for exposure-response correlation (both safety and efficacy) is not currently known (Kamath and Iyer, 2015; Khot et al., 2015; Singh et al., 2015; Wang et al., 2016). An important question is to determine key parameters involving in ADC in vivo efficacy.
Fig. 1.
(A) Deconjugation and catabolism of THIOMAB ADC. (B) Catabolism of disulfide-linked ADC. (C) Catabolite formation of methyl- and cyclobutyl-containing PBD-dimer conjugates in tissues. (D) Catabolite formation of cyclopropyl-containing PBD-dimer conjugate in tissues.
The ADC linker determines the mechanism and rate of payload release, both of which affect exposure of normal and tumor tissues to a drug payload; thus, the ADC linker is a critical part of an ADC. Recently, Pillow et al. (2016) discovered a self-immolating disulfide linker (β-mercaptoethyl-carbamate, -SCH2CH2OCO-) that can be directly attached to cysteine thiols of antibodies with cysteine residues engineered into IgG light or heavy chains (called THIOMAB) (Junutula et al., 2008, 2010; Shen et al., 2012). This linker can protect a cytotoxin from early release to circulation but could release a cytotoxin in tumors in a right form to a proper concentration. Figure 1B shows catabolism of a disulfide-linked ADC through disulfide cleavage and immolation to release a payload drug (Erickson and Lambert, 2012; Carter and Senter, 2008; Pillow et al., 2016).
Pyrrolo[2,1-c][1,4]benzodiazepine dimer (PBD-dimer) belongs to a class of DNA alkylators that covalently modify DNA minor grooves (Hartley, 2011). Recently, several ADCs using PBD analogs as toxin drugs have entered clinic trials (Jeffrey et al., 2013; Kung Sutherland et al., 2013; Saunders et al., 2015). The structural requirement for DNA binding and alkylation by the PBD-dimer (Jenkins et al., 1994; Rahman et al., 2009; Hartley, 2011) would depend on the connection site, like the N10 of PBD-dimer, and demand efficient and clean release of the payload to exert its cell-killing activities (Fig. 1, C and D). We designed conjugates and tested pharmacological activities of ADCs that link PBD-dimer to anti-CD22 through the cleavable disulfide linkers (Pillow et al., 2016). Distinct efficacy profiles were observed with the ADC molecules that contained structurally analogous linkers following administration of the related ADCs in xenograft mice. Catabolite analysis showed that these ADCs released different payloads and that intratumor catabolites had very different kinetic profiles from those of ADC species in circulation. These are good examples to demonstrate that the chemical nature and concentration of intratumor catabolites depend on the linker type and thus determine ADC efficacy. In addition, the analysis of ADC species in circulation that is routinely performed is not sufficient to explain or predict ADC efficacy outcomes. In this report, we describe the findings of in vivo efficacy, assessment of the total antibody and catabolite identification, and quantitation in the plasma and tumors of xenograft mice, and subsequently correlate these ex vivo catabolite analyses with the in vivo efficacy outcomes of related ADCs.
Materials and Methods
Materials.
Ammonium formate, formic acid, and calf thymus DNA were purchased from Sigma-Aldrich (St. Louis, MO). Human CD22 and Napi2b antibodies with two engineered cysteine residues were generated as described previously (Bhakta et al., 2013; Polson et al., 2010). Anti-CD22 conjugates (light chain-K149C-anti-CD22-PBD-dimer conjugates) of methyl-disulfide-PBD-dimer, cyclopropyl-disulfide-PBD-dimer, and cyclobutyl-disulfide-PBD-dimer, as well as the corresponding control conjugates (light chain-K149C-anti-NaPi-PBD-dimer conjugates): anti-NaPi-methyl-disulfide-PBD-dimer, anti-NaPi-cyclopropyl-disulfide-PBD-dimer, and anti-NaPi-cyclobutyl-disulfide-PBD-dimer, PBD-dimer, and cyclopropyl thiol (Fig. 1, C and D) were prepared as described previously (Zhang et al., 2016). Mice (CB-17 SCID, female, nude, and Balb/C strains) were purchased from Charles Rivers Laboratories. All animal studies were carried out in compliance with NIH guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee at Genentech, Inc.
Xenograft Studies: Efficacy and Tissue Collection.
The efficacy of the anti-CD22 antibody drug conjugates (ADCs) was investigated in a mouse xenograft model of human diffuse large B-cell lymphoma WSU-DLCL2 (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). To establish the subcutaneous xenograft model, the tumor cells [20 million cells in 0.2 ml Hank’s balanced salt solution (Hyclone/GE Healthcare and Life Sciences, Marlborough, MA)] were inoculated subcutaneously into the flanks of female CB17 SCID mice (Charles River Laboratories, Wilmington, MA). When tumors reached a desired volume, animals were divided into groups of eight mice and each received a single intravenous injection of anti-NaPi2b ADCs (nontarget controls), anti-CD22-methyl-disulfide-PBD-dimer, anti-CD22-cyclopropyl-disulfide-PBD-dimer, or anti-CD22-cyclobutyl-disulfide-PBD-dimer at 0.5 or 1 mg/kg through the tail vein (Day 0 of the study). Tumors and body weights of mice were measured one to two times a week throughout the study. Mice were promptly euthanized when body weight loss was >20% of their starting weight. All animals were euthanized before tumors reached 3000 mm3 or showed signs of impending ulceration. Tumor volume was measured in two dimensions (length and width) using calipers, and the tumor volume was calculated using the formula: Tumor size (mm3) = (Longer measurement × Shorter measurement2) × 0.5.
Selected tissues including tumor, liver, and plasma, were collected at 24 and 96 hours postdose of single i.v. 5-mg/kg dose of anti-CD22-cyclopropyl-disulfide-PBD-dimer or anti-CD22-cyclobutyl-disulfide-PBD-dimer. The plasma and tissue samples were kept frozen at –80°C until analyzed for exposure assessment of total antibody and payloads. Selected plasma samples were collected at 1, 4, and 7 days postdose of single i.v. dose of 1 mg/kg of anti-CD22-methyl-disulfide-PBD-dimer, anti-CD22-cyclopropyl-disulfide-PBD-dimer, or anti-CD22-cyclobutyl-disulfide-PBD-dimer and kept frozen at –80°C until DAR analysis.
Characterization and Quantitation of Catabolites in Tissues.
Total antibody (mAb representing all ADC species containing the antibody component such as DAR2, DAR1, and DAR0) and catabolites were characterized in tumors and other tissues after xenograft mice were dosed with anti-CD22-cyclopropyl-disulfide-PBD-dimer or anti-CD22-cyclobutyl-disulfide-PBD-dimer with a single i.v. dose of 5 mg/kg. The plasma, liver, and tumor were collected at 24 and 96 hours, homogenized in control mouse plasma, and analyzed by LC-MS/MS for catabolites following extraction with an organic solvent. Another portion of the tissues were homogenized in the phosphate-buffered saline solution buffer containing protease inhibitor cocktail (Sigma-Aldrich) and analyzed by affinity-capture LC-MS/MS for total antibody following Protein A capture and trypsin digestion.
To quantitate the concentrations of catabolites in the mouse plasma and tissues, 50 μl of plasma or approximately 50 mg of tissues (liver or tumor) were mixed with ice-cooled 150 μl blank mouse plasma. The samples were thoroughly homogenized, two cycles of 30 seconds each, by precooled beads (10 beads, –80°C) and then extracted by ice-cooled 400 μl of acetonitrile containing 10 nM internal standard (IS; 589.2/261.0). After a 15-minute centrifugation at 6500g, 10 μl of supernatant was injected to an AB Sciex Triple Quad 6500 mass spectrometer (Concord, Ontario, Canada) coupled with a Shimadzu liquid chromatography system. Peak separation was achieved using a Phenomenex Kinetex C18 column (Torrance, CA), 1.7 μm, 100 Å, 100 × 2.1 mm with mobile phase A (0.1% formic acid) and B (100% acetonitrile) using a gradient of 0–0.5 minutes 5% B, 0.5–3.5 minutes 5–90% B, 3.5–4.0 minutes 90% B, 4.0–4.5 minutes 90–5% B, 4.5–5.0 minutes 5% B at a flow rate of 0.5 ml/min (column temperature of 35°C). The retention times of PBD-dimer, cyclopropyl thiol, and IS were 2.6, 3.1, and 2.9 minutes, respectively. The multiple reaction monitor (MRM) transitions in MS were: PBD-dimer, 585.1/504.2, and cyclopropyl thiol,732.2/504.2. The compound-dependent MS parameters were 61, 10, 29–41, 14 for declustering potential (DP), entrance potential (EP), collision energy (CE), and collision cell exit potential (CXP), respectively. The MS instrument-dependent parameters were collision gas (CAD) (–3), curtain gas (CUR) (10), nebulizer gas (GS1) (90), turbo gas (GS2) (50), ionspray voltage (IS) (5500), and ionspray temperature (TEM) (500). The standard curve samples for quantitation were 0.24–3.91 and 0.24–7.81 nM for PBD-dimer and cyclopropyl thiol, respectively. The lower limits of quantification of PBD-dimer and cyclopropyl thiol were both 0.24 nM. The matrix effects of the tissue samples were minimized by homogenizing the tissues in blank mouse plasma.
The identification of compounds was done by LC-MS/MS on a Triple TOF 5600 mass spectrometer (AB Sciex) coupled with high-performance liquid chromatography (HPLC) separation. The column was a Hypersil Gold C18 column (100 × 2.1, 1.9 μM; Thermo Scientific). The compounds were eluted by a gradient of 0.1% formic acid in 10 mM ammonium formate to acetonitrile containing 0.1% formic acid in 10 mM ammonium formate. PBD-dimer was identified by the molecular ion at m/z found, 585.2708, and calculated, 585.2711, C33H36N4O6, and by major fragments at m/z 504.2144, 492.2144, 411.1570, 327.1724, 259.1096, and 246.1139. Cyclopropyl thiol was identified by molecular ion at m/z found, 733.2901, and calculated, 733.2902, C38H44N4O9S, and by major fragments at m/z 715.2814, 585.2716, 536.2040, 504.2140, and 492.2140.
An affinity capture approach using protein-A magnetic beads was used to enrich the cyclopropyl-disulfide-PBD-dimer and cyclobutyl-disulfide-PBD-dimer ADCs from the mouse-tissue homogenate in the phosphate-buffered saline solution buffer, pH 7.4. The bound ADCs were subject to “on-bead” proteolysis with trypsin following standard protein denaturation, reduction, and alkylation processing steps. Briefly, quantification of the total antibody concentration was achieved by using LC-MS/MS measurement of its surrogate peptide(s) produced by proteolytic digestion. A surrogate peptide TTPPVLDSDGSFFLYSK, generated from the human unique Fc region to allow the differentiation of cyclopropyl-disulfide-PBD-dimer and cyclobutyl-disulfide-PBD-dimer ADCs from the endogenous matrix components, was quantified by a MRM transition of 938.0/836.7. In addition, several other peptides characteristic with the human Fc region were monitored for the conformation and troubleshooting purposes as described previously (Xu et al., 2014).
The DAR was determined as described previously (Xu et al., 2011). Briefly, an appropriate volume of mouse plasma after intravenous administration of ADCs was incubated at room temperature with the biotinylated CD22 target antigen, which was coupled to the streptavidin paramagnetic beads (Invitrogen/Thermo Fisher Scientific). The bead-captured ADC analytes were washed and deglycosylated at 37°C overnight. The resulting samples in 30% acetonitrile in water containing 1% formic acid were injected onto a Triple TOF 5600 mass spectrometer (AB Sciex) coupled with HPLC using a reversed-phase HPLC column. The compounds were eluted by a gradient of mobile phase A (water with 0.1% formic acid) and mobile phase B (acetonitrile with 0.1% formic acid) at a flow rate of 5 μl/min. Positive time-of-flight (TOF) MS scan was acquired and processed. Peak deconvolution was performed to obtain the distribution profiles of DAR0, DAR1, and DAR2 species, and the corresponding peak areas were measured. Subsequently, the relative ratio of each DAR and the average DAR value at each time point were calculated.
DNA Binding.
PBD-dimer and cyclopropyl thiol were incubated at 1 μM with 1 mg/ml calf thymus DNA for 1 hour in 0.5 ml of 10 mM Bis-Tris, pH 7.1, at 37°C. Ethyl acetate (1 ml) was used to extract the reaction mixture twice by vortex (20 seconds) and centrifugation (4 minutes at 5000g). The combined organic extracts were concentrated under vacuum and reconstituted in 0.15 ml of 1:1 water/methanol before injection on LC-MS/MS. The incubation samples were analyzed by LC-MS/MS–UV on Sciex TripleTOF 5600 using a Hypersil Gold C18 column (100 × 2.1, 1.9 μM; Thermo Fisher Scientific) in a positive electrospray ionization (ESI) mode. The compounds were eluted by a gradient of buffer A (0.1% formic acid) and buffer B (0.1% formic acid in acetonitrile) (5% B 0–0.5 minutes, 5–25% B 0.5–8 minutes, 25–75% B 8–13 minutes, and 75–95% B 13–13.5 minutes, 95% B 13.5–14.5 minutes, 95–5% B 14.5–15 minutes) at a flow rate of 0.4 ml/min.
Results and Discussion
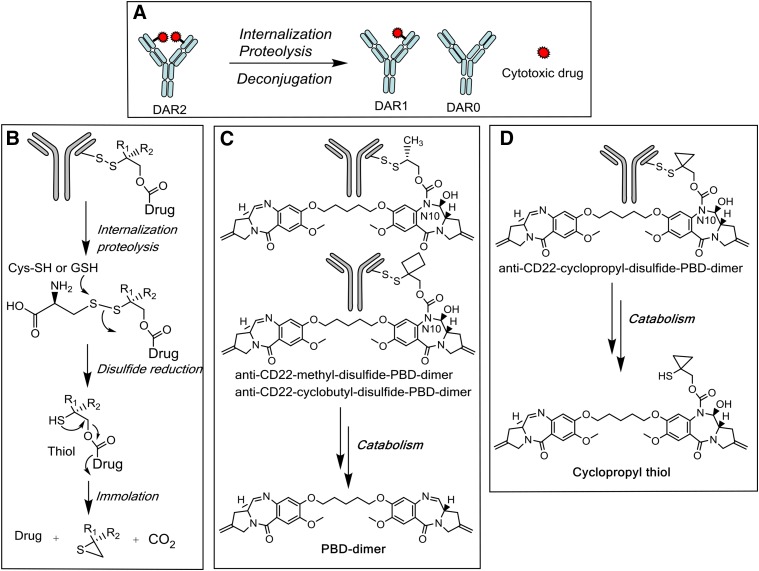
The CD22 antigen was chosen for our ADC design because of its high expression on cancers of B-cell origin and relatively low prevalence on non-B cell-related normal cells and tissues (Polson et al., 2010). The efficacy of CD22 conjugates of methyl-disulfide-PBD-dimer cyclopropyl-disulfide-PBD-dimer and cyclobutyl-disulfide-PBD-dimer was studied in vivo in a CD22-expressing WSU-DLCL2 xenograft mouse model. When dosed once intravenously at both 0.5 and 1 mg/kg, conjugates containing disulfide linkers bearing methyl and cyclobutyl substituents exhibited strong efficacy, with partial responses observed beginning in week 1 and lasting for 35 days (Fig. 2A). These entities were well tolerated in mice, as the corresponding body weight losses were within 5% in all treatment groups (Fig. 2B). The related nontarget NaPi2b (Lin et al., 2015) control conjugates displayed minimal efficacy in the same mouse model. In stark contrast, 1.0 mg/kg i.v. administration of the anti-CD22 conjugate of cyclopropyl-disulfide-PBD-dimer afforded almost no antitumor activity and all animals in this group together with the NaPi2b control group were euthanized owing to tumor growth within 2 weeks of ADC dosing (Fig. 2, A and B).
Fig. 2.
(A) Anti-tumor effects of disulfide-linked PBD-dimer ADCs in WSU-DLCL2 tumor xenograft mice. In vivo efficacy of antibody PBD-dimer conjugates of methyl-, cyclopropyl-, and cyclobutyl-containing linkers were tested in mice bearing human diffuse large B-cell lymphoma WSU-DLCL2 xenografts (n = 8). Mean (±S.E.M.) tumor volumes are plotted over time (days postdose). The control anti-Napi2b conjugates did not show efficacy in a separate experiment. (B) Tolerability of mice in the in vivo efficacy study using the antibody PBD-dimer conjugates of methyl-, cyclopropyl-, and cyclobutyl-containing linkers. The xenograft models used mice bearing human diffuse large B-cell lymphoma WSU-DLCL2 (n = 8). Mean (±S.E.M.) percent body weight changes are plotted over time (days postdose from the Day 0 baseline).
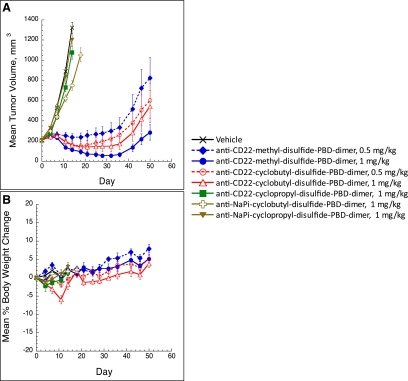
Poor systemic pharmacokinetics could be a possibility for the weak efficacy exhibited by the cyclopropyl-containing conjugate in the experiment described above. To explore this possibility, total antibody and catabolite concentrations were measured in plasma, liver, and tumors of WSU-DLCL2 xenograft mice after animals were administered a single i.v. dose of 5 mg/kg of the cyclopropyl-containing or cyclobutyl-containing disulfide-PBD-dimer ADCs. The tissue sampling was performed at relatively early time points (24 and 96 hours) to ensure that sufficient tumor quantities were available for analysis. As shown in Table 1, the measured tumor and plasma exposures of total antibody (mAb) for both entities were similar at both 24 and 96 hours after dosing, with much higher total mAb concentrations in plasma (216–370 nM) than in tumors (16–59 nM). In a separate experiment employing 1 mg/kg i.v. doses, the drug-to-antibody ratios of the two conjugates determined from plasma samples were similar and relatively stable over a 7-day period (Fig. 3). Consequently, the conjugated antibody (calculated from total antibody and DAR analyses) should also have similar concentrations between the cyclopropyl- and cyclobutyl-containing ADCs in mice. These results indicated that there was no significant pharmacokinetic (PK) and/or in vivo stability difference that could be responsible for the drastically different efficacy outcomes observed for the cyclopropyl-disulfide-PBD-dimer and cyclobutyl-disulfide-PBD-dimer ADCs.
TABLE 1 .
Total antibody (mAb) and catabolite concentrations in tissues of WSU-DLCL2 xenograft mice following a single 5-mg/kg i.v. dose of anti-CD22-cyclopropyl-disulfide-PBD-dimer, or anti-CD22-cyclobutyl-disulfide-PBD-dimer ADCs (n = 2)
Catabolites PBD-dimer and cyclopropyl-thiol and were not detected in tissues of mice dosed with vehicle. ND = not detected.
| ADC | Time | PBD-dimer |
Cyclopropyl-thiol |
mAb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasma | Liver | Tumor | Plasma | Liver | Tumor | Plasma | Liver | Tumor | ||
| h | nM | nM | nM | nM | nM | nM | nM | nM | nM | |
| Cyclobutyl-containing | 24 | 0.43 | 0.26 | 1.03 | NA | NA | NA | 370 | 24.8 | 43.5 |
| 0.55 | 0.55 | 2.09 | NA | NA | NA | 342 | 19.5 | 25.0 | ||
| 96 | <LLOQ | <LLOQ | 1.93 | NA | NA | NA | 285 | 20.9 | 25.3 | |
| <LLOQ | <LLOQ | 2.05 | NA | NA | NA | 272 | 21.1 | 56.0 | ||
| Cyclopropyl-containing | 24 | ND | ND | ND | 0.74 | 1.42 | 7.58 | 294 | 28.4 | 59.4 |
| ND | ND | ND | 0.57 | 0.63 | 6.76 | 216 | 18.6 | 27.4 | ||
| 96 | ND | ND | ND | <LLOQ | <LLOQ | 4.33 | 349 | 23.4 | 23.5 | |
| ND | <LLOQ | ND | 0.30 | 0.34 | 4.69 | 331 | 16.6 | 16.6 | ||
NA, not applicable; LLOQ, lower limit of quantitation, 0.24 nM for both analytes in homogenates (nanomolar concentration in tumor and liver was estimated on the basis of an assumption of tissue density of 1 g/ml).
Fig. 3.
In vivo stability of the methyl-, cyclopropyl-, and cyclobutyl-containing disulfide-PBD-dimer ADCs in plasma of mice following single i.v. dose of 1 mg/kg (n = 3). The DAR data were analyzed as described previously (Xu et al., 2011).
Examination of the associated catabolites present in tumors, however, did help rationalize these disparate efficacy results. In experiments employing the cyclobutyl-containing ADC, the PBD-dimer was detected in marked quantities in tumors (1.0–2.0 nM) at both 24 and 96 hours after dosing (Table 1, Fig. 1C). As anticipated for an ADC-mediated delivery, the tumor concentrations of PBD-dimer were significantly higher than those detected in plasma and liver at both time points. In addition, the concentration of PBD-dimer was maintained or slightly increased in tumors from 24 to 96 hours but decreased to a very low level in plasma. In contrast, PBD-dimer was not detected at appreciable levels in tumors or plasma in experiments that used the cyclopropyl-containing ADC. Instead, measurable levels of cyclopropyl thiol catabolite were observed in tumor (4.3–7.5 nM) as well as in plasma and liver (0.5–1.4 nM at 24 hours) that were much higher than the corresponding concentrations of PBD-dimer from the cyclobutyl-containing ADC (Table 1, Fig. 1D).
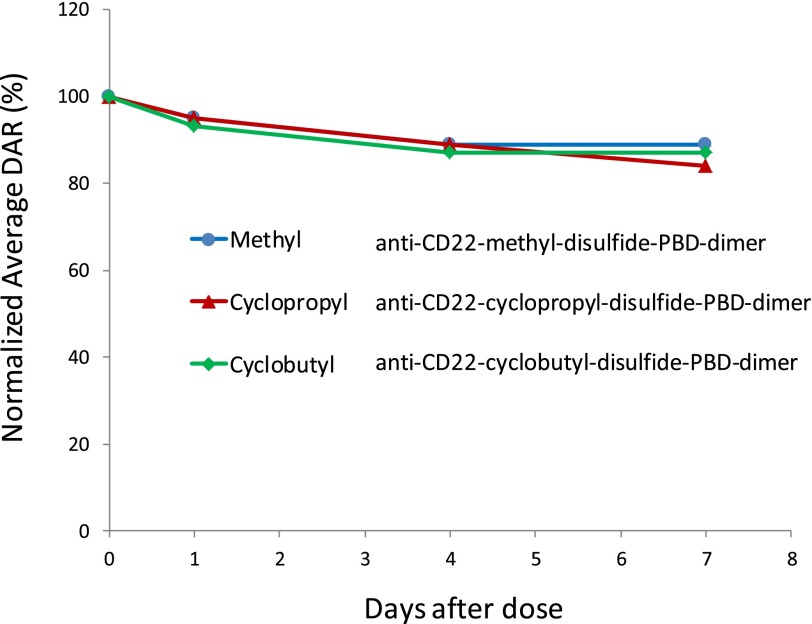
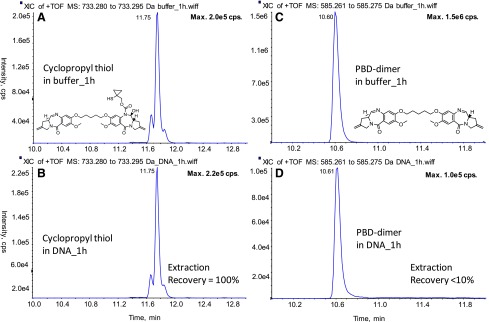
The DNA binding potential of these identified catabolites (PBD-dimer and cyclopropyl thiol) was evaluated in vitro. As shown in Fig. 4, after 1-hour incubation of PBD-dimer in a DNA solution, approximately 94% of PBD-dimer became unextractable. In contrast the cyclopropyl thiol was quantitatively recovered from similar incubations. These results suggest that cyclopropyl thiol does not bind to DNA, whereas PBD-dimer quickly binds to DNA, which supports their distinct efficacy profiles. Therefore, the lower PBD-dimer concentrations in the tumor and liver relative to cyclopropyl thiol (approximately 2.5- to 4-fold) are consistent with loss of PBD-dimer resulting from efficient DNA alkylation in tissues (Fig. 4; cyclopropyl thiol does not appreciably bind DNA). On the basis of these results from tissue analysis, an i.v. dose of 0.5 mg/kg of the cyclobutyl-disulfide-PBD-dimer ADC is predicted to generate PBD-dimer at a concentration range of 0.10–0.20 nM in tumors (representing <5% of total mAb measured in tumor) assuming linear PK, uptake, and catabolism in mice. Such PBD-dimer concentrations are consistent with the strong tumor-killing observed in the corresponding efficacy experiments performed with cyclobutyl-containing ADC given the high potency of PBD-dimer (IC50 = ∼50 pM) in target-expressing cell lines (Hartley, 2011). In addition, the PBD-dimer concentrations in tumors likely represent underestimation of actual intratumor quantities owing to DNA alkylation. Very interestingly, the similar concentrations of total antibody for the cyclopropyl-containing and cyclobutyl-containing ADCs in plasma (data routinely collected in ADC discovery stage) would predict similar efficacies for the two ADCs in vivo. However, as shown by the data presented in this work, detailed understanding of the catabolites in the tumors (which is not routinely performed), together with their DNA binding properties, are required to explain the dramatic efficacy differences exhibited by the cyclopropyl-containing and cyclobutyl-containing ADCs. Thus, the measurement of ADC-delivered payload in tumors, which is responsible for DNA binding/alkylation and ultimate efficacy, is critically important to an understanding of the driver of efficacy of a given ADC.
Fig. 4.
DNA binding potential of PBD-dimer and cyclopropyl thiol. Cyclopropyl thiol was incubated in buffer (A) or 1 mg/ml calf thymus DNA (B) at 37°C for 1 hour; PBD-dimer was incubated in buffer (C) or 1 mg/ml calf thymus DNA (D) at 37°C for 1 hour.
The different products identified in mouse tumors and tissues following intravenous administration of the cyclobutyl- or cyclopropyl-containing ADCs are consistent with those identified from chemical reduction of model small molecules of corresponding disulfide linker drugs (Zhang et al., 2016). Although the in vitro disulfide cleavage and nonimmolation of the cyclopropyl linker would predict different payload release from cyclobutyl- and cyclopropyl-containing ADCs, the possibility of formation of PBD-dimer in vivo from the cyclopropyl-containing ADC by other reactions such as hydrolysis or oxidation of the linker in addition to immolation could not be completely eliminated until catabolite analysis in tumors of mice dosed with the corresponding ADCs. Unlike in small-molecule drug discovery, in which many established in vitro models for activity and disposition can predict in vivo outcomes, the ADC discovery relies more heavily on in vivo efficacy studies, because in vitro models that can predict in vivo efficacy outcomes are often lacking. Consequently, the following are common scenarios in ADC discovery: an ADC appears to be perfectly designed but shows no activity, an ADC shows potency in vitro but shows no efficacy in vivo, or an ADC has no activities in vitro but shows efficacy in vivo. This study provides an excellent example to demonstrate that ADC efficacy is determined by the nature and concentration of intratumor catabolites that are not normally determined and that the ADC efficacy shows no correlation with the concentrations of ADC species in circulation that are routinely analyzed.
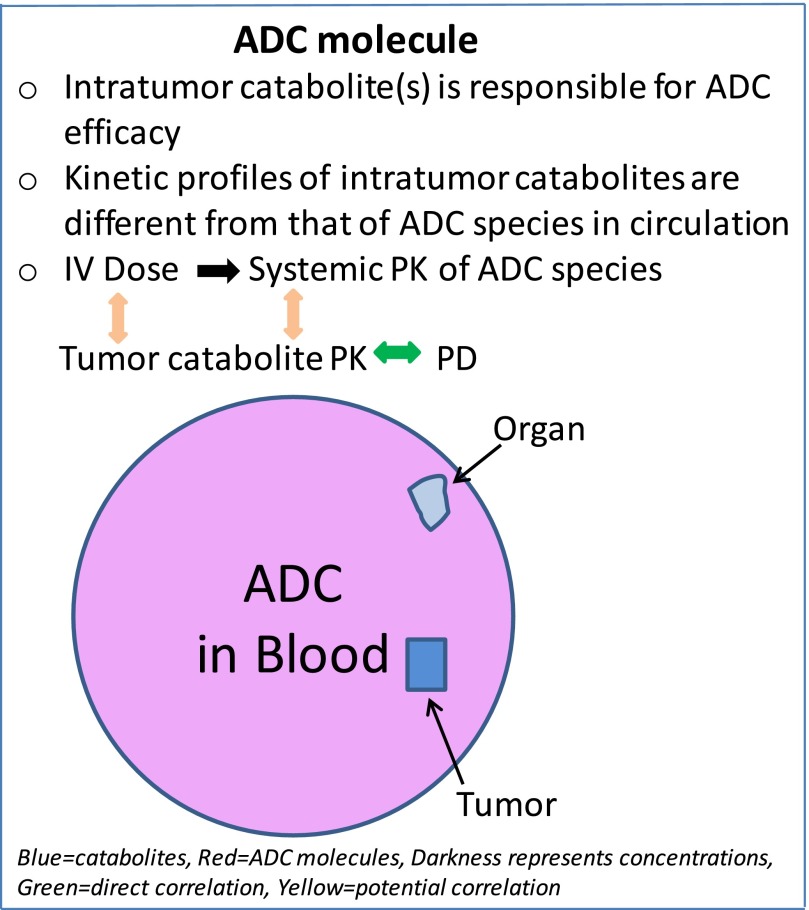
The intratumor catabolites appear to have kinetic profiles different from the circulating ADC species as demonstrated in this study for DNA alkylators and by others for antimitotic agents (Erickson et al., 2012; Leal et al., 2015; Singh et al., 2015). The intratumor catabolites are produced through biotransformation of ADC molecules that entered the tumor cells and are ultimately responsible for the efficacy of an ADC. The formation and function of ADC catabolites and related pharmacokinetic characteristics are summarized in Fig. 5. Pharmacokinetic-pharmacodynamic (PK/PD) modeling has had limited success in correlating ADC efficacy with the PK parameters of ADC species in circulation (Singh et al., 2015). Detailed modeling and simulation studies with anti-5T4 MMAE ADC showed that tumor exposure to an ADC and its payload will depend on many factors, such as payload dissociation from ADC, tumor size, quantitative processing of ADC, and payload disposition (Shah et al., 2014; Khot et al., 2015). Other important factors that will have impacts on ADC efficacy also include tumor type, antigen expression and resynthesis, linkers, site of attachment, payload permeability, and cell retention, as well as potency of payload (Polakis, 2016). Until we gain detailed mechanistic understanding of quantitative processing of a given ADC for payload release in tumor cells, any prediction of payload concentration at the site of action and the systemic PK-PD relationship for general applications may remain challenging (Singh et al., 2015). It is not an intention of this communication to discuss the best approach to identify the intratumor catabolites that are responsible for ADC efficacy or to discover an efficacious ADC. However, with a better understanding of the driver of in vivo efficacy obtained through exploration of the intratumor payload exposure-efficacy relationships (“metabolite-driven”), intratumor catabolite analysis could be performed more routinely at the preclinical stages and provide a mechanistic understanding for payload release that would support ADC efficacy or toxicity and potential PK-PD modeling of ADCs.
Fig. 5.
The formation and function of ADC catabolites and related pharmacokinetic characteristics.
The catabolite analysis of the cyclobutyl- and cyclopropyl-containing disulfide-linked PBD-dimer ADCs in xenograft mouse models demonstrates that intratumor catabolites had kinetic profiles different from the circulating ADC species, and that the payload drug needs to be released in the right form for proper concentration at the site of action in order to make an efficacious ADC. The intention of this communication is to reveal that the chemical nature and concentration of intratumor catabolites determine the ADC efficacy, and analysis of ADC species in circulation is not sufficient to explain or predict ADC efficacy outcomes.
Acknowledgments
We would like to thank Douglas Leipold, Amrita Kamath, Chunze Li, and Paul Polakis for discussions and their review of the manuscript.
Abbreviations
- ADC
antibody drug conjugate
- DAR
drug-to-antibody ratio
- HPLC
high-performance liquid chromatography
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- mAb
antibody that could include DAR2, DAR1, and DAR0 species
- MRM
multiple reaction monitoring
- PK
pharmacokinetic(s)
- PD
pharmacodynamic(s)
- PBD-dimer
pyrrolo[2,1-c][1,4]benzodiazepine
Authorship Contributions
Participated in research design: Zhang, Yu, Dragovich, Pillow, Erickson, Hop, Khojasteh.
Conducted experiments: Zhang, Ma, Liu, Del Rosario, He.
Contributed new reagents or analytic tools: Dragovich, Pillow, Sadowsky, Pei, Zhang.
Performed data analysis: Zhang, Yu, Ma, Xu, Erickson, Sadowsky, Khojasteh, Hop.
Wrote or contributed to writing of the manuscript: Zhang, Xu, Hop, Khojasteh.
Footnotes
References
- Alley SC, Anderson KE. (2013) Analytical and bioanalytical technologies for characterizing antibody-drug conjugates. [DOI] [PubMed]
- Bhakta S, Raab H, Junutula JR. (2013) Engineering THIOMABs for site-specific conjugation of thiol-reactive linkers. Methods Mol Biol 1045:189–203. [DOI] [PubMed] [Google Scholar]
- Carter PJ, Senter PD. (2008) Antibody-drug conjugates for cancer therapy. Cancer J 14:154–169. [DOI] [PubMed] [Google Scholar]
- Chari RVJ, Miller ML, and Widdison WC (2014) Antibody-drug conjugates: An emerging concept in cancer therapy. [DOI] [PubMed]
- Erickson HK, Lambert JM. (2012) ADME of antibody-maytansinoid conjugates. AAPS J 14:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HK, Lewis Phillips GD, Leipold DD, Provenzano CA, Mai E, Johnson HA, Gunter B, Audette CA, Gupta M, Pinkas J, and Tibbitts J (2012) The effect of different linkers on target cell catabolism and pharmacokinetics/pharmacodynamics of trastuzumab maytansinoid conjugates. Mol Cancer Ther 11:1133–112. [DOI] [PubMed] [Google Scholar]
- Hartley JA. (2011) The development of pyrrolobenzodiazepines as antitumour agents. Expert Opin Investig Drugs 20:733–744. [DOI] [PubMed] [Google Scholar]
- Jeffrey SC, Burke PJ, Lyon RP, Meyer DW, Sussman D, Anderson M, Hunter JH, Leiske CI, Miyamoto JB, Nicholas ND, et al. (2013) A potent anti-CD70 antibody-drug conjugate combining a dimeric pyrrolobenzodiazepine drug with site-specific conjugation technology. Bioconjug Chem 24:1256–1263. [DOI] [PubMed] [Google Scholar]
- Jenkins TC, Hurley LH, Neidle S, Thurston DE. (1994) Structure of a covalent DNA minor groove adduct with a pyrrolobenzodiazepine dimer: evidence for sequence-specific interstrand cross-linking. J Med Chem 37:4529–4537. [DOI] [PubMed] [Google Scholar]
- Junutula JR, Flagella KM, Graham RA, Parsons KL, Raab H, Bhakta S, Nguyen T, Dugger DL, Li G, Lewis Phillips GD, et al. (2010) Engineered thio-trastuzumab-DM1 conjugate with an improved therapeutic index to target human epidermal growth factor receptor 2-positive breast cancer. Clin Cancer Res 16:4769–4778. [DOI] [PubMed] [Google Scholar]
- Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al. (2008) Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol 26:925–932. [DOI] [PubMed] [Google Scholar]
- Kamath AV, Iyer S. (2015) Preclinical pharmacokinetic considerations for the development of antibody drug conjugates. Pharm Res 32:3470–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khot A, Sharma S, Shah DK. (2015) Integration of bioanalytical measurements using PK-PD modeling and simulation: implications for antibody-drug conjugate development. Bioanalysis 7:1633–1648. [DOI] [PubMed] [Google Scholar]
- Kung Sutherland MS, Walter RB, Jeffrey SC, Burke PJ, Yu C, Kostner H, Stone I, Ryan MC, Sussman D, Lyon RP, et al. (2013) SGN-CD33A: a novel CD33-targeting antibody-drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood 122:1455–1463. [DOI] [PubMed] [Google Scholar]
- Leal M, Wentland J, Han X, Zhang Y, Rago B, Duriga N, Spriggs F, Kadar E, Song W, McNally J, et al. (2015) Preclinical development of an anti-5T4 antibody-drug conjugate: pharmacokinetics in mice, rats, and NHP and tumor/tissue distribution in mice. Bioconjug Chem 26:2223–2232. [DOI] [PubMed] [Google Scholar]
- Lin K, Rubinfeld B, Zhang C, Firestein R, Harstad E, Roth L, Tsai SP, Schutten M, Xu K, Hristopoulos M, et al. (2015) Preclinical development of an anti-NaPi2b (SLC34A2) antibody-drug conjugate as a therapeutic for non-small cell lung and ovarian cancers. Clin Cancer Res 21:5139–5150. [DOI] [PubMed] [Google Scholar]
- Pillow TH, Sadowsky JD, Zhang D, Yu SF, Del Rosario G, Xu K, He J, Bhakta S, Ohri R, Kozak KR, et al. (2016) Decoupling stability and release in disulfide bonds with antibody-small molecule conjugates. Chem Sci (Camb), in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. (2016) Antibody drug conjugates for cancer therapy. Pharmacol Rev 68:3–19. [DOI] [PubMed] [Google Scholar]
- Polson AG, Williams M, Gray AM, Fuji RN, Poon KA, McBride J, Raab H, Januario T, Go M, Lau J, et al. (2010) Anti-CD22-MCC-DM1: an antibody-drug conjugate with a stable linker for the treatment of non-Hodgkin’s lymphoma. Leukemia 24:1566–1573. [DOI] [PubMed] [Google Scholar]
- Rahman KM, Thompson AS, James CH, Narayanaswamy M, Thurston DE. (2009) The pyrrolobenzodiazepine dimer SJG-136 forms sequence-dependent intrastrand DNA cross-links and monoalkylated adducts in addition to interstrand cross-links. J Am Chem Soc 131:13756–13766. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, Desai R, Escarpe PA, Hampl J, Laysang A, et al. (2015) A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 7:302ra136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senter PD, Sievers EL. (2012) The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol 30:631–637. [DOI] [PubMed] [Google Scholar]
- Shah DK, King LE, Han X, Wentland J-A, Zhang Y, Lucas J, Haddish-Berhane N, Betts A, Leal M. (2014) A priori prediction of tumor payload concentrations: preclinical case study with an auristatin-based anti-5T4 antibody-drug conjugate. AAPS J 16:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B-Q, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu S-F, Mai E, et al. (2012) Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol 30:184–189. [DOI] [PubMed] [Google Scholar]
- Singh AP, Shin YG, and Shah DK (2015) Application of pharmacokinetic-pharmacodynamic modeling and simulation for antibody-drug conjugate development. Pharm Res 32:3508–3525. [DOI] [PubMed] [Google Scholar]
- Wang J, Gu H, Liu A, Kozhich A, Rangan V, Myler H, Luo L, Wong R, Sun H, Wang B, et al. (2016) Antibody-drug conjugate bioanalysis using LB-LC-MS/MS hybrid assays: strategies, methodology and correlation to ligand-binding assays. Bioanalysis 8:1383–1401. [DOI] [PubMed] [Google Scholar]
- Xu K, Liu L, Maia M, Li J, Lowe J, Song A, Kaur S. (2014) A multiplexed hybrid LC-MS/MS pharmacokinetic assay to measure two co-administered monoclonal antibodies in a clinical study. Bioanalysis 6:1781–1794. [DOI] [PubMed] [Google Scholar]
- Xu K, Liu L, Saad OM, Baudys J, Williams L, Leipold D, Shen B, Raab H, Junutula JR, Kim A, et al. (2011) Characterization of intact antibody-drug conjugates from plasma/serum in vivo by affinity capture capillary liquid chromatography-mass spectrometry. Anal Biochem 412:56–66. [DOI] [PubMed] [Google Scholar]
- Zhang D, Pillow T, Ma Y, Cruz-Chuh J, Kozak KR, Sadowsky JD, Lewis Phillips GD, Guo J, Darwish M, Fan P, et al. (2016) Linker immolation determines cell killing activity of disulfide-linked pyrrolobenzodiazepine antibody drug conjugates. ACS Med Chem Lett, submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]